Abstract
The neurodegenerative disease Friedreich's ataxia is caused by reduced levels of frataxin, a mitochondrial matrix protein. The in vivo role of frataxin is under debate. Frataxin, as well as its yeast homologue Yfh1, binds multiple iron atoms as an oligomer and has been proposed to function as a crucial iron-storage protein. We identified a mutant Yfh1 defective in iron-induced oligomerization. This mutant protein was able to replace functionally wild-type Yfh1, even when expressed at low levels, when mitochondrial iron levels were high and in mutant strains having deletions of genes that had synthetic growth defects with a YFH1 deletion. The ability of an oligomerization-deficient Yfh1 to function in vivo suggests that oligomerization, and thus oligomerization-induced iron storage, is not a critical function of Yfh1. Rather, the capacity of this oligomerization-deficient mutant to interact with the Isu protein suggests a more direct role of Yfh1 in iron–sulphur cluster biogenesis.
Keywords: frataxin, Yfh1, mitochondria, iron, oligomerization
Introduction
Deficits in the levels of frataxin, a highly conserved protein of the mitochondrial matrix, are the cause of the neurodegenerative disease Friedreich's ataxia, which, at the cellular level, is characterized by an increase in mitochondrial iron levels and a decrease in the activity of iron–sulphur cluster (ISC)-containing proteins (Pandolfo, 2002). In Saccharomyces cerevisiae, deletion of YFH1, the yeast frataxin homologue, also causes an accumulation of iron in mitochondria (Babcock et al, 1997) and a decrease in the activity of ISC-containing enzymes (Rötig et al, 1997). The important cellular functions of frataxin/Yfh1 are a matter of continuing debate. In eukaryotes, biosynthesis of ISCs is mediated by a set of highly conserved mitochondrial proteins (Gerber & Lill, 2002). Isu proteins act as a scaffold for the assembly of ISCs before transfer to recipient apoproteins (Mühlenhoff et al, 2003). Nfs1, a cysteine desulphurase, provides the necessary sulphur for the formation of ISCs (Li et al, 1999). It has been reported that Yfh1 interacts with Isu and Nfs1 in mitochondrial extracts (Gerber et al, 2003) and that human frataxin can form a complex with Isu and mediate the transfer of iron for ISC formation on Isu (Yoon & Cowan, 2003), suggesting that Yfh1 may have an important role in ISC biogenesis.
Conversely, it has been proposed that Yfh1 serves as an iron-storage protein. Incubation of Yfh1 with ferrous iron induces the formation of oligomers having up to 48 Yfh1 subunits that bind >2,000 iron atoms (Adamec et al, 2000; Park et al, 2003), with the size of oligomers and amount of iron bound being dependent on the Yfh1:iron stoichiometry in the solution. These in vitro results have led to the hypothesis that Yfh1 functions as an iron chaperone and an ironstorage protein (Park et al, 2003). Human frataxin and CyaY, the Escherichia coli homologue, share a common structural fold (Dhe-Paganon et al, 2000; Cho et al, 2000). Mature Yfh1 has 37% amino-acid identity with human frataxin. Both frataxin and CyaY bind iron in an oligomeric state (Adinolfi et al, 2002; Cavadini et al, 2002), suggesting that iron storage is a function conserved through evolution.
As the in vivo relevance of Yfh1 oligomerization has not been assessed, we isolated YFH1 mutants encoding oligomerization-deficient proteins and tested their capacity to function in vivo. Oligomerization-deficient Yfh1 was able to rescue the YFH1 deletion phenotypes as well as wild-type (wt) protein, suggesting that neither oligomerization nor oligomerization-related iron storage is a critical cellular function of Yfh1. However, this mutant can still interact with Isu, indicating a possible direct role of Yfh1 in ISC formation.
Results
Changes in Yfh1's anionic patch affect oligomerization
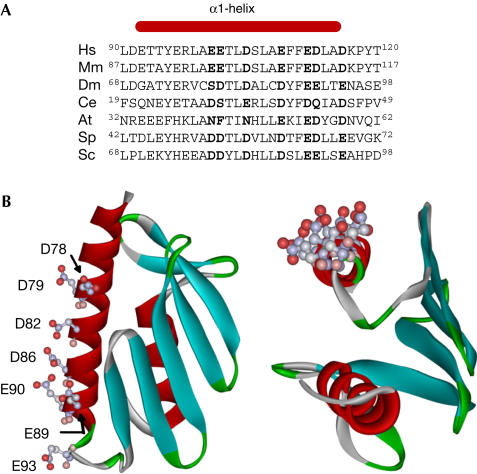
We generated a structural model of Yfh1 on the basis of its sequence similarity to the human and E. coli homologues. Negatively charged amino acids at conserved positions in the amino-terminal α-helix form a solvent-exposed acidic patch (Fig 1). Several of these residues have previously been shown to be important for oligomerization of CyaY in vitro (Adinolfi et al, 2002). To alter the oligomerization capacity of Yfh1, residues D86, E90 and E93 were substituted with their noncharged counterparts to generate 86N/90Q/93Q Yfh1.
Figure 1.
Comparison of Yfh1 and human frataxin. (A) Alignment of sequences corresponding to the α1-helix region of eukaryotic frataxins. Negatively charged residues are in bold. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. (B) Structural model of Yfh1 generated using SWISS-MODEL program (Schwede et al, 2003). Side chains of solvent-exposed conserved, negatively charged residues in α1-helix are shown.
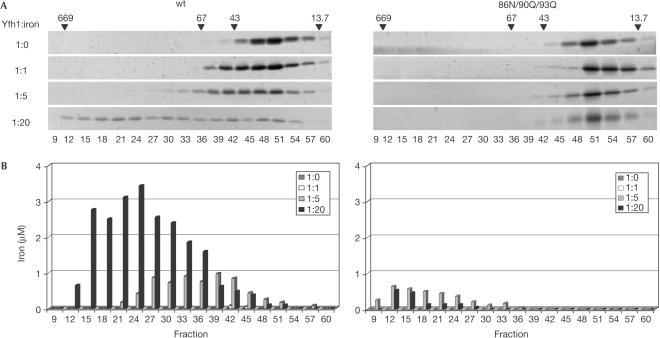
The ability of wt and 86N/90Q/93Q Yfh1 to oligomerize was tested in vitro. Incubation of wt Yfh1 with increasing amounts of ferrous sulphate promoted its oligomerization into sequentially larger complexes (Fig 2A). At a 1:20 protein to iron ratio, less than 30% of Yfh1 remained as a monomer, whereas the remaining 70% was present as oligomers up to ∼650 kDa. In contrast, no 86N/90Q/93Q Yfh1 was detected in fractions corresponding to oligomeric species, regardless of the protein to iron ratio used, either by SYPRO-Ruby staining or western blot analysis (Fig 2A; data not shown).
Figure 2.
Oligomerization of wt and 86N/90Q/93Q Yfh1 in the presence of ferrous iron. Purified wt or 86N/90Q/93Q Yfh1 was incubated with Fe(NH4)2(SO4)2 at indicated Yfh1:iron molar ratios. Samples were subjected to size-exclusion chromatography and fractions analysed. (A) SDS–PAGE stained with SYPRO-Ruby. Positions of thyroglobulin (669 kDa), BSA (67 kDa), ovalbumin (43 kDa) and ribonuclease A (13.7 kDa) are indicated. (B) Iron content measured by atomic absorption spectrometry.
Measurements using atomic absorption spectrometry indicated that at protein to iron ratios of 1:5 and 1:20, iron was present in fractions corresponding to oligomeric Yfh1 complexes, with the larger oligomers containing a higher content of iron per monomer than smaller oligomers (Fig 2B). On the basis of these measurements, ∼400 atoms of iron are present in the larger multimers that contain 40–50 subunits. When 86N/90Q/93Q Yfh1 was incubated with iron at ratios of 1:5 or 1:20, low amounts of iron were present in fractions corresponding to high-molecular-weight components, even though no Yfh1 protein was detected. Similar levels of iron were detected in these fractions when either bovine serum albumin (BSA) or ovalbumin was incubated with iron under the same conditions (data not shown). These controls suggest that the presence of iron in these fractions can be attributed to oxidation of the more soluble ferrous iron to the relatively insoluble ferric ion in the absence of an iron binding protein. Furthermore, the presence of ∼30 ng of Yfh1 would be required to bind the amount of iron present in these fractions, an amount substantially above the detection limit of both assays. We conclude that 86N/90Q/93Q Yfh1 is oligomerization defective.
Oligomerization-deficient Yfh1 is functional in vivo
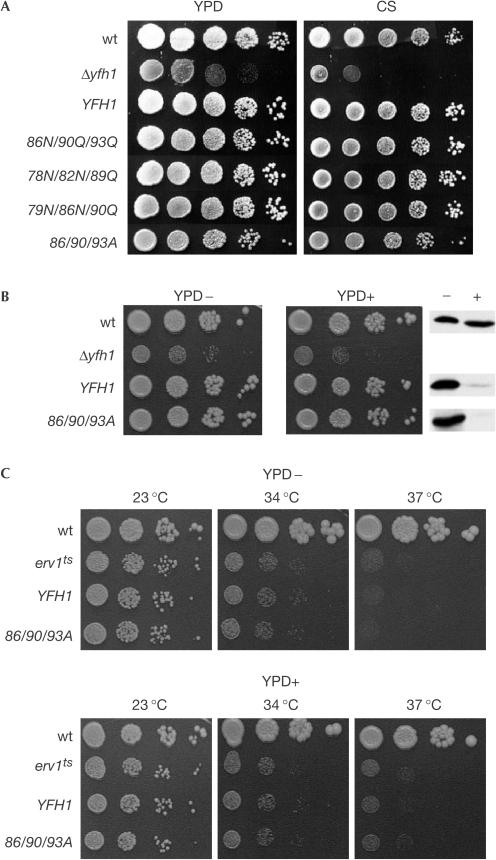
YFH1 is not an essential gene but its deletion (Δyfh1) severely compromises cell growth (Fig 3A). To test whether oligomerization is necessary for Yfh1 to carry out its cellular function, the ability of 86N/90Q/93Q Yfh1 expressed from a plasmid to rescue the growth defect of Δyfh1 cells was tested. The 86N/90Q/93Q mutant rescued the growth defect of Δyfh1 cells as well as wt YFH1 in both rich and minimal media containing glucose over a range of temperatures and in rich media containing galactose or glycerol as a carbon source (Fig 3A; data not shown).
Figure 3.
Mutations that alter the oligomerization capacity of Yfh1 do not affect cell growth even at low protein expression or high mitochondrial iron content. (A) Tenfold serial dilutions of cell suspensions of wt, Δyfh1 and Δyfh1 carrying a plasmid expressing either wt or indicated YFH1 mutant plated on rich (YPD) or minimal synthetic (CS) media. (B) Left and centre panels: Tenfold serial dilutions of wt, Δyfh1 and Δyfh1 containing wt or 86/90/93A YFH1 in a plasmid under the control of the tetO promoter. Right panel: Cells grown on YPD plate were harvested, cell extracts prepared and immunoblot analysis using Yfh1specific antibody was carried out. (C) Tenfold serial dilutions of wt, erv1ts and Δyfh1erv1ts containing either wt or 86/90/93A YFH1 on a plasmid under the control of the tetO promoter. Cells were grown at 30°C or indicated temperatures for 2 days. YPD− and YPD+ indicate plates lacking (−) or containing (+) 25 μg ml−1 doxycycline.
Because of the lack of phenotypic effect of the mutant YFH1, we constructed additional mutants: two of them, 78N/82N/89Q and 79N/86N/90Q, combined substitutions of different negatively charged residues of the same α-helix and a third changed residues D86, E90 and E93 to Ala (86/90/93A). These mutants also completely rescued the growth defect of the Δyfh1 strain under all conditions tested (Fig 3A; data not shown).
We confirmed that 86/90/93A, similar to 86N/90Q/93Q, was oligomerization defective in vitro (data not shown). This mutant was used in the experiments described below, because its substitutions, which altered both charge and size, had the potential to be more deleterious. The 86/90/93A allele was integrated into the chromosome and was shown to rescue the growth phenotype of Δyfh1 (data not shown). In addition, when mitochondrial iron levels and aconitase activity were measured, no differences were observed between wt and 86/90/93A mitochondria (1.3 compared with 1.1 pmol Fe μg−1; 916 compared with 933 nmol aconitate mg−1 min−1), whereas Δyfh1 mitochondria have at least tenfold higher iron and 7.5-fold lower aconitase levels (Babcock et al, 1997; Rötig et al, 1997; data not shown).
Because the mutant proteins rescued Δyfh1 phenotypes under normal conditions, we decided to test the function of Yfh1 under more stringent conditions. As Yfh1 cellular levels can be significantly reduced before adverse effects on cell growth are detected (Karthikeyan et al, 2003), wt and mutant Yfh1 proteins were placed under the control of the tetracycline-regulatable promoter (tetO) (Garí et al, 1997). In the absence of the drug, expression of mutant and wt protein was similar to normal levels; in the presence of the drug, expression was reduced by more than 90% (Fig 3B). Cells expressing wt and mutant protein grew similarly even when expressed at these disparate levels.
In vitro oligomerization of Yfh1 is mediated by iron (Adamec et al, 2000). We reasoned that if oligomerization was important in vivo, this requirement might be accentuated when mitochondrial iron levels were increased. To obtain high mitochondrial iron levels, we used a temperaturesensitive mutation in the ERV1 gene (erv1ts) (Lisowsky, 1994). Inactivation of ERV1 leads to an increase in mitochondrial iron levels, but has no effect on ISC assembly in the mitochondrion itself (Lange et al, 2001). In our strain background, erv1ts mitochondria accumulated tenfold higher iron levels than normal after 7 h of growth at the nonpermissive temperature of 37°C. A Δyfh1erv1ts strain containing wt or 86/90/93A YFH1 on a plasmid under the control of the tetO promoter was analysed. At the permissive temperature (23°C), the erv1ts strain grew nearly as well as wt, with no difference between cells expressing wt or mutant YFH1, either at normal or low levels of expression. At 34, 35.5 and 37°C, the growth defect of the temperaturesensitive mutant was significant, but cells expressing the 86/90/93A mutant were no more compromised for growth than cells expressing wt YFH1 (Fig 3C; data not shown), indicating that the deficiency in oligomerization of Yfh1 had no deleterious effect on cell growth under conditions of mitochondrial iron accumulation.
Genetic interactions between YFH1 and ISC genes
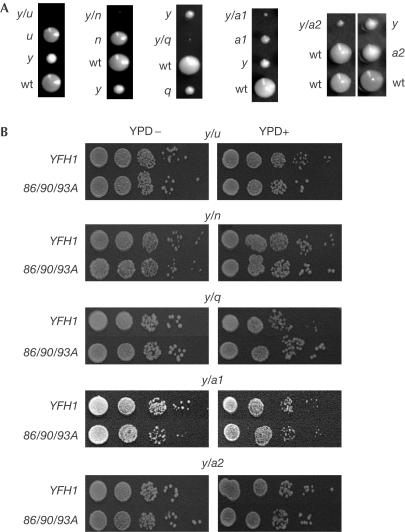
In a continuing effort to uncover deleterious effects of the 86/90/93A mutant in vivo, we undertook a search for genetic interactions between YFH1 mutants and deletions of other genes known to be involved in ISC biosynthesis. As expected (Ramazzotti et al, 2004), deletion of YFH1 and ISU1 resulted in a synthetic-lethal interaction, as no strain containing both deletions was recovered. Deletion of YFH1 in combination with deletions of NFU1, SSQ1, ISA1 or ISA2 showed previously unreported genetic interactions. Although all these double mutants were viable, they grew considerably slower than a Δyfh1 strain (Fig 4A).
Figure 4.
Genetic interaction between YFH1 and genes involved in ISC biosynthesis. (A) Haploids of indicated genotypes resulting from analysis of tetrads of indicated strains. The genotype of the inviable y/u spore was deduced from the genotype of the three viable spores. (B) Tenfold serial dilutions of indicated double-deletion strains expressing wt or 86/90/93A YFH1 in a plasmid under the control of the tetO promoter. y/u and y/n, 30°C for 2 days; y/q, 34°C for 3 days; y/a1 and y/a2, 30°C for 4 days. y, u, n, q, a1 and a2 indicate deletion of YFH1, ISU1, NFU1, SSQ1, ISA1 and ISA2, respectively. (−) and (+) indicate absence and presence of doxycycline (25 μg ml−1), respectively.
The ability of 86/90/93A YFH1, compared with wt YFH1, to overcome these genetic interactions was tested. Plasmids carrying wt or 86/90/93A YFH1 under the control of the tetO promoter were transformed into the heterozygous diploids; double-deletion haploids containing the plasmids were recovered in all cases. No combination of 86/90/93A with the deletion of ISU1, NFU1, SSQ1, ISA1 or ISA2 produced any different phenotypic effect than the same combinations with wt YFH1, whether expressed at normal or low levels (Fig 4B). This result indicates that the ability to oligomerize is not an important function of Yfh1 in its genetic interaction with other ISC biosynthetic genes.
Oligomerization-deficient mutant can interact with Isu
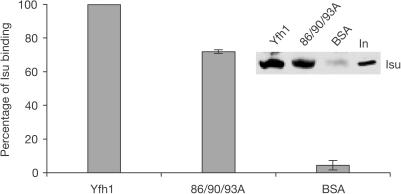
The physical as well as genetic interaction between Yfh1 and Isu1 (Gerber et al, 2003; Ramazzotti et al, 2004) suggested a direct role of Yfh1 in ISC formation rather than iron storage. We reasoned that if Isu interaction is critical for Yfh1 function, 86/90/93A mutant should retain the capacity to interact with Isu because of its functionality in vivo. To test the Isu:Yfh1 interaction, wt or 86/90/93A Yfh1 containing a carboxy-terminal polyhistidine tag was bound to Ni resin and incubated with lysates from wt mitochondria. 86/90/93A Yfh1 bound Isu nearly as well as wt (Fig 5). Binding of Isu with mutant Yfh1 was not a result of interaction with wt Yfh1 present in the mitochondrial lysates, as no detectable wt Yfh1 was bound to the resin in the presence of 86/90/93A Yfh1 (data not shown).
Figure 5.
wt and 86/90/93A Yfh1 interact with Isu. His-tagged wt or His-tagged 86/90/93A Yfh1 bound to Ni resin was incubated with wt mitochondrial extracts. Binding of Isu was detected by immunoblot analysis using antibodyspecific for Isu. Binding of Isu in two independent experiments was quantified; binding of wt Yfh1 indicated as 100%. BSA, control for nonspecific binding; In, 5% of total input of mitochondrial extract.
Discussion
Because of the vigorous debate concerning its role in Yfh1 function, we have sought to determine the in vivo importance of the iron-induced oligomerization. Here, we report that YFH1 mutants encoding proteins completely defective in oligomerization in vitro showed no in vivo phenotypes, even when protein expression was markedly reduced and mitochondrial iron levels were elevated. These results indicate that oligomerization of Yfh1 is not required for its in vivo roles. To argue otherwise, Yfh1 86/90/93A would need to have sufficient residual oligomerization activity when in the in vivo environment to meet all the challenges of the different growth conditions and genetic backgrounds tested here.
Whether Yfh1 oligomerizes in vivo remains a matter of debate. We attempted to analyse the effect of the amino-acid alterations on in vivo oligomerization, as the presence of Yfh1 in high-molecular-weight complexes has been reported in vivo (Adamec et al, 2000). However, in agreement with other reports (Mühlenhoff et al, 2002; Gerber et al, 2003), we did not observe wt or mutant Yfh1 oligomers of the size observed in vitro in lysates of either wt or mutant mitochondria, even from those having high iron levels (supplementary information online). Failure to observe oligomers in vivo raises the question as to whether oligomerization and thus iron binding of Yfh1 only occur in vitro under particular experimental conditions (Adinolfi et al, 2002).
We cannot completely exclude the possibility that oligomerization, and perhaps an iron-storage function, is important under some condition that we did not test. However, the fact that the amount of Yfh1 in mitochondria necessary for normal growth is several hundred-fold less than the amount of iron in mitochondria that is not sequestered in haeme or ISCs (data not shown) severely challenges the idea that iron storage is a vital function of Yfh1. We conclude that if iron storage is a function of Yfh1, this function is certainly secondary to other more critical functions. The genetic interactions reported here with genes known to be involved in ISC biogenesis point to a role in this process. Indeed, recent reports have suggested a role for Yfh1/frataxin in ISC cluster biogenesis, perhaps by acting as an iron donor for assembly of a transient cluster on the Fe/S scaffold Isu (Gerber et al, 2003; Yoon & Cowan, 2003; Ramazzotti et al, 2004). The characteristics of the mutant proteins that we have studied do not preclude this possibility, as it is unresolved as to whether monomeric Yfh1 binds iron transiently and functions directly in delivery of iron to Isu. However, Yfh1 may have a role in ISC biogenesis independent of any direct interaction with iron. Further biochemical and genetic analysis will be required to determine the precise role of Yfh1/frataxin in ISC biogenesis.
Methods
Yeast strains, plasmids, media and chemicals. YFH1 mutant plasmids were generated by PCR mutagenesis using pRS316 YFH1 plasmid as template (Voisine et al, 2000) and then subcloned into the indicated vectors. Similarly, erv1ts, F124S (Lisowsky, 1994), was generated after amplification of the wt gene from genomic DNA.
All strains are in the W303 genetic background. Strains carrying YFH1 genes on plasmids were created by sporulation and dissection of YFH1/Δyfh1 diploids (Voisine et al, 2000) transformed with an appropriate centromeric plasmid. wt or mutant YFH1 were integrated into the LEU2 locus of Δyfh1 using standard integration procedures.
To construct erv1ts, first ERV1 was replaced by ERV1:TRP1 to create ERV1/Δerv1. The erv1ts mutant was then integrated into the LEU2 locus of the ERV1/Δerv1 strain. To create haploid double-deletion strains, a haploid Δyfh1 strain carrying either wt or 86/90/93/A mutant in a plasmid under the control of the tetO promoter (Garí et al, 1997) was first crossed to the haploid deletions Δssq1:HPH, Δnfu1:TRP1, Δisu1:LEU2, Δisa1:MET2 or Δisa2:HPH (Schilke et al, 1999; this study).
Yeast were grown in rich (YPD) and complete synthetic (CS) media. All reagents, unless stated otherwise, were purchased from Sigma.
Protein expression and purification. The mature form of Yfh1 (amino acids 52–174) with an N-terminal His tag and a thrombin cleaving site in between was cloned into pET-3a vector (Novagen, Madison, WI, USA). Protein overexpression was induced for 4 h at 37°C in BL21(DE3) E. coli by adding isopropyl-β-D-galactopyranoside at a final concentration of 1 mM. Cells were harvested and lysed following standard lysozyme treatment. Cell lysate was subjected to His-Bind Resin (Novagen) chromatography and protein (over 90% pure) was eluted with 200 mM imidazole in the binding buffer (50 mM Tris–HCl (pH 8), 50 mM NaCl). The His tag was cleaved using biotinylated thrombin, and thrombin was removed by streptavidin agarose (Novagen). The His tag was removed and digestion buffer was changed to 10 mM HEPES–KOH (pH 7.4) using Centriprep-10 concentrators (Millipore, Billerica, MA, USA).
In vitro oligomerization and iron binding assay. Purified Yfh1 was diluted to a concentration of 8 mM in 10 mM HEPES–KOH (pH 7.4), and Fe(NH4)2(SO4)2 was added at the indicated molar ratios. The mixture was aerobically incubated for 1 h at 30°C; after centrifugation at 20,817g, 4°C for 5 min, the supernatant was fractionated on a Superdex-200 column (Amersham Biosciences, Little Chalfont, UK) equilibrated in 10 mM HEPES–KOH (pH 7.4) and 100 mM NaCl. One-fifth of each fraction was analysed by SDS–PAGE and protein was detected with SYPRO-Ruby Protein Gel Stain (Molecular Probes, Eugene, OR, USA). Iron quantification of the same fractions was performed in a Perkin-Elmer 3030 Graphite Furnace Atomic Absorption Spectrometer.
Pull-down assay. His-Bind Resin (Novagen) equilibrated in 50 mM Tris–HCl (pH 8) and 50 mM NaCl was saturated with His-tagged wt or His-tagged 86/90/93A Yfh1 (BSA for control reactions) for 1 h at 4°C. In parallel, 500 μg of wt mitochondria (Gambill et al, 1993) was lysed for 5 min on ice in 50 mM Tris–HCl (pH 7.5), 80 mM KCl, 0.1% Triton X-100, 50 μM pyridoxal phosphate, 1 mM ascorbic acid, 10 mM imidazole and complete EDTA-free protein inhibitor cocktail (Roche, Basel, Switzerland) buffer. Membrane debris was removed by centrifugation at 16,000g, 4°C for 10 min. The supernatant was incubated with previously prepared protein beads for 1 h at 4°C. Beads were collected by centrifugation and washed twice with 500 μl of lysis buffer without Triton X-100 and with 60 mM imidazole.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Dr R. McClain for his inestimable help with the atomic absorption spectrometer. This work was supported by National Institutes of Health grants GM27870 (E.A.C.) and 5T326M08349 (A.A.). K.A. was a recipient of a Postdoctoral Fellowship from the Basque Country Government.
References
- Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G (2000) Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am J Hum Genet 67: 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adinolfi S, Trifuoggi M, Politou AS, Martin S, Pastore A (2002) A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum Mol Genet 11: 1865–1877 [DOI] [PubMed] [Google Scholar]
- Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Cavadini P, O'Neill HA, Benada O, Isaya G (2002) Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum Mol Genet 11: 217–227 [DOI] [PubMed] [Google Scholar]
- Cho SJ, Lee MG, Yang JK, Lee JY, Song HK, Suh SW (2000) Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc Natl Acad Sci USA 97: 8932–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Shigeta R, Chi YI, Ristow M, Shoelson SE (2000) Crystal structure of human frataxin. J Biol Chem 275: 30753–30756 [DOI] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol 123: 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Gerber J, Lill R (2002) Biogenesis of iron–sulfur proteins in eukaryotes: components, mechanism and pathology. Mitochondrion 2: 71–86 [DOI] [PubMed] [Google Scholar]
- Gerber J, Mühlenhoff U, Lill R (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep 4: 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan G, Santos JH, Graziewicz MA, Copeland WC, Isaya G, Van Houten B, Resnick MA (2003) Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum Mol Genet 12: 3331–3342 [DOI] [PubMed] [Google Scholar]
- Lange H, Lisowsky T, Gerber J, Mühlenhoff U, Kispal G, Lill R (2001) An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep 2: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, Dancis A (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron–sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem 274: 33025–33034 [DOI] [PubMed] [Google Scholar]
- Lisowsky T (1994) ERV1 is involved in the cell-division cycle and the maintenance of mitochondrial genomes in Saccharomyces cerevisiae. Curr Genet 26: 15–20 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R (2002) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet 11: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfo M (2002) Iron metabolism and mitochondrial abnormalities in Friedreich ataxia. Blood Cells Mol Dis 29: 536–547 [DOI] [PubMed] [Google Scholar]
- Park S, Gakh O, O'Neill HA, Mangravita A, Nichol H, Ferreira GC, Isaya G (2003) Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J Biol Chem 278: 31340–31351 [DOI] [PubMed] [Google Scholar]
- Ramazzotti A, Vanmansart V, Foury F (2004) Mitochondrial functional interactions between frataxin and Isu1p, the iron–sulfur cluster scaffold protein, in Saccharomyces cerevisiae. FEBS Lett 557: 215–220 [DOI] [PubMed] [Google Scholar]
- Rötig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P (1997) Aconitase and mitochondrial iron–sulphur protein deficiency in Friedreich ataxia. Nat Genet 17: 215–217 [DOI] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisine C, Schilke B, Ohlson M, Beinert H, Marszalek J, Craig EA (2000) Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol Cell Biol 20: 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon T, Cowan JA (2003) Iron–sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe–2S] clusters in ISU-type proteins. J Am Chem Soc 125: 6078–6084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data