Summary
Rational exploitation of interactions between microbes and plants can help to transform agriculture
One major challenge for the twenty-first century will be the production of sufficient food—the United Nations Population Fund estimates that the global human population may well reach 10 billion by 2050 (www.unfpa.org). This means increasing agricultural productivity of food crops, as plants form the basis of every food chain. However, agriculture in developed countries already creates a range of serious environmental problems through the use of pesticides and herbicides, salinization and the depletion of water resources. Furthermore, agricultural production in many African, Asian and South American countries cannot be increased without further destroying more areas by turning them into arable land, thus threatening global biodiversity, which is already under stress from human action. If global food production is to keep pace with an increasingly urbanized and growing population, while formulating new food production strategies for developing countries, the great challenge for modern societies is to boost plant productivity in an environmentally sustainable manner.
A major effort in plant biotechnology is therefore the development of new crop varieties with enhanced disease and pest resistance, greater drought and salt tolerance and better nutritional value through the introduction of desirable traits either by conventional breeding or genetic modification. However, these efforts have traditionally focused on plant phenotypes. What has been largely ignored is the important role of microbial communities that interact with plants to influence plant health, productivity and biodiversity. The impact of the microbial world on plants is evident: worldwide each year, microbial diseases cost crop producers billions of Euros. Similarly, the important role of nitrogen fixation by rhizobia and other bacteria for plant growth has been known for decades. What is less appreciated, and less well understood, is the pervasive influence that other microbes have on plant health and growth; they enhance stress tolerance, provide disease resistance, aid nutrient availability and uptake, and promote biodiversity. Although major advances in genomic technologies and in situ studies of beneficial plant–microbe interactions have produced a large amount of knowledge and given insights into the mechanisms of these interactions, their application in biotechnology and agriculture has yet to be exploited. A greater understanding of how plants and soil microbes live together and benefit each other can therefore provide new strategies to improve plant productivity, while helping to protect the environment and maintain global biodiversity.
The most intense interactions between microbes and plants take place at the rhizosphere, which is the interface between plant roots and the soil. The concept of the rhizosphere was broached 100 years ago by the German biologist Lorenz Hiltner (Fig 1), who first proposed that the area around the roots is a region of high microbial activity (Hiltner, 1904). Despite a large amount of work and research on the biology and biochemistry of the rhizosphere, there is still very little known about the detailed mechanisms that take place in the soil. Although most plant biodiversity research focuses on what is visible above the ground, researchers have demonstrated a clear link between 'above-ground' and 'below-ground' diversity (Helgason et al, 1998; Marcel et al, 1998). The biological and ecological details that underpin this phenomenon remain elusive, but these findings suggest that maintenance of a high diversity of plant species requires a correspondingly high level of diversity in the soil microbial community (Wardle et al, 2004).
If global food production is to keep pace with an increasingly urbanized and growing population [...] the great challenge for modern societies is to boost plant productivity in an environmentally sustainable manner
Figure 1.
Lorenz Hiltner: pioneer of the rhizosphere concept. Courtesy of the Bavarian State Institute for Agriculture in Freising, Germany
The influence of plants on microbial population structure and function in the rhizosphere has important ecological implications for soil function, including biogeochemical cycles. Similarly, soil microbes have a tremendous influence on plant health and productivity (Bloemberg & Lugtenberg, 2001). One straightforward and visible benefit for the plant is a better supply of and access to nutrients. The role of mutualistic nitrogen-fixating rhizobia has been well documented for decades, but recent data detail the intimate exchange of nutrients during the symbiosis of plant roots and bacteria (Lodwig et al, 2003). The plant attracts nitrogen-fixating bacteria to invade the cells in the root and provides them with carbohydrates as a food source while the bacteria reduce nitrous compounds in the soil that are then used by the plant. Similarly, interactions between plants and fungi can also provide nutrients for the plant. Arbuscular mycorrhizal fungi, which form an intricate internal symbiosis with the roots of most flowering plants, are associated with the provision of phosphorous to the plant in exchange for organic carbohydrates (Smith & Read, 1997). Microbes also indirectly aid nutrient uptake—bacteria of the Azospirillum genus promote increased root mass and more efficient nitrogen uptake from the soil in response to the plant hormone indole-3-acetic acid. Using these bacteria and fungi could provide significant environmental benefits as they would allow a reduction in the application of nitrogen and phosphorous fertilizers. The overuse of such fertilizers has become a major concern because they cause nitrate contamination of soil and groundwater by leachates and because microbial denitrification converts residual nitrogen into the greenhouse gas nitrous oxide (Nosengo, 2003; Reay, 2004). Equally, excess phosphorous compounds leach into groundwater, rivers and streams, where they promote algal growth and other environmental problems. Although commercial fertilizer products based on rhizobia and Azospirillum are already available, their wider application is restricted by inconsistent performance and limitations of host range. A better understanding of plant–microbe symbiosis could help to overcome at least some of these problems.
In addition to enhancing the nutrient supply of plants, microbes also confer a degree of protection against plant diseases. In particular, various bacteria and fungi—especially of the genera Pseudomonas, Bacillus and Trichoderma—produce a range of metabolites against other phytopathogenic fungi (Bloemberg & Lugtenberg, 2001; Walsh et al, 2001; Raijaamkers et al, 2002). Such biocontrol agents are already efficiently used in the field, but they have not yet attained the degree of efficacy and consistency that is needed for largescale commercialization. But there is the potential for improvement and, with development, such microbes could become a realistic alternative to the heavy fungicide regimens used in agriculture at present. A reduction in the use of these chemicals would lead to obvious environmental benefits and it would also appeal to consumers who seek more natural produce.
What has been largely ignored is the important role of microbial communities that interact with plants to influence plant health, productivity and biodiversity
In addition to these direct effects on plant growth, rhizobacteria exert another health-promoting effect on the plants with which they interact. This phenomenon is known as induced systemic resistance and arises when interactions with non-pathogenic bacteria confer better disease resistance on plants (van Loon et al, 1998). Induced systemic resistance differs from acquired systemic resistance in plants; the latter occurs in response to localized microbial attack. Induced resistance is triggered by microbial interactions in the rhizosphere, but also enhances resistance of remote aerial plant parts against pathogens. This systemic change in plant physiology bears conceptual similarities to other phenotypes, such as improved stress, drought or disease resistance, that have been linked to plant–fungal associations. Little is known about why bacteria induce this condition, which is clearly beneficial to the plant. Although induced systemic resistance is generally ascribed to interactions with bacteria, there is emerging evidence that some fungi, particularly endophytic fungi, may also be able to induce a similar response in plants (Arnold et al, 2003; Harman et al, 2004; Clay, 2004).
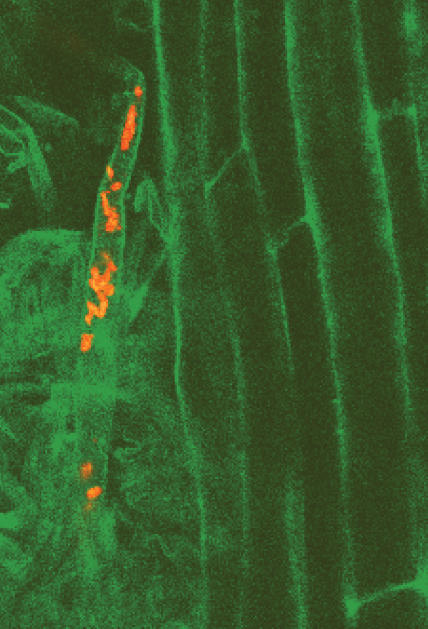
The interactions of rhizosphere microbes with plants depend on the establishment of intimate associations between the two partners (Fig 2). Research on some of these interactions, such as those between symbiotic rhizobia and legumes, has demonstrated that this intimate cooperation between plant and bacteria displays a high level of host specificity. There is also a growing body of evidence suggesting that many other associations between plants and microbes show similar degrees of specificity; different plant species, and even different cultivars of the same plant species, establish distinct microbial populations in their rhizospheres when grown in the same soil. The formation of these communities depends, at least in part, on the activation of specific programmes of gene expression in the microbe in response to chemical signals secreted from the plant. A pertinent example is the induction of nodulation genes in receptive rhizobia, which are triggered by the production and secretion of particular flavonoids by the plant. In the case of the rhizobia–legume interaction, the plant also responds to bacterial signals, and it is likely that this type of chemical cross-talk is typical of other microbe–plant interactions. Other examples of plant-derived signals that influence microbial gene expression include phenolics exuded from plant wounds, which induce expression of virulence genes in pathogenic Agrobacterium spp., and compounds that mimic the quorum sensing signals used by bacteria to regulate gene expression (Loh et al, 2002; Newton & Fray, 2004). In general, however, there is only scant knowledge of signalling interactions between beneficial microbes and plants. Understanding how microbes respond to plant signals in terms of growth and gene expression, and the role that plant signalling has in determining interaction specificity or driving population selection is central to reaping the benefits of plant–microbe interactions.
The influence of plants on microbial population structure and function in the rhizosphere has important ecological implications for soil function, including biogeochemical cycles
Figure 2.
Intimate associations between bacteria and plants: fluorescently labelled bacteria visualized in situ growing on the root surface (unpublished figure provided by Estibaliz Larrainzar)
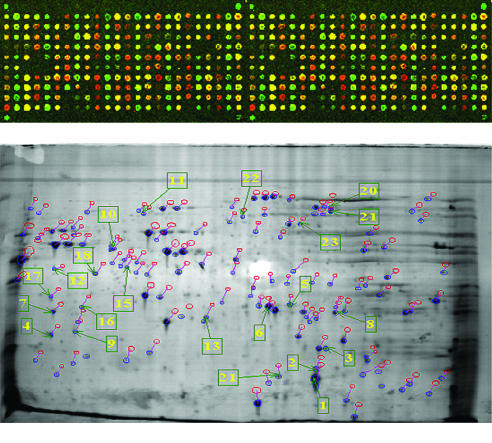
Before the advent of genomic technologies, scientists had only limited options to investigate these interactions in detail, particularly with a view to their commercial exploitation. This situation has changed considerably, now that the genomes of more than ten plant-associated bacteria have been sequenced, and the sequencing of another 35 relevant genomes is already under way (Puhler et al, 2004). A detailed investigation of the molecular basis of pathogenic, symbiotic and associative plant–microbe interactions, both at the levels of comparative and of functional genomics is now possible. So far, most data have come from comparative analyses with a particular emphasis on mutualistic and pathogenic interactions. Although comparative genomics focuses on genetic potential rather than gene expression, these studies do raise some interesting questions about the distinction between pathogenic and non-pathogenic bacteria. For instance, investigations have already yielded a surprising insight regarding type-III protein secretion systems. These are ordinarily associated with pathogenicity in bacteria and are possibly also involved in non-pathogenic associations (Puhler et al, 2004). In the future, transcriptome profiling and functional genomics (Fig 3) are likely to produce more information about microbial responses to plant signals and the contribution of specific gene products to the establishment of an interaction with the host. An important question is whether plant metabolites exuded from roots induce microbial type-III protein secretion systems, and, if so, how? Pseudomonas fluorescens, for instance, is chemotactic towards components of root exudates (de Weert et al, 2002), but the global effect of these exudates on gene expression in the bacterium is still unknown. By profiling the complete genetic response to root exudates, it will be possible to assemble a full picture of how gene expression, and thereby function, is modulated in the bacterium after perception of plant signals. It will facilitate studies of how different bacterial species and subspecies respond to particular plants and how a bacterium responds differentially to signals from different plant species or varieties. This, in turn, will lead to a better understanding of the basis of host specificity and host selection during microbe–plant interactions. It may also be possible to describe general principles of plant–microbe communication that define or distinguish associative, mutual and pathogenic interactions of microbes with plants.
Figure 3.
The '-omics' technologies: proteomics and genomics are revolutionizing the study of interactions between plants and microbes (unpublished figure provided by Olive Gleeson and Gordon McAlester)
In addition to the economic effects of plant disease, microbial interaction with plants can also have serious and direct consequences for human health. One notorious case is the reputed association between the consumption of ergot-contaminated products and the hysteria that characterized the Salem witch trials in seventeenth century USA. A hypothesis has been put forward that psychoactive alkaloids related to LSD, produced by the fungus Claviceps purpurea during rye infection, caused the symptoms seen during that period, although this can never be unequivocally proven. Another example of microbial toxins in the food chain are fungal aflatoxins, produced by certain strains of Aspergillus flavus and A. parasiticus growing on foods and feeds, which are a serious health concern. The effects of microbes on plant physiology itself have received less attention, although it is known that many food crops naturally produce toxic metabolites; potatoes and tomatoes, for example, can accumulate high levels of toxic steroidal alkaloids (Friedman, 2002; Korpan et al, 2004). The possible role of microbes in inducing toxin production, or in modifying metabolites or metabolic pathways within the plant, remains largely unexplored. There are clear precedents for this premise, for instance the production of secondary metabolites by plants, such as phytoalexins, in response to pathogenic attack and fungal modification of plant saponins (Morrissey & Osbourn, 1999; Bouarab et al, 2002). Although it is clearly a complex issue, the combination of plant and microbial functional genomics with metabolome analysis provides a route to start addressing these questions.
Understanding how microbes respond to plant signals [...] is central to reaping the benefits of plant–microbe interactions
Future biotechnological developments in the agricultural sector—whether based on gene modification technology or on traditional breeding—should recognize the importance of plant–microbe associations. The 'traditional' plant biotechnology sector is based around plant breeding and the selection of varieties with desired traits, and it pays little attention to plant–microbial ecology. But the expression of desirable traits, such as disease resistance, or drought and salt tolerance, could equally be driven by interactions between a particular plant variety and the colonizing microbial flora. Conversely, particular plant genotypes may attract a microbial flora with undesirable traits. Understanding the genetic basis of plant–microbe interactions in the context of how the plant selects its microbial population in the soil may allow 'conditioning' of the rhizosphere to promote desirable traits in the plant, which is, in fact, the basis of natural disease-suppressive soils. Similarly, it is also an attractive proposition to influence or modulate the microbes that interact with the plant to generate improved productivity or health for the latter. Furthermore, bacteria could be genetically engineered to confer increased disease resistance or growth promotion that is only activated when the bacterium is associated with its host plant. From the perspective of developing nations, these are exciting strategies that may help to increase yields while avoiding some of the costs and environmental problems that come with the use of fertilizers, pesticides, herbicides and fungicides. However, most of the microbial biodiversity in soil remains unexplored and much work remains to be done to first identify and then characterize microorganisms that could be used in such applications. Furthermore, such approaches require a detailed knowledge of the molecular signalling that takes place between plants and microbes to drive expression of desirable traits and suppress unwanted effects in a controlled manner. Exploiting plants and microbes by using such an integrated approach requires a systems biology strategy to understand the degree and complexity of plant–microbe interactions through the application of modern '-omics' technologies.
One could describe the evolution of technology as passing through three phases: the low-tech stage, the advanced stage and the high-tech stage. The low-tech phase is, as the name implies, characterized by the use of basic technology; for instance, before the advent of vaccines and antibiotics, care and hygiene were the only means to help patients to cope with infectious diseases. The advanced phase uses modern technologies but is clunky and intrusive, such as the infamous iron lung that was used in the 1950s and 1960s to help polio victims breathe. The high-tech stage finally provides elegant, efficient and seemingly simple solutions on the basis of detailed scientific knowledge—by understanding the problem and addressing the cause rather than the symptoms. In the fight against polio, it was the Salk and Sabin vaccines that eventually helped the medical community to combat the disease. Modern agriculture has gone through similar phases in recent history. The first agricultural revolution in the eighteenth century introduced crop rotation to take advantage of and manipulate microbial populations in the soil, although at that time it was not known why this benefited plant health and growth. The second revolution, which began in the 1960s and is sometimes described as the 'green revolution', was based on improved plant-breeding techniques and the development of hybrid varieties; it now includes genetic engineering of plants but also a heavy reliance on the use of chemicals. We may now be at the cusp of a third stage, which will combine both approaches in a more holistic and elegant strategy. Applying knowledge about beneficial plant–microbe interactions in the rhizosphere to plant-breeding and genetic-engineering technologies may allow us to increase food production while reducing stress on the environment and on global biodiversity.
Applying knowledge about beneficial plant–microbe interactions [...] may allow us to increase food production while reducing stress on the environment and on global biodiversity




Acknowledgments
Research in the authors' laboratories is supported by research grants from Science Foundation Ireland (02/IN.1/B1261; 03/IN3/B373), the HEA PRTLI programmes, Enterprise Ireland (SC/02/0420; SC/02/517), the European Union (QLK3-2001-00101; QLK5-CT-2002-00914) and the Health Research Board (RP76/2001).
References
- Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (2003) Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci USA 100: 15649–15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg GV, Lugtenberg BJ (2001) Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 4: 343–350 [DOI] [PubMed] [Google Scholar]
- Bouarab K, Melton R, Peart J, Baulcombe D, Osbourn A (2002) A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418: 889–892 [DOI] [PubMed] [Google Scholar]
- Clay K (2004) Fungi and the food of the gods. Nature 427: 401–402 [DOI] [PubMed] [Google Scholar]
- de Weert S, Vermeiren H, Mulders IH, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJ (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15: 1173–1180 [DOI] [PubMed] [Google Scholar]
- Friedman M (2002) Tomato glycoalkaloids: role in the plant and in the diet. J Agric Food Chem 50: 5751–5780 [DOI] [PubMed] [Google Scholar]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2: 43–56 [DOI] [PubMed] [Google Scholar]
- Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW (1998) Ploughing up the wood-wide web? Nature 394: 431. [DOI] [PubMed] [Google Scholar]
- Hiltner L (1904) Uber neue Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie. Arbeiten der Deutschen Landwirtschaftgesellschaft 98: 59–78 [Google Scholar]
- Korpan YI, Nazarenko EA, Skryshevskaya IV, Martelet C, Jaffrezic-Renault N, El'skaya AV (2004) Potato glycoalkaloids: true safety or false sense of security? Trends Biotechnol 22: 147–151 [DOI] [PubMed] [Google Scholar]
- Lodwig EM, Hosie AH, Bourdes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS (2003) Amino-acid cycling drives nitrogen fixation in the legume–Rhizobium symbiosis. Nature 422: 722–726 [DOI] [PubMed] [Google Scholar]
- Loh J, Pierson EA, Pierson LS III, Stacey G, Chatterjee A (2002) Quorum sensing in plant-associated bacteria. Curr Opin Plant Biol 5: 285–290 [DOI] [PubMed] [Google Scholar]
- Marcel G, Van Der Heijden A, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller R, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72 [Google Scholar]
- Morrissey JP, Osbourn AE (1999) Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol Mol Biol Rev 63: 708–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JA, Fray RG (2004) Integration of environmental and host-derived signals with quorum sensing during plant–microbe interactions. Cell Microbiol 6: 213–224 [DOI] [PubMed] [Google Scholar]
- Nosengo N (2003) Fertilized to death. Nature 425: 894–895 [DOI] [PubMed] [Google Scholar]
- Puhler A, Arlat M, Becker A, Gottfert M, Morrissey JP, O'Gara F (2004) What can bacterial genome research teach us about bacteria–plant interactions? Curr Opin Plant Biol 7: 137–147 [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, Vlami M, de Souza JT (2002) Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81: 537–547 [DOI] [PubMed] [Google Scholar]
- Reay DS (2004) Fertiliser 'solution' could turn local problem global. Nature 427: 485. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (1997) Mycorrhizal Symbiosis 2nd edn. San Diego, CA, USA: Academic [Google Scholar]
- van Loon LC, Bakker PA, Pieterse CM (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36: 453–483 [DOI] [PubMed] [Google Scholar]
- Walsh UF, Morrissey JP, O'Gara F (2001) Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr Opin Biotechnol 12: 289–295 [DOI] [PubMed] [Google Scholar]
- Wardle DA, Bardgett RD, Klironomos JN, Setala H, van der Putten WH, Wall DH (2004) Ecological linkages between above ground and below ground biota. Science 304: 1629–1633 [DOI] [PubMed] [Google Scholar]