Abstract
We describe a method for microscopic identification of DNA-synthesizing cells in bacterioplankton samples. After incubation with the halogenated thymidine analogue bromodeoxyuridine (BrdU), environmental bacteria were identified by fluorescence in situ hybridization (FISH) with horseradish peroxidase (HRP)-linked oligonucleotide probes. Tyramide signal amplification was used to preserve the FISH staining during the subsequent immunocytochemical detection of BrdU incorporation. DNA-synthesizing cells were visualized by means of an HRP-labeled antibody Fab fragment and a second tyramide signal amplification step. We applied our protocol to samples of prefiltered (pore size, 1.2 μm) North Sea surface water collected during early autumn. After 4 h of incubation, BrdU incorporation was detected in 3% of all bacterial cells. Within 20 h the detectable DNA-synthesizing fraction increased to >14%. During this period, the cell numbers of members of the Roseobacter lineage remained constant, but the fraction of BrdU-incorporating Roseobacter sp. cells doubled, from 24 to 42%. In Alteromonas sp. high BrdU labeling rates after 4 to 8 h were followed by a 10-fold increase in abundance. Rapid BrdU incorporation was also observed in members of the SAR86 lineage. After 4 h of incubation, cells affiliated with this clade constituted 8% of the total bacteria but almost 50% of the visibly DNA-synthesizing bacterial fraction. Thus, this clade might be an important contributor to total bacterioplankton activity in coastal North Sea water during periods of low phytoplankton primary production. The small size and low ribosome content of SAR86 cells are probably not indications of inactivity or dormancy.
As awareness of the importance, diversity, and complexity of marine microbial communities develops, it is increasingly recognized that an understanding of the ecology of the various bacterial (and archaeal) populations requires simultaneous information about both the identity and the activity of individual cells in environmental samples (8, 27, 49). Consequently, a considerable amount of recent research effort has focused on the development of techniques that permit estimation of various activities of single bacterial cells (16, 35). In addition, cultivation-independent approaches have been developed for quantification of the sizes of populations of individual bacterial taxa, such as fluorescence in situ hybridization (FISH) with oligonucleotide or polynucleotide rRNA-targeted probes (1, 9, 10, 17). FISH is an increasingly popular tool for microscopic visualization of individual groups of bacteria and archaea in the marine environment (7, 9, 14, 22, 29, 31). Furthermore, recent technical advances have greatly increased the sensitivity of this staining technique (9, 29, 30), so that the low ribosome content of many planktonic bacteria no longer limits the applicability of FISH to productive or coastal regions (30). One of the challenges of present-day microbial ecology is to combine FISH with techniques that allow researchers to determine defined microbial activities (e.g., microautoradiography) (8).
The introduction of tritiated thymidine (TdR) as a tracer has made it possible to estimate both community DNA synthesis rates by bulk uptake measurement methods (15) and the fraction of TdR-incorporating cells by microautoradiography (28). Bromodeoxyuridine (BrdU) is a halogenated nucleotide analogue of TdR that is also incorporated into newly synthesized DNA. The efficiency of BrdU incorporation into genomic DNA is not known and may be different in different species. Binnie and Coote (4) reported that TdR was replaced by BrdU at a rate of 25% in Bacillus subtilis DNA.
Pulse-labeling of DNA with BrdU and subsequent immunocytochemical (ICC) detection of labeled DNA are extensively used in histochemistry and cytochemistry to study the proliferation of eukaryotic cells (25, 34), whereas this technique has received comparatively little attention in microbial ecology. A keyword search of the Institute of Scientific Information database returned more than 5,200 citations containing the term “bromodeoxyuridine” for the past 10 years, but only 4 of these citations are relevant to environmental microbiology (5, 42, 45, 48). So far, BrdU has been used as a nonradioactive alternative to TdR to measure bacterioplankton growth rates (42), to separate DNA of growing and inactive bacteria (5, 45, 48), and for immunochemical labeling of marine bacterial isolates in pure cultures (45).
One major drawback of the ICC approach is that at this time the available highly sensitive methods for detection of BrdU incorporated into whole cells cannot be employed for microscopic visualization of individual bacterial cells from natural assemblages (38). Cell walls and membranes need to be permeabilized for antibody penetration, which usually results in a high level of species-specific cell loss (29). In addition, difficulties with cell preparation arise because the anti-BrdU antibody reacts only with single-stranded DNA (25). Here we describe a permeabilization and DNA denaturation protocol for ICC detection of BrdU incorporated into the DNA of individual bacterioplankton cells, and we describe simultaneous identification of individual DNA-synthesizing populations by FISH. To increase the sensitivity of the techniques, both FISH and BrdU detection were coupled with signal amplification by catalyzed reporter deposition (CARD) (29, 41).
MATERIALS AND METHODS
Bacterial isolates, growth conditions, and postincubation processing.
Twenty marine bacterial isolates (Table 1) were precultured overnight in 10 ml of autoclaved North Sea water amended with yeast extract (5 mg liter−1). Stationary-phase cultures were then inoculated into 10 ml of fresh medium supplemented with TdR (33 nM; Sigma-Aldrich, Seelze, Germany) and 5-bromo-2′-deoxyuridine (BrdU) (20 μM; Roche Diagnostics, Mannheim, Germany) and grown at room temperature (RT) in the dark for 21 h. Addition of TdR at trace concentrations blocked de novo synthesis of this compound (45). Negative controls without BrdU were included for each strain. The isolates were affiliated with the gamma subgroup of the Proteobacteria, the Cytophaga-Flavobacterium group, and the Actinobacteria. BrdU uptake by members of the alpha subgroup of the Proteobacteria has been demonstrated previously by Urbach et al. (45). A stationary-phase culture of Escherichia coli was transferred to fresh Luria broth amended with nalidixic acid (50 mg liter−1) to block de novo DNA synthesis and was incubated at 37°C. Controls were incubated in Luria broth without nalidixic acid. Thirty minutes after inoculation, cultures were pulsed with BrdU (20 μM) and TdR (33 nM) for 1.5 h and subsequently fixed as described below. Subsamples (0.1 to 5 ml) of the cell suspensions were filtered onto white polycarbonate membrane filters (type GTTP; pore size, 0.2 μm; diameter, 25 mm; Millipore, Eschborn, Germany), washed with 10 ml of deionized, particle-free water (MilliQ; Millipore), and stored at −20°C until further processing.
TABLE 1.
Bacterial strains that were tested for the ability to incorporate BrdU
| Group | Strain | Accession no. | Closest relativea | Labeling index (%) |
|---|---|---|---|---|
| Cytophaga-Flavobacterium-Bacteroides clade | 0234A | AF235124 | Flavobacterium columnare | >90 |
| 02ds22 | AF235114 | Cytophaga uliginosa | 100 | |
| 11ds02 | AF235111 | Cytophaga marinoflava | 100 | |
| 0803 | AF235117 | Flavobacterium salegense | 100 | |
| γ-Proteobacteria | 11ds10 | AF239707 | NOR6 clade | 100 |
| KT71 | AY007676 | NOR5 clade | >90 | |
| 0246 | AF173966 | Shewanella sp. | 100 | |
| 0919 | AF173964 | Colwellia psychrophila | >90 | |
| 0903 | AF235119 | Pseudoalteromonas atlantica | 100 | |
| 0910 | AF173963 | Pseudoalteromonas haloplanktis | 100 | |
| KT15 | Polaribacter sp. | 80-90 | ||
| 0924 | Oceanospirillum sp. | >90 | ||
| 0232 | AF235125 | Alteromonas sp. | >90 | |
| 1111 | AF173968 | Halomonas sp. | >90 | |
| 0901 | AF172840 | Vibrio splendidius | >90 | |
| 0248 | AF235127 | Photobacterium | 100 | |
| 11ds07 | AF235112 | NOR1 clade | 100 | |
| 1114 | AF235108 | NOR2 clade | 100 | |
| Actinobacteria | 1115 | AF2315113 | Micrococcus sp. | 30-40 |
| 1110 | AF239706 | NOR7 clade | 100 | |
| E. coli | ATCC 11775T | E. coli | 100 |
Closest relative as determined by 16S ribosomal DNA analysis.
Study site, sample preparation, and bacterial abundance.
Water samples were taken on 24 September 2001 from a depth of 1 m at the Helgoland Roads sampling site (54°09′N, 7°52′E) near the island of Helgoland, which is situated 23 miles offshore in the German Bight of the North Sea. Seawater was prefiltered through 1.2-μm-pore-size cellulose nitrate membrane filters (Sartorius, Göttingen, Germany) within 1 h after samples were collected, and subsamples (700 ml) were placed in acid-prewashed glass bottles. The filtrates were supplemented either with BrdU (20 μM) and TdR (33 nM) or with only TdR as controls. Duplicate preparations were incubated in the dark at the in situ temperature (10°C) with mild agitation. Subsamples (50 ml) were taken at four times (0, 4, 8, and 20 h) and fixed at 4°C for 20 h in a particle-free formaldehyde solution (final concentration in H2O, 2% [vol/vol]). The fixed samples were filtered onto white polycarbonate membrane filters (type GTTP; pore size, 0.2 μm; diameter, 47 mm; Millipore), washed with 10 ml of MilliQ H2O, and stored at −20°C until further processing. Total numbers of bacteria were determined by epifluorescence microscopy after staining with 4′,6′-diamidino-2-phenylindole (DAPI).
Sample preparation for FISH and ICC analyses.
A detailed protocol that includes all the sample-processing steps used for CARD-FISH and ICC analyses is shown in Table 2. To avoid cell loss during cell wall permeabilization and epitope retrieval, filters were dipped in low-gelling-point agarose (0.2% [wt/vol] in MilliQ H2O; MetaPhor, Bioproducts, Rockland, Maine), dried on glass slides at 46°C, and subsequently dehydrated in 96% (vol/vol) ethanol for 1 min (29). For DNA denaturation the filters were (i) incubated in preheated epitope retrieval buffer (95% [vol/vol] formamide, 1× SSC [15 mM sodium citrate plus 150 mM sodium chloride; pH 7.0], 0.5% [vol/vol] Triton X-100) at 60°C for 15 min and subsequently washed three times in excess MilliQ H2O at RT; (ii) dipped for 5 s in preheated HCl (1 M, 60°C) and then washed three times in excess MilliQ H2O at RT; and (iii) incubated in prewarmed permeabilization buffer (0.1 M Tris HCl, 0.1 M EDTA, 1% [vol/vol] Triton X-100) at 60°C for 5 min for nonenzymatic permeabilization and then washed in MilliQ H2O at RT, dehydrated in 96% (vol/vol) ethanol for 1 min, and air dried. Next, a second agarose embedding step was performed as described above. The air-dried and double-embedded filters were subsequently incubated in a lysozyme solution (10 mg of lysozyme per ml of 0.1 M Tris-0.05 M EDTA) at 37°C for 60 min and then washed in MilliQ H2O, dehydrated in 96% (vol/vol) ethanol for 1 min, and air dried.
TABLE 2.
Summary of steps used for FISH and BrdU detection in marine bacteria
| Step | Procedure |
|---|---|
| Embedding | Prepare subsamples on membrane filters |
| Dip filters in 0.2% low-gelling-point agarose, place filters face up on glass slides and let air dry at 46°C | |
| Dehydrate in 96% ethanol (1 min, RT), air drya | |
| Permeabilization and epitope unmasking | Incubate in epitope retrieval buffer (60°C, 15 min), wash in MilliQ H2O |
| Wash in HCl (1 M) (60°C, 5 s), wash in MilliQ H2O | |
| Incubate in permeabilization buffer (60°C, 5 min), wash in MilliQ H2O, wash in 96% ethanol, let air dry | |
| Repeat embedding step | |
| Incubate with lysozyme (10 mg/ml in 0.1 M Tris-HCl-0.05 M EDTA) (37°C, 60 min) | |
| Wash in MilliQ H2O, wash in 96% ethanol, air drya | |
| Hybridization | Place filter sections in reaction vial (1.5 ml, 10 to 20 sections per vial) and cover filter sections with 1,000 μl of hybridization buffer (0.5 ng of probe/μl), incubate at 35°C for 2 h |
| Wash filters in prewarmed washing buffer (5 min, 37°C) | |
| Tyramide signal amplification | Incubate in PBST (50 ml, RT, 15 min), dab filter on blotting paper, but do not let run dry |
| Incubate filter sections in freshly prepared substrate mixture (1 part of Alexa488-tyramide, 100 parts of amplification buffer) (37°C, 10 min, in the dark) | |
| Wash in PBST (RT, 5 min, in the dark), wash in MilliQ H2O (RT, 1 min), wash in ethanol (96%, RT, 1 min), air drya | |
| Antibody reaction | Bleach peroxidases with 0.01 M HCl (RT, 10 min), wash three times in MilliQ H2O |
| Place filter sections on Parafilm and cover with 500 μl of anti-BrdU reaction mixture (1.5 U of anti-BrdU-HRP per ml, 1 × PBS, 1% blocking reagent, 20 U of HAEIII per ml), seal with Parafilm and incubate at 37°C for 2.5 to 3 h | |
| Wash filter sections in PBST (10 min, RT, in the dark) | |
| Tyramide signal amplification | Dab filter sections on blotting paper but do not let run dry |
| Incubate in freshly prepared substrate mixture (1 part of Alexa536-tyramide, 200 parts of amplification buffer) (37°C, 10 min, in the dark) | |
| Dab filter sections on blotting paper, wash in PBST (5 min, RT), wash in MilliQ H2O (RT, 1 min), wash in ethanol (96%, RT, 1 min), air drya | |
| Embed samples and visualize |
Preparations may be stored at −20°C for several days to weeks without apparent loss of signal.
Synthesis of fluorescently labeled tyramides.
Tyramide HCl (Fluka, Taufkirchen, Germany) was labeled with succinimidyl esters of the fluorescent dyes Alexa488, Alexa546, and Alexa350 (Molecular Probes, Leiden, The Netherlands) as described previously for other fluorophores (21). A tyramide stock solution was prepared by dissolving 10 mg of tyramide HCl in 1 ml of dimethylformamide (DMF) containing 10 μl of triethylamine. Active dye esters were dissolved in DMF (10 mg ml−1) and then were added in 1.1-fold molar amounts to the tyramide stock solution and incubated at RT in the dark for 2 h on a rotation shaker (Dynal MTV; Dynal Biotech, Oslo, Norway) at 10 rpm. All stock solutions had to be freshly prepared (<10 min) prior to the labeling reaction. The synthesized tyramide conjugates were diluted with absolute ethanol to obtain a concentration of 1 mg ml−1. Lyophilized (freeze-dried) aliquots were stored at −20°C. For the signal amplification reaction, dye-conjugated tyramides were freshly dissolved in DMF, and the preparations could be stored at −20°C for at least 2 months without any reduction in reactivity.
FISH with enzyme-labeled oligonucleotide probes.
The pretreated filters were cut into 16 to 20 equal sections. Up to 25 individual filter sections were put into a 1.5-ml reaction vial (Eppendorf, Hamburg, Germany) and covered with 1,000 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris HCl [pH 7.5], 10% [wt/vol] dextran sulfate, 0.02% [wt/vol] sodium dodecyl sulfate, 55% [vol/vol] formamide [Fluka], 1% [wt/vol] blocking reagent [Boehringer, Mannheim, Germany], horseradish peroxidase [HRP]-labeled oligonucleotide probes [final concentration, 0.5 ng μl−1; Interactiva, Ulm, Germany] prepared as described previously [29]). The vial was incubated at 35°C for 2 h on a rotation shaker (10 rpm). Next, the filter sections were removed from the hybridization mixture and incubated in 50 ml of prewarmed washing buffer (3 mM NaCl, 5 mM EDTA [pH 8.0], 20 mM Tris HCl [pH 7.5], 0.01% [wt/vol] sodium dodecyl sulfate) at 37°C for 5 min. The FISH probes were targeted to Bacteria (EUB338), Roseobacter sp. (ROS537) (14), the SAR86 clade (SAR86-1249), Oceanospirillum sp. (OCE232), Alteromonas sp. (ALT1413), and Pseudoalteromonas sp. (PSA184) (12).
In situ detection of HRP-conjugated oligonucleotide probes.
To equilibrate the probe-delivered HRP, sections were incubated in 50 ml of PBST (1× phosphate-buffered saline [PBS], 0.05% Triton X-100) for 10 to 15 min at RT. To remove excess buffer, the filter sections were dabbed onto blotting paper, immediately transferred to a substrate mixture containing 1 part of tyramide-Alexa488 (or Alexa350 for isolates) and 100 parts of freshly prepared amplification buffer (1× PBS, 0.0015% H2O2), and incubated for 10 to 15 min at 37°C in the dark. Filter sections were again briefly put on blotting paper and washed for 5 to 10 min at RT in the dark in 50 ml of PBST. To decrease background fluorescence, sections were subsequently washed in MilliQ H2O and 96% ethanol (1 min each). At this stage, the preparations were either embedded in DAPI-amended mountant (11 parts of Citifluor AF1 [Citifluor Ltd., London, United Kingdom], 2 parts of VectaShield [Vector Laboratories, Burlingame, Calif.], 1 part of 1× PBS amended with DAPI [final concentration, 1 μg/μl]) for FISH counting or processed further for BrdU detection.
ICC detection of incorporated BrdU.
Filters were incubated in 0.01 M HCl for 10 min at RT in the dark in order to inactivate the FISH probe-delivered peroxidases and were subsequently washed three times in excess MilliQ H2O. Filter sections were then placed onto Parafilm and covered with 500 μl of antibody mixture (anti-BrdU-HRP, Fab fragments, clone BMG-6H8 [1.5 U ml−1; Roche Diagnostics, Mannheim, Germany], 1× PBS, 1% [wt/vol] blocking reagent, DNA restriction enzyme HaeIII [20 U ml−1; New England Biolabs, Beverly, Mass.]). The samples were then sealed with Parafilm, placed in a petri dish, incubated at 37°C for 2.5 to 3 h, and subsequently washed for 10 min at RT in 50 ml of PBST. Signal amplification (1 part of tyramide-Alexa546, 200 parts of amplification buffer) was carried out as described above for FISH. The filters were then air dried and stored at −20°C until further processing (within 3 days)
Microscopic evaluation.
Filter sections were covered with DAPI-amended mountant and evaluated with an Axioplan II microscope (Carl Zeiss, Jena, Germany) equipped with an HBO 100-W Hg vapor lamp, appropriate filter sets for Cy3 (Alexa546), fluorescein isothiocyanate (Alexa488), and DAPI (Alexa350) fluorescence (18), and a 100× Plan Apochromat objective. Between 600 and 800 DAPI-stained objects per filter were counted in a hybridized sample without ICC staining. In double-stained FISH and ICC preparations the fractions of BrdU-incorporating cells of all hybridized cells were quantified by using 10 individual microscopic fields.
RESULTS
Optimization of sample pretreatment and ICC.
Since heat (39), DNase (11, 44), detergents (47), and HCl (2) are all known to be useful in recovering immunoreactivity of BrdU epitopes in formalin-fixed tissues and cell preparations, it was necessary to investigate the effects of a variety of agents in combination with each other and at different temperatures on BrdU cell-labeling indices (i.e., the percentages of hybridized cells that showed BrdU labeling). The best results (highest labeling indices) were achieved by using a combination of several permeabilizing and epitope-unmasking steps (Table 2). Epitope retrieval was performed before hybridization, because of potential FISH signal loss during treatment with hot acid and formamide. In addition, samples were incubated with lysozyme, which is required for CARD-FISH permeabilization (29, 37). Filters that were not treated for epitope retrieval produced only a very few anti-BrdU-HRP signals.
Although BrdU incorporated into DNA was readily detected after only heat, formamide, and acid treatment, repeated staining with anti-BrdU-HRP resulted in different signal intensities and labeling indices (data not shown) (33). Furthermore, preliminary results suggested that the use of DNase I before the antibody reaction sometimes resulted in substantial loss of the target DNA. Therefore, we used a restriction enzyme to relax the partially denatured DNA. Since no restriction enzymes that specifically recognize T-containing sites could be used, a GG/CC cutter (HaeIII) was used (32). Nuclease digestion was carried out simultaneously with antibody binding. This strategy has been employed previously (11, 42) and is the recommended protocol in some commercially available cell proliferation assays (Boehringer Mannheim). For subsequent quantification of labeling indices of isolates and of DNA-synthesizing North Sea bacterioplankton populations by FISH and ICC, we used the optimal set of conditions determined in our preceding experiments (Table 2).
We also investigated whether the various permeabilization procedures resulted in loss of cells from the filters. Initially, we sometimes observed detachment of pieces of the agarose cover after the lysozyme incubation step, possibly caused by acid and heat during epitope retrieval. Therefore, the embedding step was repeated before treatment with lysozyme. This effectively restored the protective properties of the agarose cover during all subsequent incubations. The mean bacterial cell numbers (DAPI counts) in the bacterioplankton samples were 5.4 × 105 ± 0.6 × 105 cells ml−1 (mean ± 1 standard deviation) for the untreated samples and 5.1 × 105 ± 0.6 × 105 cells ml−1 after all permeabilization steps. In summary, no significant decrease in cell number was observed as a result of the pretreatment procedures (P > 0.05, as determined by a Wilcoxon matched pair test [n = 10]).
Bacterial isolates.
All of the strains investigated, including the E. coli strains, were able to incorporate BrdU into DNA (Table 1). Because the DAPI signal is partially lost during ICC staining, probably due to the partially denatured DNA, the relative amount of active bacteria was always expressed as a percentage of the cells that hybridized with EUB338-HRP. The majority of bacterial cultures that had been supplemented with BrdU showed labeling indices that were greater than 90% of the cells that hybridized with EUB338-HRP. Using our protocol, we did not detect anti-BrdU-HRP-positive cells in samples that were not supplemented with BrdU. After BrdU pulse-labeling of E. coli cultures, we observed no staining of cells with anti-BrdU-HRP in preparations amended with nalidixic acid.
Quantification of bacterioplankton by FISH and ICC.
During 20 h of incubation with no substrate addition, the total numbers of North Sea bacterioplankton cells determined by EUB338 FISH counting did not change or increased very little for the various treatments (Fig. 1). The bacterial cells visualized by FISH with EUB338-HRP accounted for the majority (mean, 97.9%; range, 90 to 100%) of the DAPI-stained cells (Table 3). Therefore, the total amounts of BrdU-positive bacteria were expressed as the percentages of all hybridized cells. The fractions of cells that hybridized with probes OCE232 and PSA184 were less than 1% of the DAPI-stained cells, and therefore these cells were excluded from further analysis.
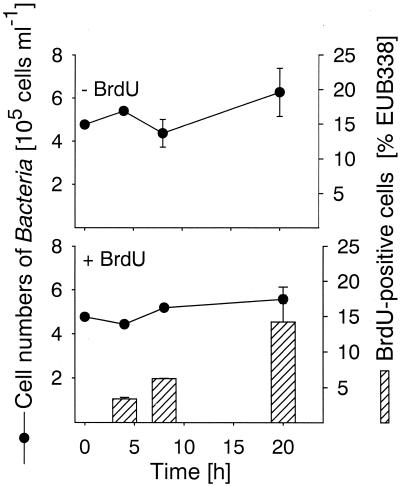
FIG. 1.
Numbers of bacterioplankton cells in filtrates (pore size, 1.2 μm) of North Sea surface water detected by probe EUB338-HRP during 20 h of incubation. The bars indicate the percentages of anti-BrdU-HRP-positive Bacteria cells, and the error bars indicate the total ranges for duplicates.
TABLE 3.
Percentages of DAPI-stained cells detected by probe EUB338-HRP in filtrates of North Sea surface water during 20 h of incubation
| Time (h) | % of DAPI-stained cells
|
|||
|---|---|---|---|---|
| Without BrdU
|
With BrdU
|
|||
| Sample I | Sample II | Sample I | Sample II | |
| 0 | 94.13 | 94.13 | 94.14 | 94.14 |
| 4 | 90.42 | 98.66 | 93.69 | 99.21 |
| 8 | 93.47 | 99.62 | 97.68 | 99.25 |
| 20 | 100.00 | 100.00 | 100.00 | 100.00 |
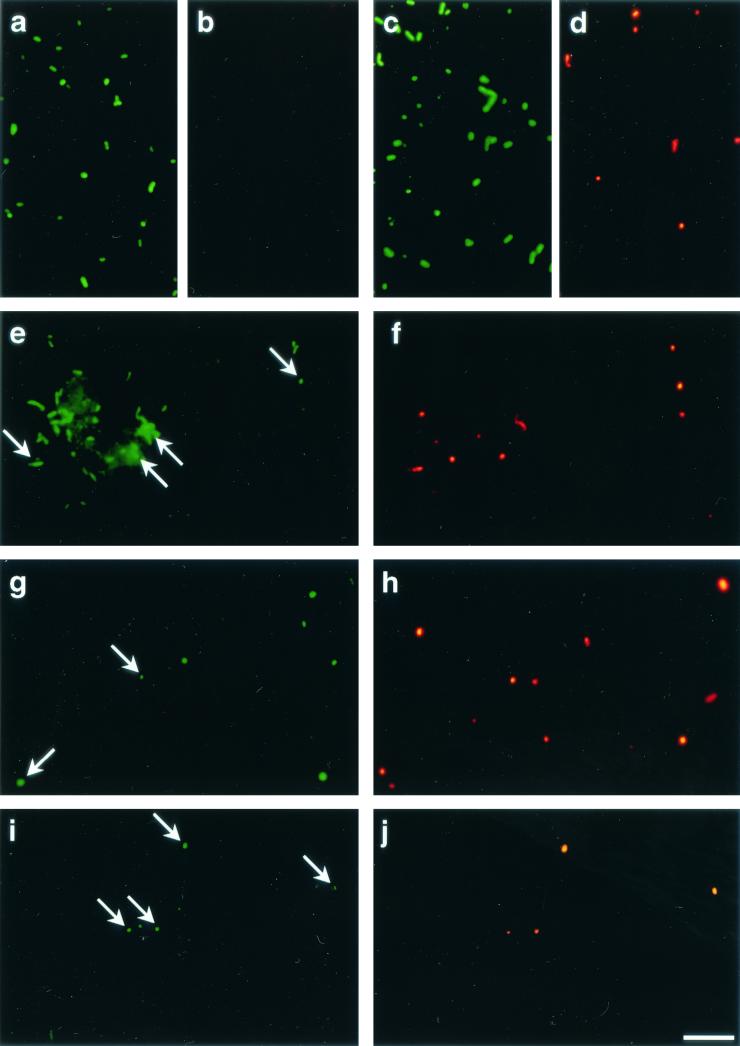
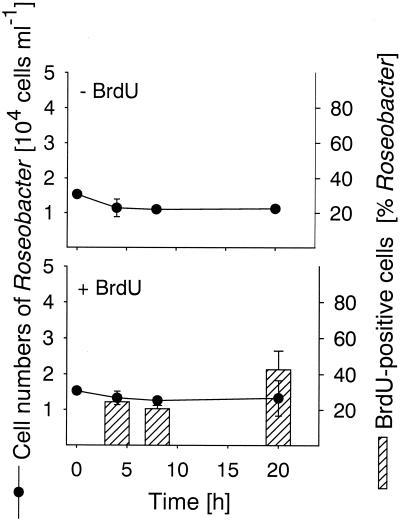
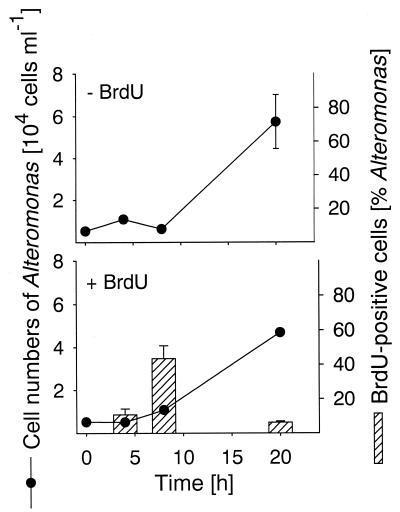
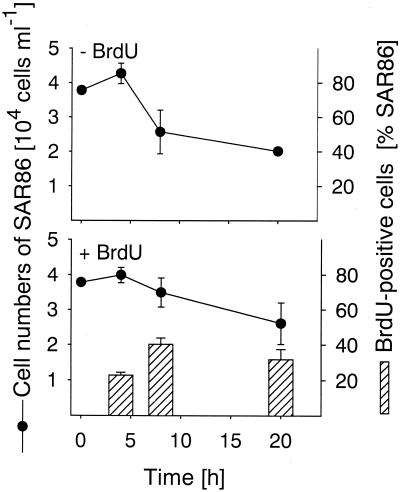
In all three bacterial populations studied BrdU-incorporating cells could be readily visualized by BrdU antibody staining and CARD (Fig. 2). The total detection rates with the anti-BrdU-HRP antibody increased from 3.4% of the cells that hybridized with EUB338-HRP (range, 3.2 to 3.6%) after 4 h of incubation to 14.2% of the cells that hybridized with EUB338-HRP (range, 9.3 to 19.2%) after 20 h (Fig. 1). We found no anti-BrdU-HRP-positive cells in the zero-time sample and in samples that were not supplemented with BrdU. There was no clear change in cell number for bacteria affiliated with the genus Roseobacter during 20 h of incubation (Fig. 3), and the detection rates for probe ROS537-HRP ranged between 1.4 and 3.3% of the DAPI-stained cells (mean, 2.4%). The BrdU labeling indices for Roseobacter increased from 24.4% of the cells detected with probe ROS537 (range, 20.6 to 28.1%) after 4 h of incubation to 42.7% (range, 32.3 to 53.0%). The numbers of Alteromonas sp. cells increased 10-fold during 20 h of incubation, from 5.2 × 103 to 4.9 × 104 cells ml−1 (Fig. 4). Although the relative amounts of the cells hybridizing with probe ALT1413 rose from 1.0 to 8.8% of the DAPI-stained cells (range, 8.1 to 9.5%), the change was too small to be clearly reflected in the total bacterial cell numbers (Fig. 1). The percentages of anti-BrdU-HRP-positive cells were highest (mean, 43.6% of the cells that hybridized with ALT1413; range, 36 to 51%) at the onset of growth after 8 h of incubation and declined thereafter (6.3% of the cells that hybridized with ALT1413; range, 5.4 to 7.1%). Members of the SAR86 clade constituted 8.1% of the total cell counts but represented almost 50% of all DNA-synthesizing bacteria after 4 h of incubation with BrdU. During incubation the concentration of SAR86 cells decreased from 3.8 × 104 cells ml−1 at zero time to 2.2 × 104 cells ml−1 (range, 2.0 × 104 to 3.2 × 104 cells ml−1) (Fig. 5). Concomitantly, a decline in the FISH intensities of hybridized cells was observed after 20 h despite CARD signal amplification. The percentage of anti-BrdU-HRP-positive SAR86 cells was highest after 8 h of incubation (40.2%; range, 36.6 to 43.8%) and remained constant or even declined slightly (mean, 31.7%; range, 25.9 to 37.5%) after 20 h.
FIG. 2.
Photomicrographs of FISH- and anti-BrdU-HRP-stained marine bacteria. Each pair of panels shows the FISH signal (green) (left) and the anti-BrdU-HRP signal (red) (right). (a and b) Probe EUB338-HRP and anti-BrdU-HRP staining of preparations incubated without BrdU; (c and d) probe EUB338-HRP and anti-BrdU-HRP staining of preparations supplemented with BrdU; (e and f) probe ALT1413-HRP, specific for members of the Alteromonas lineage, and anti-BrdU-HRP staining; (g and h) probe ROS537-HRP, specific for members of the Roseobacter lineage, and anti-BrdU-HRP staining; (i and j) probe SAR86-1249-HRP, specific for members of the SAR86 clade, and anti-BrdU-HRP staining. The arrows indicate cells that show both FISH staining and anti-BrdU-HRP staining. Bar, 10 μm.
FIG. 3.
Cell numbers of members of the Roseobacter lineage detected by probe ROS537-HRP. The bars indicate the percentages of anti-BrdU-HRP-positive Roseobacter cells, and the error bars indicate the total ranges for duplicates.
FIG. 4.
Cell numbers of members of the Alteromonas lineage detected by probe ALT1413-HRP. The bars indicate the percentages of anti-BrdU-HRP-positive Alteromonas cells, and the error bars indicate the total ranges for duplicates.
FIG. 5.
Cell numbers of members of the SAR86 clade detected by probe SAR86. The bars indicate the percentages of anti-BrdU-HRP-positive SAR86 cells, and the error bars indicate the total ranges for duplicates.
DISCUSSION
We successfully developed a protocol for ICC detection of BrdU incorporation into whole bacterial cells from environmental samples. In spite of a low ambient temperature (10°C) and the typically low DNA synthesis rates of slowly growing bacterioplankton during autumn (40), a population of BrdU-positive cells could be readily visualized after 4 h of incubation (Fig. 1). Similar estimates of the DNA-synthesizing fraction of free-living bacterioplankton cells were obtained by microautoradiography of tritiated TdR ([3H]TdR) in a brackish water environment (28). It is nevertheless possible that only the most rapidly DNA-synthesizing bacteria are visualized by our method. We can envisage two potential shortcomings of the protocol for ICC detection of DNA synthesis: (i) the signal intensity sets a threshold for detection and (ii) not all pelagic bacteria are able to incorporate BrdU.
Sensitivity of the BrdU assay.
CARD amplification of the BrdU signal was required to reduce the risk of underestimating the number of proliferating cells. The use of HRP-labeled antibodies for detection of BrdU results in a significant increase in signal intensity compared to the weak and heterogeneous signals of directly fluorochrome-labeled antibodies (46). To our knowledge, this is the first report of using a directly HRP-labeled antibody Fab fragment for ICC staining of BrdU. Such a fragment is probably more suitable for antigen detection inside bacterial cells than a whole antibody because of its smaller size (50 kDa, compared to approximately 160 kDa for a whole immunoglobulin G antibody).
BrdU labeling indices are affected by many factors, and incorporation rates can be underestimated, depending on the fixative and/or pretreatment used (24). We cannot exclude the possibility that other pretreatment strategies might result in even more sensitive detection of BrdU incorporation. For example, it has been shown that the influence of formalin on tissue antigenicity depends on the total time of fixation. This means that immunostaining results can be compared only when the fixative, the time of fixation, and the method of epitope retrieval are standardized. In our experiment, we used formaldehyde fixation, since to our knowledge this is the only strategy which is useful if subsequent permeabilization with lysozyme is required. Massive cell loss was observed after lysozyme treatment if samples were fixed with ethanol (A. Pernthaler, unpublished data).
Selective uptake and potential toxicity of BrdU.
Twenty years ago, microbiologists claimed that wild-type bacteria do not take up BrdU to any significant extent (6). However, the E. coli strain used in this study and all 20 marine bacterial isolates tested were able to incorporate BrdU into DNA. The majority of the isolates showed BrdU labeling indices of >90% of the cells that hybridized with EUB338-HRP, indicating that our protocol is adequate for detection of growth for a variety of marine bacteria. In contrast to a previous report (45), several isolates belonging to the Cytophaga-Flavobacterium lineage could also be readily stained by our assay. This finding is important, because a large fraction of the bacterioplankton in coastal North Sea surface water consists of Cytophaga-related microbes during the spring and summer months (13). Judging from our results obtained with both isolates and uncultured populations (Table 1 and Fig. 3 and 5), it is thus likely that members of the various phylogenetic lineages common in marine bacterioplankton are able to take up and incorporate BrdU.
Several assumptions underlie the use of TdR and BrdU as tracers for the study of DNA synthesis patterns in microbial communities (38). For example, it is assumed that [3H]TdR is either incorporated into DNA by cells or lost (i.e., that the tritium label is not transferred to cellular constituents other than DNA as a result of TdR catabolism). It is also assumed that cells do not have large endogenous TdR pools and thus incorporate exogenous [3H]TdR into DNA if the labeled material is present during the period of DNA synthesis. Two advantages of BrdU over [3H]TdR as a marker are its probable greater stability and perhaps greater specificity. During immunodetection, the specificity of the antibody for BrdU presumably ensures that brominated degradation products, whether they are produced during storage or by cell metabolism, are not detected if they are incorporated into other compounds. Moreover, previous studies have established that TdR added to samples at a trace concentration (33 nM) is sufficient to inhibit the activity of thymidylate synthase, an enzyme required for de novo synthesis of dTMP, in bacterial strains capable of importing TdR (26). This inhibition forces dependence on imported nucleotides, resulting in increased incorporation of exogenously supplied BrdU.
In many eukaryotic cells BrdU selectively affects processes associated with cell differentiation but has no significant effect on cell growth or overall RNA and protein synthesis (36). At high concentrations, BrdU negatively affected the growth rate, viability, and sporulation of a TdR-requiring Bacillus subtilis strain (4). In contrast, the growth kinetics of marine bacterial cultures treated with BrdU and TdR were found to be similar to those of cultures treated with TdR alone (45), suggesting that the level of toxicity of BrdU is low when it is used at a concentration of 20 μM. Preferential incorporation of TdR over BrdU has been observed in E. coli (20), but again the effect was found to decline with increasing BrdU/TdR ratios in the medium. In summary, BrdU appears to be adequate as a marker of DNA synthesis for pulse-labeling and short-term incubation of bacterioplankton.
Simultaneous in situ identification and activity staining.
Although there are several techniques for detection of growth-related features in individual cells (16, 23, 35), these techniques are not designed to distinguish between individual active bacterial populations. It is increasingly recognized that such a black-box approach needs to be supplemented by simultaneous information concerning bacterial identity (8). Our method was specifically developed for studying growth of different members of the bacterioplankton. Based on CARD signal amplification, this method was sensitive enough for detection of DNA synthesis in cells of the SAR86 clade that could not be visualized by FISH with monolabeled probes because of their low ribosome content (29).
All groups investigated, including Roseobacter, Alteromonas, and SAR86, were capable of incorporating BrdU, but they showed different patterns for abundance and BrdU labeling indices. The numbers of cells of members of the Roseobacter clade did not change during the 20 h of incubation, but the fraction of BrdU-positive Roseobacter cells approximately doubled (Fig. 3). Since DNA synthesis occurs before cell division, Roseobacter showed a pattern typical of the onset of cell division. Thus, the incubation conditions resulted in activation of one or several subpopulations belonging to this lineage. This agrees with the finding that some Roseobacter spp. and members of related genera are readily culturable (13, 19).
Members of the Alteromonas lineage also synthesized DNA during incubation, as indicated by BrdU labeling after 8 h of incubation. The high ICC labeling index at that time and the subsequent increase in Alteromonas abundance (Fig. 4) provide evidence that a large fraction of BrdU-positive cells indeed precedes cell multiplication. Therefore, our assay could be a sensitive measure of the in situ growth potential of bacterial populations. Interestingly, the fraction of anti-BrdU-HRP-positive Alteromonas cells was lower during cell multiplication than during the onset of growth. Some strains of this lineage might have selectively decreased the uptake of BrdU, as has been demonstrated for mutant B. subtilis strains (6), or might have catabolized the incorporated nucleotide. Moreover, BrdU is known to undergo a tautomeric shift with higher frequency than TdR. In this case its normal hydrogen bonding is disrupted, and it binds to guanine instead of adenine (43). In the second round of DNA replication, the polymerase recognizes the mismatch and removes one of the two bases. Thus, due to the potential long-term mutagenicity of BrdU and its eventual removal from de novo synthesized genomes, we suggest that labeling of DNA with BrdU should be carried out in short-term incubation experiments.
After 4 h of incubation the SAR86 population constituted 8.1% of the total bacterial population but approximately 50% of all DNA-synthesizing bacteria. This indicates that during a season in which there is generally declining bacterial activity (13) SAR86 may represent an important component of the active fraction of the bacterioplankton in coastal North Sea surface water. It also illustrates that a cell size of <1 μm and a low ribosome content (12) do not imply nongrowth or dormancy (16) in this bacterial lineage. Although the SAR86 population was active in the original filtrates, the cell numbers and signal intensities of hybridized cells decreased during incubation (Fig. 5). This indicates that SAR86 was deactivated by the treatments. Proteorhodopsin genes from environmental genome fragments of marine bacteria related to SAR86 have been described (3), implying that there is potential for light-driven energy generation in this lineage. Since incubation was carried out in the dark (to avoid BrdU phototoxicity), the enrichment conditions might have been detrimental for SAR86 cells. Furthermore, the possible phototrophy of SAR86 is consistent with our finding that this group constituted an important fraction of all DNA-synthesizing cells during a season when there was low input of organic carbon from phytoplankton primary production.
While BrdU incorporation in combination with FISH can indicate that specific populations of bacteria in the plankton are growing, this method cannot exclude the possibility that there is DNA synthesis in a particular species in the environment, unless the species has been shown to incorporate BrdU in pure culture. Nevertheless, our approach could provide information about the growth activity of different uncultured microbial populations even during a period when microbial activity is expected to be low. Thus, we hope that the combination of in situ identification and detection of DNA synthesis in individual cells may provide new insight into the ecological roles of different bacterial lineages in the marine plankton.
Acknowledgments
The Biologische Anstalt Helgoland is acknowledged for providing guest research facilities. We thank C. Alonso for help with sampling and N. Neese for advice on fluorescent staining.
This work was supported by the German Ministry of Education and Research (grant BMBF 01 LC0021/TP4) and by the Max Planck Society.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bak, P. M., and R. J. Panos. 1997. Protease antigen recovery decreases the specificity of bromodeoxyuridine detection in formalin-fixed tissue. J. Histochem. Cytochem. 45:1165-1170. [DOI] [PubMed] [Google Scholar]
- 3.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 4.Binnie, C., and J. G. Coote. 1986. Inhibition of sporulation in Bacillus subtilis by bromodeoxyuridine and the effect on DNA replication. J. Gen. Microbiol. 132:493-502. [DOI] [PubMed] [Google Scholar]
- 5.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specified stimuli. Appl. Environ. Microbiol. 65:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coote, J. G., and C. Binnie. 1986. Tolerance to bromodeoxyuridine in a thymidine-requiring strain of Bacillus subtilis. J. Gen. Microbiol. 132:481-492. [DOI] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic Archaea and Bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 11.Dinjens, W. N. M., J. Tenkate, M. Lenders, E. P. M. Vanderlinden, and F. T. Bosman. 1992. Bromodeoxyuridine (BrdU) immunocytochemistry by exonuclease-III (exo-III) digestion. Histochemistry 98:199-205. [DOI] [PubMed] [Google Scholar]
- 12.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 16.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and A. Hagstrom. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton composition in lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanawalt, P. C. 1967. Preparation of 5-bromouracil-labeled DNA. Methods Enzymol. 12:702-708. [Google Scholar]
- 21.Hopman, A. H. N., F. C. S. Ramaekers, and E. J. M. Speel. 1998. Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for in situ hybridization using CARD amplification. J. Histochem. Cytochem. 46:771-777. [DOI] [PubMed] [Google Scholar]
- 22.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 23.Lebaron, P., P. Servais, H. Agogue, C. Courties, and F. Joux. 2001. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 67:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGinley, J. N., K. K. Knott, and H. J. Thompson. 2000. Effect of fixation and epitope retrieval on BrdU indices in mammary carcinomas. J. Histochem. Cytochem. 48:355-362. [DOI] [PubMed] [Google Scholar]
- 25.Moran, R., Z. Darzykiewicz, L. Staiano-Coico, and M. R. Melamed. 1985. Detection of 5-bromodeoxyuridine (BrdUrd) incorporation by monoclonal antibodies: role of the denaturation step. J. Histochem. Cytochem. 33:821-827. [DOI] [PubMed] [Google Scholar]
- 26.Moriarty, D. J. W. 1986. Measurement of bacterial growth rates in aquatic systems from rates of nucleic acid synthesis. Adv. Microb. Ecol. 9:245-292. [Google Scholar]
- 27.Ouverney, C. C., and J. A. Fuhrman. 2001. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 67:1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedros-Alio, C., and S. Y. Newell. 1989. Microautoradiographic study of thymidine uptake in brackish waters around Sapelo Island, Georgia, USA. Mar. Ecol. Prog. Ser. 55:83-94. [Google Scholar]
- 29.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. A comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 32.Petruska, J., and D. Horn. 1983. Sequence-specific responses of restriction endonucleases to bromodeoxyuridine substitution in mammalian DNA. Nucleic Acids Res. 11:2495-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raap, A. K., J. G. J. Marijnen, J. Vrolijk, and M. Van der Ploeg. 1986. Denaturation, renaturation, and loss of DNA during in situ hybridization procedures. Cytometry 7:235-242. [DOI] [PubMed] [Google Scholar]
- 34.Rizzoli, R., N. M. Maraldi, A. Galanzi, N. Zini, M. Falconi, M. Vitale, and G. Mazzotti. 1988. High-sensitivity detection of DNA-synthesis by immunolocalization of bromodeoxyuridine. Inst. Phys. Conf. Ser. 93:551-552. [Google Scholar]
- 35.Rodriguez, G., D. Phipps, K. Ishiguro, and H. Ridgway. 1992. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl. Environ. Microbiol. 58:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutter, W. J., R. C. Pictet, and M. P. W. 1973. Toward molecular mechanisms of development processes. Annu. Rev. Biochem. 42:601-664. [DOI] [PubMed] [Google Scholar]
- 37.Schönhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro, H. M. 1995. Practical flow cytometry, 3rd ed. Wiley-Liss, Inc., New York, N.Y.
- 39.Shi, S.-R., M. E. Key, and K. L. Kalra. 1991. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 39:741-748. [DOI] [PubMed] [Google Scholar]
- 40.Shiah, F. K., and H. W. Ducklow. 1995. Multiscale variability in bacterioplankton abundance, production, and specific growth rate in a temperate salt-marsh tidal creek. Limnol. Oceanogr. 40:55-66. [Google Scholar]
- 41.Speel, E. J. M., F. C. S. Ramaekers, and A. H. N. Hopman. 1997. Sensitive multicolor fluorescence in situ hybridization using catalyzed reporter deposition (CARD) amplification. J. Histochem. Cytochem. 45:1439-1446. [DOI] [PubMed] [Google Scholar]
- 42.Steward, G. F., and F. Azam. 1999. Bromodeoxyuridine as an alternative to H-3-thymidine for measuring bacterial productivity in aquatic samples. Aquat. Microb. Ecol. 19:57-66. [Google Scholar]
- 43.Stryer, L. 1994. Biochemie. Spektrum Akademischer Verlag, Heidelberg, Germany.
- 44.Takagi, S., M. L. McFadden, R. E. Humphreys, B. A. Woda, and T. Sairenji. 1993. Detection of 5-bromo-2-deoxyuridine (Brdurd) incorporation with monoclonal anti-Brdurd antibody after deoxyribonuclease treatment. Cytometry 14:640-648. [DOI] [PubMed] [Google Scholar]
- 45.Urbach, E., K. L. Vergin, and S. J. Giovannoni. 1999. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl. Environ. Microbiol. 65:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Heusden, J., P. de Jong, F. Ramaekers, H. Bruwiere, M. Borgers, and G. Smets. 1997. Fluorescein-labeled tyramide strongly enhances the detection of low bromodeoxyuridine incorporation levels. J. Histochem. Cytochem. 45:315. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, D. M., and C. Bianchi. 1999. Improved immunodetection of nuclear antigens after sodium dodecyl sulfate treatment of formaldehyde-fixed cells. J. Histochem. Cytochem. 47:1095-1100. [DOI] [PubMed] [Google Scholar]
- 48.Yin, B., D. Crowley, G. Sparovek, W. J. De Melo, and J. Borneman. 2000. Bacterial functional redundancy along a soil reclamation gradient. Appl. Environ. Microbiol. 66:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zubkov, M. V., B. M. Fuchs, P. H. Burkill, and R. Amann. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]