Abstract
Degradation of oil on beaches is, in general, limited by the supply of inorganic nutrients. In order to obtain a more systematic understanding of the effects of nutrient addition on oil spill bioremediation, beach sediment microcosms contaminated with oil were treated with different levels of inorganic nutrients. Oil biodegradation was assessed respirometrically and on the basis of changes in oil composition. Bacterial communities were compared by numerical analysis of denaturing gradient gel electrophoresis (DGGE) profiles of PCR-amplified 16S rRNA genes and cloning and sequencing of PCR-amplified 16S rRNA genes. Nutrient amendment over a wide range of concentrations significantly improved oil degradation, confirming that N and P limited degradation over the concentration range tested. However, the extent and rate of oil degradation were similar for all microcosms, indicating that, in this experiment, it was the addition of inorganic nutrients rather than the precise amount that was most important operationally. Very different microbial communities were selected in all of the microcosms. Similarities between DGGE profiles of replicate samples from a single microcosm were high (95% ± 5%), but similarities between DGGE profiles from replicate microcosms receiving the same level of inorganic nutrients (68% ± 5%) were not significantly higher than those between microcosms subjected to different nutrient amendments (63% ± 7%). Therefore, it is apparent that the different communities selected cannot be attributed to the level of inorganic nutrients present in different microcosms. Bioremediation treatments dramatically reduced the diversity of the bacterial community. The decrease in diversity could be accounted for by a strong selection for bacteria belonging to the alkane-degrading Alcanivorax/Fundibacter group. On the basis of Shannon-Weaver indices, rapid recovery of the bacterial community diversity to preoiling levels of diversity occurred. However, although the overall diversity was similar, there were considerable qualitative differences in the community structure before and after the bioremediation treatments.
Although accidental releases account for only a small percentage of the oil released into the marine environment, large oil spills attract much public attention and cause environmental concern. Spill incidents have prompted research on cost-effective, environmentally benign cleanup strategies. Physical and, on rare occasions, chemical methods are capable of rapidly removing the majority of beached oil, but they are rarely completely successful (33). Natural degradative processes aid in removing the remaining oil.
Bacteria are considered to represent the predominant agents of hydrocarbon degradation in the environment (27), and hydrocarbon-degrading bacteria are ubiquitous. More than 20 genera of marine hydrocarbon-degrading bacteria, distributed over several (sub)phyla (α-, β-, and γ-proteobacteria; Gram positives; Flexibacter-Cytophaga-Bacteroides) have been described so far (5, 13, 16-18, 22, 48). As a single species typically is capable of degrading only a limited number of the compounds found in crude oil, a consortium composed of many different bacterial species is usually involved in oil degradation.
Because of the high carbon content of oil and the low level of other nutrients essential for microbial growth, the rate and extent of degradation are, in general, limited by the low availability of nitrogen and phosphorus (2, 33). Consequently, growth of hydrocarbon-degrading bacteria and hydrocarbon degradation can be strongly enhanced by fertilization with inorganic N and P. This has proven an effective bioremediation treatment on several types of shorelines (4, 40, 41, 46).
Bioremediation studies have, in general, been dominated by an empirical approach, and optimum nutrient amendment levels are often informed by laboratory incubations. In the field, care must be taken in supplying optimum concentrations of inorganic nutrients. Too high concentrations may result in eutrophication, and too low concentrations may result in suboptimal biodegradation. A better understanding of the systematic effects of nutrient amendment on biodegradative microbial populations and the progress of bioremediation would assist the development of more rational bioremediation strategies (21, 37). Therefore, laboratory beach microcosm experiments were performed to determine the effects of different levels of N and P supply on oil degradation and bacterial community dynamics. Microbial communities were characterized by using cultivation-independent molecular techniques (16S rRNA gene [rDNA] PCR-based denaturing gradient gel electrophoresis [DGGE] [31] and clone libraries). Numerical analysis (34) was used to assess changes in microbial community structure and the effects of bioremediation treatments.
MATERIALS AND METHODS
Field site.
Sediment for microcosms was obtained from the upper part of the intertidal zone of Stert Flats, Somerset, United Kingdom (51°12.3′ N, 3°3.9′ W). The upper 10 to 15 cm of this beach is mixed by tidal action and waves, and sediment was collected from this depth interval. Sediment was transported to the laboratory in a cool box at ambient temperature (<10°C) and used for microcosm experiments within 1 day of collection. The sediment was a fine sand (80% of the particles were in the range of 125 to 180 μm) containing 3.2% mud (sediment particles of <63 μm), with a density of 1.6 g/cm3 (wet weight) and a moisture content of 34% (vol/vol) at the time of collection at low tide. The average total organic matter content of 10 sediment samples was 0.6% ± 0.12%. The slope of the beach was 4% at the sampling point. On the basis of this slope and tide information, a tidal regimen for the laboratory microcosms was set at a mean vertical flow rate of seawater of 1.6 m/h and a submergence of the sediment for 1 h in every 12 h.
Microcosm experiments.
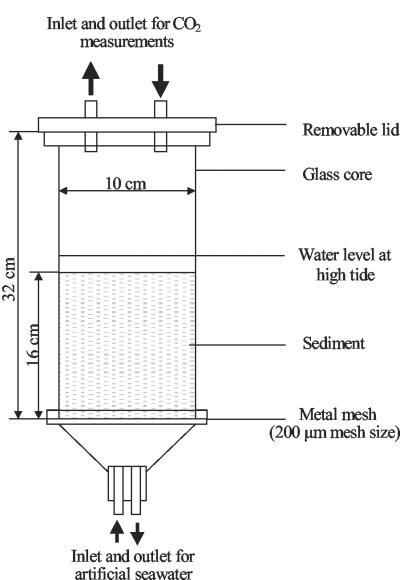
Sediment was homogenized with synthetic seawater (Instant Ocean; Aquarium Systems Inc.) in the laboratory, and 2.0 kg of wet sediment was placed into a microcosm (Fig. 1) and held at 20 ± 3°C. Fresh synthetic seawater provided each beach microcosm with two tidal cycles each day (12 h apart, 1 liter of seawater per cycle), with the ebb tide waters drained from each microcosm into 20-liter acid-washed plastic bottles. To allow equilibration of physical and chemical parameters, the microcosms were subjected to six tidal cycles before the addition of oil. Oil spilled at sea and washed onto a beach was simulated by weathering forties crude oil by distillation at 250°C in order to remove low-molecular-weight volatile hydrocarbons (e.g., <nC11 and BTEX [benzene, toluene, ethylbenzene, xylene]). Total resolvable hydrocarbons (TRH), nC11-to-nC35 alkanes, and aromatic hydrocarbons contributed, respectively, 31.2% ± 3.8%, 12.7% ± 0.6%, and 1.9% ± 0.2% of the total petroleum hydrocarbons (TPH) in the weathered oil. The weathered oil was mixed vigorously with synthetic seawater (25%, vol/vol), to form a stable water-in-oil emulsion. The emulsion was added to the microcosms at high tide at a level of 3.7 kg/m2 by pouring it onto the seawater. Inorganic nutrient solutions were added 24 h after oil addition and also after 7, 14, and 21 days. Each nutrient addition (50 ml in tap water) was calculated to provide a defined percentage of N (as sodium nitrate) and P (as potassium dihydrogen phosphate) relative to the total oil mass on a weight basis. The different treatments applied to the microcosms are shown in Table 1. Since the microcosms were labor intensive and resource demanding, only a few treatments were replicated (Table 1). Beach sediment treated with fertilizer alone was used as a control rather than untreated sediments because previous experiments had shown that nutrient-amended beach sediments produced DGGE profiles identical to those of untreated beach sediments and that the profiles obtained were stable over a 108-day sampling period (42).
FIG. 1.
Design of the microcosms used in this study.
TABLE 1.
Overview of treatments of laboratory beach sand microcosms and molecular analyses of samples taken from the microcosms
| Treatment code | No. of microcosmsa | Oil addition (kg/m2) | NaNO3 addition (% N [wt/wt] in oil) | KH2PO4 addition (% P[wt/wt] in oil) | Samples (days after fertilization) selected for:
|
|
|---|---|---|---|---|---|---|
| DGGE analysis | Clone librariese | |||||
| FOb,c | 3 | 0 | 2 | 0.2 | 0, 2, 6, 13, 20, 26 | 0 (51), 6 (40), 26 (41) |
| 0% Nc | 3 | 3.7 | 0 | 0 | 0, 2, 6, 13, 20, 26 | 0 (50), 6 (45), 26 (78) |
| 0.25% N | 1 | 3.7 | 0.25 | 0.025 | 0, 2, 6, 13, 20, 26 | |
| 0.5% N | 1 | 3.7 | 0.5 | 0.05 | 0, 2, 6, 13, 20, 26 | |
| 0.75% Nd | 3 | 3.7 | 0.75 | 0.075 | 6, 20, 26 | |
| 1% N | 1 | 3.7 | 1 | 0.1 | 0, 2, 6, 13, 20, 26 | |
| 2% N | 1 | 3.7 | 2 | 0.2 | ||
| 4% N | 1 | 3.7 | 4 | 0.4 | 0, 2, 6, 13, 20, 26 | 0 (66), 6 (67), 26 (60) |
| 6% N | 1 | 3.7 | 6 | 0.6 | ||
| 10% N | 1 | 3.7 | 10 | 1.0 | 0, 2, 6, 13, 20, 26 | |
Number of independent microcosms prepared.
FO, fertilizer only.
Molecular analyses of one microcosm from the triplicate.
Molecular analyses of all three replicates.
Sampling time in days; the value in parentheses is the number of clones screened.
Sampling from microcosms.
For all oiled microcosms, it was noted that the simulated tidal cycles mixed the oil into the upper 3 to 4 cm of the sediment. Only the oiled portion of the sediment was sampled. Triplicate samples (ca. 10 g) were taken for molecular analysis on the day that the microcosms were set up (referred to as day 0) and 2, 6, 13, 20, and 26 days after initial nutrient addition. Samples were collected by scraping the upper 2 to 4 mm of the sediment with a flame-sterilized spatula. At the start and end of the experiment, triplicate samples of each microcosm were removed for oil chemistry analysis. Samples were stored at −20°C until analysis. Samples (30 ml) of the tidal seawater leaving the microcosms were taken from the acid-washed plastic bottles for nutrient analysis and stored at −20°C.
Carbon dioxide measurements.
The evolution rate of carbon dioxide from each microcosm was determined before oil was added and daily thereafter at the same point in the tidal cycle. The headspace of a sealed microcosm was circulated through the cell of an infrared gas analyzer (Servomex) by a method detailed by Swannell et al. (39). The device was calibrated by using a CO2-free-air standard and a compressed-air cylinder containing 335 ppm (by volume) CO2. The stability of the analyzer over the duration of the measurements (<5 min) was determined by circulating the headspace of an empty glass bottle through the sample cell. The drift was always less than 4 ppm.
Nutrient analysis.
As an indirect measure of nutrient concentrations in pore water in the microcosms, inorganic nitrogen concentrations in the seawater collected after the simulated tidal cycles were determined by using a Technicon autoanalyzer system (28).
Oil chemistry.
Hydrocarbons in oiled sediments (10 g) spiked with squalane and 1,1-binaphthyl standards to determine extraction efficiency were extracted, analyzed by gas chromatography with flame ionization detection and mass spectrometric detection, and quantified as described previously (39). The efficiency of recovery of the added standards was, on average, 83%. Replicate analyses showed that the variability of measured values was always less than 10%. To distinguish between physical removal and biodegradation, TPH, total GC resolvable hydrocarbons, nC11 to nC35 alkanes, and polycyclic aromatic hydrocarbons (PAH) were expressed relative to 17α(72),25β(72)-hopane, a degradation-resistant compound present in crude oil (4). The percentage biodegradation was calculated by dividing the concentration of individual compounds relative to that of 17α(72),25β(72)-hopane at the end of the experiment by the concentrations relative to that of 17α(72),25β(72)-hopane at the start of the experiment.
Statistical analysis of data on oil chemistry, inorganic nutrients, and carbon dioxide evolution.
Statistical analysis (parametric two-way analysis of variance, Pearson correlation) was performed by using Systat 7.0 (SPSS Inc.).
DNA extraction, PCR, and DGGE analysis.
DNA was extracted from 0.5-g samples (Table 1) by using the bead beating method described by Curtis and Craine with a Mikrodismembrator-U (B. Braun Biotech) (10). PCR was performed in a total volume of 50 μl containing 0.2 μM primer Vf-GC (corresponding to positions 341 to 358 of the Escherichia coli 16S rRNA [31]), 0.2 μM primer Vr (corresponding to positions 534 to 517 of the E. coli 16S rRNA [31]), 0.2 mM deoxynucleoside triphosphates, 1 U of BioTaq enzyme and the buffer supplied with the enzyme (Bioline, London, United Kingdom), and 1 μl of template DNA. Amplification was performed by using a Hybaid Omnigene Thermocycler as follows: 95°C for 3 min, followed by 30 cycles of 94°C for 0.5 min, 55°C for 1 min, and 72°C for 1 min, with a final elongation of 72°C for 10 min. DGGE was performed with the Bio-Rad DCode system. The PCR product was loaded onto 1-mm-thick 10% (wt/vol) polyacrylamide (37.5:1 acrylamide-bisacrylamide) gels containing a 30 to 60% linear denaturing gradient. One hundred percent denaturant is 7 M urea and 40% (vol/vol) deionized formamide. Gels were run in 1× TAE buffer (40 mM Tris-acetate, 1 mM Na-EDTA, pH 8.0) at 60°C and 200 V for 3.5 h. Gels were stained in 1× TAE buffer containing SYBR Green I (diluted 1:10,000; Sigma) and photographed under UV transillumination.
Statistical analysis of DGGE tracks.
Scanned negatives were analyzed by using Quantity One 4.1 software (Bio-Rad, Hercules, Calif.), and data were exported to Excel and analyzed in Systat 7.0 (SPSS Inc.). Similarities between tracks were calculated by using the Dice coefficient (band based) or the Pearson product-moment correlation coefficient (whole densitometric curve based) (34). Since cluster analysis of the resulting similarity matrices does not allow conclusions to be drawn regarding the statistical significance of differences between clusters or groups of samples (38), similarity coefficients from matrices were assigned to different groups and subsequently tested to determine whether their means were significantly different. Nonparametric analysis of variance (Mann-Whitney U test) was applied since similarity coefficients are not necessarily normally distributed (38).
Cloning, sequencing, and phylogenetic analysis of 16S rDNA.
Almost full-length 16S rRNA gene fragments were amplified by using primers pA and pH′ (12). Except for the primers, the PCR conditions were similar to those described above. PCR products were cloned by using the AdvanTAge kit (Clontech, Palo Alto, Calif.), and the 16S rRNA gene libraries were screened by amplified ribosomal DNA restriction analysis (ARDRA). The number of clones screened from each library is given in Table 1. E. coli clones were categorized into different ARDRA types based upon the pattern obtained on simultaneous digestion (3 h, 37°C) with the restriction enzymes RsaI and HaeIII (5 U of each). Sequencing of several clones corresponding to dominantly occurring ARDRA types revealed that these ARDRA groups were internally homogeneous (data not shown). ARDRA types were named X-Yd-Z, in which X refers to the type of microcosm (Table 1), Y is the day of sampling (0, 6, or 26), and Z is the number of a representative clone in the clone library of the particular microcosm and sampling date. The distribution of ARDRA types present in different clone libraries was determined and used to calculate the Shannon-Weaver index {H = −Σ[ni · log(ni)], where ni is the relative contribution of clone type i to the whole library}. ARDRA types occurring more than once in a library were selected for sequence analysis. Sequence data were obtained with a single primer (pD′, E. coli positions 534 to 518 [12]), generating 0.5 kb of sequence data. For a selection of clones, the nearly complete 16S rDNA was sequenced in both directions. Sequences were compared to sequences deposited in the GenBank DNA database by using the BLAST algorithm (1). Alignments were performed by using ClustalW and corrected manually. Distance analysis using the Jukes and Cantor correction (24) and bootstrap resampling (100 times) was done with the TREECON package (45), and the distance matrix was used to construct a tree via neighbor joining (35). Parsimony analysis was done by using DNAPARS from the PHYLIP package (14).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AF432269 to AF432344.
RESULTS
Nutrient levels and degradation of oil in nutrient-amended microcosms.
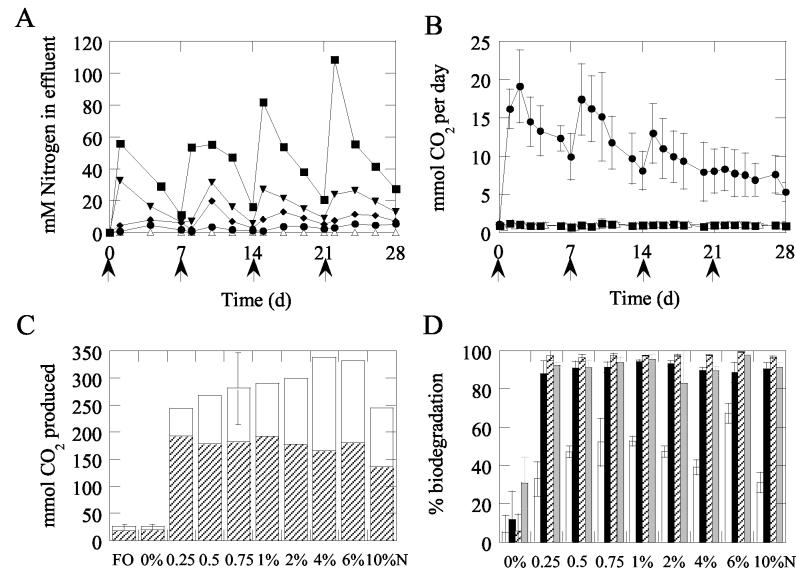
Throughout the incubation, nitrogen concentrations in the effluent (and hence the sediment pore water) were significantly related to the amount of nutrient added (Fig. 2A; average correlation of 0.90 [P < 0.05], with all correlations greater than 0.61), even though most of the nitrogen was removed by the tide directly after nutrient addition.
FIG. 2.
Chemical data from microcosms. (A) amounts of nitrogen in residing seawater of beach microcosms over time. Symbols: ▵, 0% N; •, 1% N; ♦, 2% N; ▾, 4% N; ▪, 10% N. Arrows indicate addition of nutrients. (B) Daily carbon dioxide production, averaged over three independent runs, with bars indicating standard deviation, for microcosms. Symbols: ▵, FO; ▪, 0% N; •, 0.75% N. For the FO and 0% N microcosms, standard deviation symbols were often smaller than the data symbols. Arrows indicate addition of nutrients. (C) Cumulative carbon dioxide production in microcosms. The open portion of the column indicates production during the first 7 days; carbon dioxide production during the last 21 days of the experiment is shown by the hatched portion of the column. Standard deviations are shown by error bars for the microcosms prepared in triplicate (FO, 0% N, and 0.75% N). (D) Percentage of biodegradation of TPH (open), TRH (black), n11-to-n33 alkanes (hatched), and PAH (grey) per treatment. Error bars indicate standard deviations.
The rate and extent of oil degradation were examined by measuring carbon dioxide production over time and the oil composition at the start and end of the experiment (28 days). In all oiled microcosms treated with inorganic nutrients, the rate of carbon dioxide production strongly increased relative to that of the control treated with oil only (0% N), directly after fertilization (Fig. 2B and C). After 28 days, the amount of carbon dioxide produced in the fertilized microcosms was significantly higher than that in the untreated 0% N microcosms (P < 0.001). Daily carbon dioxide production slowly declined over time, with nutrient amendments temporarily stimulating carbon dioxide production (Fig. 2B and data not shown). In general, the initial rate of carbon dioxide production appeared to increase with the amount of fertilizer added; only the 10% N treatment deviated strongly (Fig. 2C). A significant positive correlation (r = 0.74 to 0.96, P < 0.05) between the amount of nutrient added and daily carbon dioxide production, corrected for the background production in the 0% N microcosm, was observed only between days 2 and 7, when the 10% N microcosm was not considered. During the last 21 days, no correlation (r = −0.182, P > 0.05) was found between nutrient treatment and the total carbon dioxide produced in this period (Fig. 2C). For all other nutrient-amended microcosms, the amount of carbon dioxide produced after 28 days fell within the average ± the standard deviation of the 0.75% N microcosms (Fig. 2C).
Figure 2D shows the percentages of biodegradation of TPH, TRH, nC11-to-nC35 alkanes, and PAH. Significantly greater biodegradation of these compound classes was obtained with all of the bioremediation treatments than with the oil-only control (P < 0.001). TRH, nC11-to-nC35 alkanes, and aromatic hydrocarbons were almost completely removed (91% ±2%, 98% ± 1%, and 92% ± 4%, respectively). No significant differences in TRH, n-alkane, and aromatic hydrocarbon biodegradation were observed between the various nutrient amendments (P > 0.05). However, TPH degradation was significantly greater in the 6% N microcosm than in the 0.25% N, 4% N, and 10% N microcosms (P < 0.05).
Carbon dioxide production in the oiled, untreated controls (0% N) was similar (P > 0.05) to that in the unoiled microcosms treated with fertilizer (FO; Fig. 2B and C), in agreement with the absence of significant biodegradation of TPH, TRH, and alkanes (Fig. 2D).
Effect of nutrient amendment on bacterial community structure and dynamics.
Effect of bioremediation treatments on bacterial community structure was determined by using 16S rDNA-based PCR-DGGE for a number of treatments and sampling times (Table 1). In order to determine relationships between the community fingerprints, similarities were calculated on the basis of the absence or presence of bands (Dice coefficient) and on the basis of whole-track curve densitometric information (Pearson product-moment coefficient). Results based on the two coefficients were comparable; therefore, only the band-based similarities will be described. Analysis of triplicate samples taken from a single microcosm at a single sampling time showed that microbial communities were highly similar (95% ± 5%), indicating low spatial heterogeneity within the microcosms. However, obvious differences in community structure were evident both in time (Fig. 3 and 4) and between different treatments (Fig. 5 and 6). No obvious changes in the composition of the predominant bacterial communities occurred over time in the microcosm treated with fertilizer but no oil (FO). Although DGGE analysis of replicate microcosms for the FO treatment was not conducted in this study, data from a related field experiment that incorporates a fully replicated randomized block design indicate that DGGE profiles from the same beach sediments used in this study and treated with fertilizer only are highly similar over time and between replicate blocks (data not shown). The largest changes occurred in the microcosms remediated with 1 and 4% N (Fig. 4); the average similarity relative to day 0 was significantly lower for these two microcosms than in the other three oiled, nutrient-amended microcosms (P < 0.05). The most dramatic changes in community structure occurred during the first 6 days following oil addition, and profiles between days 6 and 26 were significantly more similar to each other than to profiles obtained from day 0 samples (Fig. 4; P < 0.05), suggesting that, following an initial significant change in community composition, the bacterial community structure remained relatively stable. Nevertheless, for all oiled microcosms, community changes after 6 days were significantly larger than in the microcosm that was not treated with oil (FO) over the same time period (P < 0.05).
FIG. 3.
Bacterial community dynamics during oil spill bioremediation in beach microcosms. Examples of changes in 16S rDNA PCR-DGGE profiles over time in microcosms; unoiled microcosms (FO), oiled, untreated microcosms (0% N); and oiled microcosms amended with 0.25 and 10% nitrogen. The values above the lanes indicate the numbers of days elapsed since nutrient addition.
FIG. 4.
Similarities (Dice coefficient) in DGGE profiles from microcosms receiving different treatments. Open columns show average similarities of profiles observed at days 6, 13, 20, and 26, relative to day 0, for each treatment. The average values were obtained from pairwise comparisons of DGGE profiles obtained at each time point relative to the time zero sample for each individual treatment. Thus, the data for the FO treatment, for example, represent the mean similarity obtained from four pairwise comparisons (the mean of the day 0 profile compared to the day 6 profile, the day 0 profile compared to the day 13 profile, the day 0 profile compared to the day 20 profile, and the day 0 profile compared to the day 26 profile). Error bars (n = 4) indicate standard deviations. Hatched columns show average similarity between profiles observed at days 6, 13, 20, and 26 for each treatment. In this case, the mean values were obtained by averaging the pairwise similarities of the day 6 profile compared to the day 13 profile, the day 6 profile compared to the day 20 profile, the day 6 profile compared to the day 26 profile, the day 13 profile compared to the day 20 profile, the day 13 profile compared to the day 26 profile, and the day 20 profile compared to the day 26 profile. Error bars (n = 6) indicate standard deviations. These data indicate that most of the change in the DGGE profiles occurred between days 0 and 6, since the mean similarities for all pairwise comparisons from day 6 onward show little variation.
FIG. 5.
Comparison of the effects of nutrient amendment on bacterial community structure at particular sampling times. Examples of 16S rDNA PCR-DGGE profiles from microcosms receiving different nutrient amendments, showing profiles at days 13 and 26. The values above the lanes indicate treatments (described in Table 1).
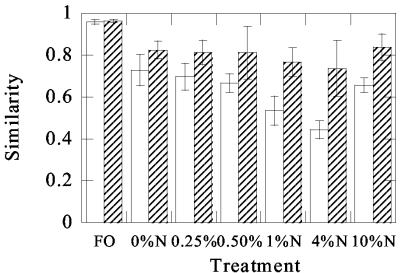
FIG. 6.
Similarities (Dice coefficient) in DGGE profiles between different treatments at each time point. Shown are average similarities of profiles observed for oiled, nutrient-amended microcosms (0.25, 0.5, 1, 4, and 10% N) relative to the oiled, unamended control (○) and unoiled, nutrient-amended control (♦), respectively; average similarity between the different oiled, nutrient-amended microcosms (▪); and average similarity between DGGE profiles from replicate microcosms receiving the same nutrient amendment (0.75% N) (□). Error bars indicate standard deviations. To aid in visualization, symbols representing different comparisons have been slightly offset. For each time point, the average values were obtained from pairwise comparisons of DGGE profiles obtained for each treatment relative to the oiled, unamended control. Thus, the data for the day 6 time point, for example, represent the mean similarity obtained from five pairwise comparisons (the mean of the 0.25% N profile compared to the 0% N profile, the 0.5% N profile compared to the 0% N profile, the 1.0% N profile compared to the 0% N profile, the 4.0% N profile compared to the 0% N profile, and the 10% N profile compared to the 0% N profile). Similar comparisons were made between the fertilizer-only control and the other treatments. For microcosms that received oil and different nutrient amendments, all possible pairwise similarities were averaged to obtain the mean value. These data indicate that, at each time point other than day 0, the variation in DGGE profiles between replicate microcosms (□) is as great as the variation between different treatments (▪).
The different nutrient treatments led to the development of clearly different microbial communities (Fig. 5). As expected, communities were highly similar at the start of the experiment (Fig. 6). The largest differences between treatments, as indicated by the lowest average similarities, were observed after 6 days (Fig. 6). The average similarity between the different treatments was low (0.58 to 0.71) and not significantly different from the similarity of each individual treatment relative to either the unfertilized, oiled control (0% N) or the unpolluted control (FO) (P > 0.05). This indicated that a unique microbial community was selected in each microcosm. However, three independent microcosms were prepared for one of the bioremediation treatments (0.75% N). The average similarity between the DGGE profiles of these three independently operated microcosms, which were subjected to the same nutrient treatment, was slightly higher than the average similarity between those of microcosms treated with different levels of inorganic nutrients (Fig. 6). However, statistical analysis showed that differences in community structure between the true replicate microcosms receiving the same nutrient treatment were not significantly different from the differences measured between the communities in microcosms receiving different nutrient treatments (P > 0.05).
Phylogenetic analysis and diversity of bacterial communities.
To obtain a more specific picture of which bacterial taxa were stimulated by the oil and bioremediation treatments, phylogenetic analysis of cloned 16S rRNA genes was performed. The microcosms and sampling times indicated in Table 1 were selected because DGGE analysis revealed the greatest differences in the predominant populations. Clone libraries were screened by ARDRA, and representatives of ARDRA profiles occurring more than once in a library were partially sequenced (Table 2). It should be noted that the samples from the 0 and 4% N microcosms taken at day 0 were frozen immediately after oil addition and, although treated with oil, the bacterial populations would not have had time to respond to the oil addition and, in effect, represent bacterial populations not yet affected by oiling. For the five libraries from microcosms not affected by oiling (day 0 libraries of the FO, 0% N, and 4% N microcosms and day 6 and day 26 libraries of the FO microcosm), phylogenetic analysis of the clones occurring more than once in the libraries indicated that they belonged to a wide range of phylogenetic groups, with none dominating the clone libraries (Table 2 and Fig. 7). Almost half of the clones were unique, meaning that their ARDRA type was only encountered once in a clone library. The purpose of this study was not to catalogue the wider diversity of the bacterial populations present in the beach sediments, and these were not analyzed further. The high diversity was also indicated by the fact that, of the 49 clones sequenced, clones with similar ARDRA profiles and >99% sequence identity were encountered only twice in different clone libraries (indicated by the letters A and B in Table 2). In contrast, oiling and bioremediation resulted in strong dominance by a few ARDRA/sequence types (Table 2 and Fig. 7). This decrease in diversity was also illustrated by the calculation of the Shannon-Weaver index for the clone libraries (Fig. 8). Diversity remained high in the microcosm treated with fertilizer only. However, oiling strongly decreased the bacterial community diversity during the first 6 days of the experiment. The strongest effect was observed for the bioremediation treatment. Nevertheless, between days 6 and 26, diversity began to recover to preoiling levels (Fig. 8).
TABLE 2.
Overview of ARDRA types occurring more than once in a clone library, their relative contributions to the clone library, and the closest relative in the GenBank database with similarity to the partially sequenced clone (0.4 kb)a
| ARDRA type | % Clones | Closest relative in GenBank database (accession no.) | % Similarity | Phylogenetic division | Degrader |
|---|---|---|---|---|---|
| FO-0d-15 | 9.8 | Unidentified proteobacterium strain OM27 (U70713) | 90.5 | α-Proteobacteria | |
| FO-0d-8 | 7.8 | Cytophaga sp. strain JTB244 (AB015262) | 92.4 | CFBd | |
| FO-0d-32 (A) | 5.9 | Sulfur-oxidizing bacterium ODIII6 (AF170422) | 93.4 | γ-Proteobacteria | |
| FO-0d-41 | 5.9 | Desulfotalea psychrophila LSv54 (AF099062) | 94.1 | δ-Proteobacteria | |
| FO-0d-31 | 5.9 | Planococcus alkanoclasticus (AF029364) | 97.7 | Gram positive | Ref. 13 |
| FO-0d-43 (B) | 3.9 | β-Proteobacterium A0640 (AF236010) | 97.4 | β-Proteobacteria | |
| FO-0d-1 | 3.9 | Cytophaga sp. strain KT02ds22 (AF235114) | 97.5 | CFB | |
| FO-0d-57 | 3.9 | Cytophaga sp. strain KT0803 (AF235117) | 95.3 | CFB | |
| FO-0d-9 | 3.9 | Unidentified proteobacterium BD2-4 (AB015534) | 93.2 | Gram positive | |
| FO-6d-109 | 7.5 | Uncultured Cytophagales CE55 (AF211292) | 96.4 | CFB | |
| FO-6d-82 | 7.5 | Flavobacterium sp. strain 5N-3 (AB017597) | 96.6 | CFB | |
| FO-6d-101 | 5.0 | Thioalcalovibrio denitrificans (AF126545) | 91.8 | γ-Proteobacteria | |
| FO-6d-134 | 5.0 | Lucina floridana gill symbiont (L25707) | 94.1 | γ-Proteobacteria | |
| FO-6d-51 | 5.0 | Oleiphilus messinensis ME102 (AJ295154) | 94.9 | γ-Proteobacteria | |
| FO-6d-103 | 5.0 | Amphora delicatissima chloroplast (U96445) | 99.4 | Chloroplast | |
| FO-6d-98 | 5.0 | Uncultured marine eubacterium HstpL32 (AF159635) | 98.4 | Chloroplast | |
| FO-26d-18 | 7.3 | Uncultured bacterium GR-Sh1-209 (AJ296579) | 89.9 | CFB | |
| FO-26d-1 | 4.9 | Sulfitobacter brevis (Y16425) | 97.6 | α-Proteobacteria | |
| FO-26d-41 | 4.9 | α-Proteobacterium egg clone D37 (AF034931) | 89.9 | α-Proteobacteria | |
| FO-26d-70 (B) | 4.9 | β-Proteobacterium A0640 (AF236010) | 95.1 | β-Proteobacteria | |
| FO-26d-66 (A) | 4.9 | Sulfur-oxidizing bacterium ODIII6 (AF170422) | 94.3 | γ-Proteobacteria | |
| FO-26d-59 | 4.9 | Uncultured δ-proteobacterium Sva0485 (AJ241001) | 94.4 | δ-Proteobacteria | |
| FO-26d-5 | 4.9 | Uncultured Cytophagales (AF141559) | 93.6 | CFB | |
| FO-26d-6 | 4.9 | Uncultured bacterium No. 0319-6J10 (AF234076) | 95.2 | CFB | |
| FO-26d-53 | 4.9 | Uncultured bacterium clone LAH9 (AF392755) | 93.8 | Green nonsulfur | |
| FO-26d-31 | 4.9 | Marine eubacterial sp. (aggregate agg8) (L10942) | 95.1 | Planctomycetes | |
| FO-26d-21 | 4.9 | Unidentified bacterium wb1_A23 (AF317748) | 88.2 | Unidentified | |
| 0%N-0d-26 | 8.0 | Flexibacter sp. strain D8 (AF125323) | 98.5 | CFB | |
| 0%N-0d-3 | 6.0 | Uncultured marine eubacterium HstpL2 (AF159654) | 96.0 | α-Proteobacteria | |
| 0%N-0d-49 | 6.0 | Aquaspirillum delicatum (AF078756) | 96.7 | β-Proteobacteria | |
| 0%N-0d-7 | 6.0 | Uncultured clone 97 (AF369719) | 95.5 | γ-Proteobacteria | |
| 0%N-0d-23 | 6.0 | Flexibacter sp. strain D8 (AF125323) | 98.6 | CFB | |
| 0%N-0d-48 | 4.0 | Uncultured δ proteobacterium Sva0485 (AJ241001) | 95.2 | δ-Proteobacteria | |
| 0%N-0d-44 | 4.0 | Flexibacter sp. strain (U64013) | 88.1 | CFB | |
| 0%N-0d-5 | 4.0 | Unknown organism (X84633) | 93.5 | Gram positive | |
| 0%N-0d-51 | 4.0 | Uncultured marine eubacterium HstpL32 (AF159635) | 98.4 | Chloroplast | |
| 0%N-0d-45 | 4.0 | Benzene-mineralizing consortium clone SB-34 (AF029049) | 91.3 | Green nonsulfur | |
| 0%N-6d-28, 44b (C) | 35.8 | Alcanivorax sp. strain ME104 (AJ302704) | 99.0-99.3 | γ-Proteobacteria | Ref. 48 |
| 0%N-6d-43 | 8.9 | γ-Proteobacterium HTB021 (AB010859) | 97.7 | γ-Proteobacteria | |
| 0%N-6d-7 | 4.4 | Marinobacter hydrocarbonoclasticus (AB021372) | 93.8 | γ-Proteobacteria | Ref. 17 |
| 0%N-6d-94 | 4.4 | Uncultured γ-proteobacterium B2M60 (AF223299) | 95.0 | γ-Proteobacteria | |
| 0%N-6d-3 (D) | 4.4 | Alcanivorax borkumensis (Y12579) | 99.8 | γ-Proteobacteria | Ref. 48 |
| 0%N-6d-20 | 4.4 | Unidentified bacterium BD2-6 (AB015536) | 93.5 | Gram positive | |
| 0%N-6d-1 | 4.4 | Uncultured bacterium Sva0725 (AJ241003) | 91.8 | Holophaga | |
| 0%N-26d-6, 48b,c (D) | 32.0 | Alcanivorax borkumensis (Y12579) | 99.8-99.9 | γ-Proteobacteria | Ref. 48 |
| 0%N-26d-20 | 6.9 | α-Proteobacterium ISHR1 (AB013442) | 98.6 | α-Proteobacteria | |
| 0%N-26d-47 | 5.6 | Halomonas sp. strain Eplumel.A1 (AF212202) | 97.9 | γ-Proteobacteria | /PICK> |
| 0%N-26d-1 | 4.2 | Unidentified bacterium BD4-11 (AB015561) | 98.9 | γ-Proteobacteria | |
| 0%N-26d-88 | 4.2 | Erythrobacter sp. strain AS-45 (AJ391206) | 95.9 | α-Proteobacteria | |
| 0%N-26d-9 | 2.8 | Marine γ-proteobacterium AS-16 (AJ391179) | 96.7 | γ-Proteobacteria | |
| 0%N-26d-54 | 2.8 | Marine γ-proteobacterium AS-16 (AJ391179) | 98.4 | γ-Proteobacteria | |
| 0%N-26d-61 | 2.8 | Cycloclasticus oligotrophus (AF148215) | 98.9 | γ-Proteobacteria | Ref. 6 |
| 0%N-26d-67 | 2.8 | Planococcus sp. strain SOS Orange (AF242541) | 95.3 | Gram positive | |
| 4%N-0d-74 | 7.6 | Unidentified bacterium ANG.clone 1a (AF022393) | 97.3 | α-Proteobacteria | |
| 4%N-0d-39 | 6.1 | Uncultured marine eubacterium HstpL35 (AF159636) | 97.7 | Chloroplast | |
| 4%N-0d-90 | 6.1 | Uncultured low-G+C, gram-positive strain MT35 (AF211297) | 93.6 | Gram positive | |
| 4%N-0d-18 | 6.1 | Microscilla sp. strain Nano 1 (AB015937) | 94.7 | CFB | |
| 4%N-0d-29 | 4.5 | Uncultured marine bacterium (AF159684) | 88.6 | γ-Proteobacteria | |
| 4%N-0d-46 | 3.0 | Unidentified α-proteobacterium (AB015247) | 95.1 | α-Proteobacteria | |
| 4%N-0d-73 | 3.0 | Methylophaga sulfidovorans (X95461) | 94.4 | γ-Proteobacteria | |
| 4%N-0d-8 | 3.0 | Colwellia sp. strain KAT2 (AF025315) | 90.1 | γ-Proteobacteria | |
| 4%N-0d-65 | 3.0 | Uncultured Cytophagales (AF141559) | 91.5 | CFB | |
| 4%N-0d-10 | 3.0 | Microscilla sp. strain Nano 1 (AB015937) | 95.1 | CFB | |
| 4%N-0d-7 | 3.0 | Cytophaga sp. strain KT02ds22 (AF235114) | 97.6 | CFB | |
| 4%N-0d-11 | 3.0 | Odontella sinensis chloroplast (Z67753) | 94.4 | Chloroplast | |
| 4%N-6d-95−109b,c (C) | 73.1 | Alcanivorax borkumensis (Y12579) | 96.9-97.0 | γ-Proteobacteria | Ref. 48 |
| 4%N-6d-94 | 3.0 | Uncultured marine eubacterium HstpL35 (AF159636) | 98.2 | Chloroplast | |
| 4%N-26d-7, −27b,c | 30.0 | Erythrobacter citreus (AF118020) | 98.1-98.2 | α-Proteobacteria | |
| 4%N-26d-5 | 20.0 | Erythrobacter sp. strain MBIC3019 (AB012062) | 97.3 | α-Proteobacteria | |
| 4%N-26d-12 | 3.3 | Sphingomonas sp. (U63956) | 100.0 | α-Proteobacteria | |
| 4%N-26d-32 | 3.3 | Roseobacter sp. strain PRLIST02 (Y15339) | 97.2 | α-Proteobacteria | |
| 4%N-26d-25 | 3.3 | DMSPe-degrading bacterium LFR (L15345) | 95.5 | α-Proteobacteria | |
| 4%N-26d-20 | 3.3 | Uncultured marine eubacterium HstpL24 (AF159662) | 92.2 | α-Proteobacteria | |
| 4%N-26d-91 | 3.3 | Nocardioides plantarum (AF005008) | 93.7 | Gram positive |
ARDRA types are named after the sample from which the library was constructed (indicated by the treatment code (Table 1) and the day (d) on which the sample was taken) and the representative clone sequenced. A letter in parentheses indicates ARDRA types encountered in more than one clone library. When the closest relative to a sequence is known to degrade oil, this is indicated by a reference (Ref.).
Two clones with similar ARDRA types sequenced.
Nearly completely sequenced (>1.4 kb).
CFB, Cytophaga-Flexibacter-Bacteroides (Phylum Bacteroidetes).
DSMP, dimethylsulfoniopropionate.
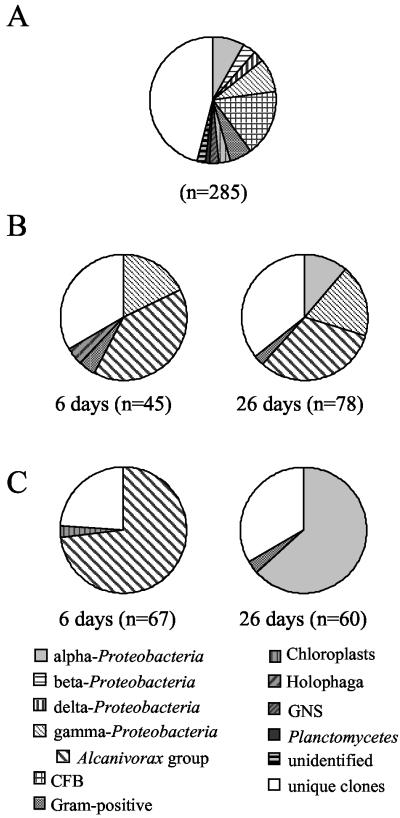
FIG. 7.
Relative contributions of different phylogenetic groups to microbial communities. (A) Microcosms not affected by oiling and remediation treatment (composed of five clone libraries [FO days 0, 6, and 26 and day 0 of the 0% N and 4.0% N microcosms]). (B) Oiled, untreated control (0% N) at days 6 and 26. (C) Oiled, 4% N-treated microcosm at days 6 and 26. CFB, Cytophaga-Flexibacter-Bacteroides; GNS, green non-sulfur bacteria.
FIG. 8.
Effect of oiling and bioremediation on changes in microbial diversity over time. The Shannon-Weaver index was calculated from the distribution of clones categorized on the basis of different ARDRA profiles. Symbols: ▵, FO; ▪, 0% N; •, 4% N.
The day 6 libraries from microcosms treated with oil only (0% N) and the bioremediated microcosm (4% N) were dominated by γ-proteobacteria (62.2% of the clones in the 0% N microcosm and 73.1% of the clones in the 4% N microcosm; Fig. 7). γ-Proteobacteria (50.6% of the clones) continued to dominate the library from the microcosm treated with oil alone (0% N) after 26 days, but in the bioremediated microcosm (4% N), α-proteobacteria became dominant (63.3%). The nearly complete cloned 16S rDNA of representatives of the dominantly occurring ARDRA types was sequenced. The dominant α-proteobacterial clones (4%N-26d-5, -7, and -27) belonged to the aerobic anoxygenic phototrophic bacteria and were phylogenetically most closely related to Erythrobacter longus and E. citreus (Table 2 and Fig. 9A). ARDRA profiles indicative of these sequences were only found in the sample taken 26 days after 4% N fertilizer treatment and represented 50.0% of the clones. A different ARDRA type (0% N-26d-88) most closely related to Erythrobacter spp. was found in the day 26 library of the 0% N library, constituting 4.2% of the library (Table 2). Two dominant ARDRA types, found in several clone libraries (indicated by C and D in Table 2 and Fig. 9B) represented the alkane-degrading Alcanivorax/Fundibacter group (5, 48) of the γ-proteobacteria. Sequences of ARDRA type C were distinct from Alcanivorax borkumensis and Fundibacter jadensis but formed a distinct group with these sequences (Fig. 9B). Type D sequences were almost identical to A. borkumensis (>99.8%). ARDRA profiles and sequences indicative of type C comprised 73 and 36% of the libraries constructed from the 4% N-amended microcosm at day 6 and the oil-only control 0% N microcosm at day 6, respectively. However, they were not detected in any other library. ARDRA profiles and sequences indicative of type D were only found in the libraries from microcosms treated with oil only (0% N) and comprised 4 and 32% of clones analyzed after 6 and 26 days, respectively. A small number of other clones most closely related to microorganisms known to degrade components of crude oil were also detected and are indicated in Table 2. All of these sequences constituted less than 6% of the clones in a library.
FIG. 9.
Phylogenetic tree based on almost complete 16S rDNA gene sequences of α-proteobacterial (A) and γ-proteobacterial (B) clones occurring as dominant sequences in clone libraries of oiled microcosms. A neighbor-joining analysis with Jukes-and-Cantor correction was performed. Only bootstrap values of greater than 50% are shown.
DISCUSSION
Trends in carbon dioxide evolution and oil chemistry results clearly indicated that the addition of N and P significantly improved oil degradation, thus showing, as expected (21), that oil degradation was limited by the supply of N and P. Upon repeated fertilization, a temporary increase in carbon dioxide evolution was observed, indicating that, at these time points, N and P were still limiting and other factors did not become limiting during the experiments. The absence of changes in carbon dioxide production in the microcosm that received inorganic nutrients but not oil argues against significant metabolism of C sources other than oil components in the oil-contaminated, nutrient-amended microcosms. Bragg et al. (4) noted a positive relationship between the rate of oil biodegradation and nitrogen concentration in beach sediment pore water, demonstrating that the pore water nutrient content was the most significant factor controlling the rate of oil degradation. Our results concur with this only partly. Nutrient addition did significantly increase the oil biodegradation rate. However, even though the various nutrient amendments resulted in different nutrient levels in the microcosms and initially carbon dioxide production rates correlated with the amount of added inorganic nutrients, over the full period of the experiment, the rates and extent of oil degradation were comparable. Experiments conducted with lower oil concentrations also demonstrated that, above a particular level of nutrient addition, nutrient levels did not further enhance oil biodegradation (3). Furthermore, Lee et al. (29) obtained similar results and showed no significant difference in oil degradation in a field experiment using low and high concentrations of fertilizers, although degradation was higher than in oiled, unamended plots. Reasons for these differences are unknown but may relate to differences in the geology of the beach and therefore different retention characteristics of the inorganic nutrients within the sediment; Bragg et al. (4) studied oil degradation on a cobble beach, while the study by Lee et al. (28) and our study were performed on sandy beach material. Nevertheless, the results indicate that bioremediation should be effective despite the usual patchy oil deposition on a heterogeneous beach after a spill and similarly uneven distribution of inorganic nutrients supplied during bioremediation.
N and P were supplied at different concentrations, which, according to resource ratio theory (44), should select different microbial communities dominated by the organisms most capable of utilizing the inorganic nutrients at the levels added to the polluted habitat. In principle, this could offer opportunities to direct oil degradation (21, 37). Clear-cut differences in bacterial community structure were found in microcosms treated with different levels of inorganic nutrients, which appears consistent with the predictions of resource ratio theory. However, statistical analysis of DGGE profiles revealed that the average similarities between DGGE profiles from three independently prepared microcosms receiving the same amount of inorganic nutrients (0.75% N) were not significantly different from the similarities between microcosms subjected to different nutrient amendments. Therefore, it cannot be concluded that the different communities selected resulted from differences in nutrient amendment alone. It is unlikely that heterogeneity within the microcosms, as the result of stratification or poor mixing of oil and sediment/water, contributed significantly to the observed highly variable composition in bacterial community structure between microcosms (only 65% ± 6% similarity). Replicate samples taken at the same time from a single microcosm, which would be subject to the same constraints on mixing and stratification as separate microcosms, were highly reproducible (95% ± 5%). Also, the community profiles between days 6 and 26, after major changes in bacterial communities had occurred, were similar for single microcosms (80% ± 4%).
Nevertheless, the results revealed that communities with highly different compositions (similarities of only 58 to 71%) showed similar rates and extents of oil degradation at different nutrient concentrations. This observation may be due largely to the strong selection for a few members of the Alcanivorax/Fundibacter group, which are capable of degrading alkanes (5, 48), a major oil component. Comparable observations have been made for a functionally stable methanogenic reactor fed with glucose (15). This reactor revealed considerable dynamics in microbial communities over a 605-day period, despite minimal differences in performance over time.
Bioremediation treatments are aimed at stimulating pollutant-degrading microorganisms to speed the recovery of contaminated ecosystem to a prepollution state in terms of biodiversity and ecosystem function. In this study, changes in the predominant bacterial populations occurred in all of the microcosms, except that which received inorganic nutrients but not oil. Oiling and especially bioremediation led to a strong decrease in bacterial community diversity at day 6, but a rapid recovery to near preoiling levels of diversity occurred subsequently. Still, despite having a similar level of biodiversity, the component organisms contributing to that diversity were somewhat different from the original community, as revealed by DGGE analysis and clone libraries. Following bioremediation, α-proteobacteria were dominant by day 26 of the experiment, whereas prior to oil contamination, they had only represented 8% of the clones in the libraries. Sequences from α-proteobacteria most closely related to Erythrobacter spp. were most commonly encountered. Erythrobacter spp. belong to the aerobic anoxygenic phototrophic bacteria (49). They metabolize organic substrates, with light enhancing their growth. Recently, it was revealed that aerobic anoxygenic phototrophic bacteria play a critical role in the carbon cycle in the ocean (26). Aerobic anoxygenic phototrophic bacteria grow well in high-nutrient media and are often isolated from environments with a high level of organic matter content (49), as well as from intertidal beach sediments (36). Recently, a PAH-degrading bacterium most closely related to Erythrobacter spp., Lutibacterium anuloederans LC8, was isolated from intertidal beach sediments (8). The rRNA gene of L. anuloederans shows 95.3 to 96.1% similarity to our Erythrobacter-like sequences. An α-proteobacterial sequence related to Erythrobacter spp. was also detected in beach sediments from an oil spill bioremediation field experiment conducted in Delaware (30).
Perhaps the most dramatic observation made in the present study was the rapid and strong selection for γ-proteobacteria in oil-treated microcosms. The γ-proteobacteria persisted in clone libraries from sediment samples treated with oil alone taken on day 26, while they were replaced by α-proteobacteria in microcosms treated with oil and inorganic nutrients. This may reflect slow ongoing alkane degradation in the microcosms not treated with inorganic nutrients and selection of organisms growing on residual PAH and secondary products of hydrocarbon degradation in the bioremediated microcosms. A field experiment on shoreline sediments in the Norwegian Arctic (19) revealed an increase in γ-proteobacteria in oil-contaminated beach sediments, while the microbial community of beached oil paste after the Nakhodka oil spill accident in the Japan Sea was also dominated by γ-proteobacteria (25). These results are consistent with our observation of a strong dominance by γ-proteobacteria, and this may be a characteristic of bacterial communities associated with recently spilled oil. In our case, it was mainly members of the Alcanivorax/Fundibacter group that were selected. Alcanivorax borkumensis (48) and Fundibacter jadensis (5) were described recently. Both are capable of using only a few organic substrates, especially alkanes (5, 48) and the alkyl groups of n-alkylbenzenes and n-alkylcycloalkanes (11). Alkanes are major oil components, explaining why Alcanivorax/Fundibacter-like sequences made a significant contribution (up to 73%) to clone libraries early in the bioremediation process and for a longer period when the hydrocarbon degradation rate was lower. Because of biases associated with PCR and cloning (47), these clone percentages probably do not directly represent Alcanivorax cell numbers, but since all samples were analyzed in the same fashion and the predominance of Alcanivorax-like sequences in the clone libraries was so dramatic, it is likely that our results do reflect a genuine increase in the relative abundance of this group of bacteria. Furthermore, fluorescence in situ hybridization analyses have shown that Alcanivorax constituted more than 90% of the entire microbial community in laboratory incubations of seawater supplemented with oil (43).
Although in the contaminated microcosms without nutrient treatment (0% N), no significant oil degradation occurred over the 28-day incubation period, remarkably, the bacterial communities changed considerably and the predominance of Alcanivorax/Fundibacter-like sequences in the corresponding clone library was apparent. This may be explained by the release of nitrogen and phosphorus from biomass killed by the oil, supporting limited hydrocarbon degradation. Interestingly, hydrocarbon pollutants can induce prophages, resulting in lysis of a large proportion of the bacterial community (9, 23), and this may explain changes in community composition independent of extensive oil degradation. Even in the absence of significant hydrocarbon degradation, Alcanivorax/Fundibacter-like sequences constituted about 40% of the clone libraries constructed from samples taken at days 6 and 26 from the unamended, oil-contaminated control microcosm. To assess if this is a consequence of relatively very low rates of hydrocarbon degradation by Alcanivorax/Fundibacter-like bacteria under these conditions, information on their activity would be required, for example, measurement of mRNA that encodes enzymes involved in biodegradation.
The members of the Alcanivorax/Fundibacter group appear to have a cosmopolitan distribution, as their presence has been noted in coastal waters and on beaches of the United Kingdom (this study), Germany (5, 48), Italy (GenBank accession numbers AB0302701 to -4), Singapore (GenBank accession number AF062642), Japan (20, 25), and the United States (7, 32). They have been detected mainly in laboratory enrichments with oil components (5, 7, 20, 48), but more importantly, their occurrence in oil paste and seawater has been described following an oil spill in the Japan Sea (25). The worldwide distribution of Alcanivorax spp. indicates that they may be of considerable global significance in marine hydrocarbon degradation, and a project to sequence the genome of this important organism has recently been instigated (http://www.uni-bielefeld.de/presse/pm/engpm31.htm).
Acknowledgments
We gratefully acknowledge the funding of this work by the U.K. Maritime Coastguard Agency, the U.K. Department of Environment, Food and Rural Affairs, the Environment Agency, the BBSRC, and the Canadian Department of Fisheries and Oceans.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M., and R. Bartha. 1972. Degradation and mineralization of petroleum in seawater: limitation by nitrogen and phosphorus. Biotech. Bioeng. 14:309-318. [DOI] [PubMed] [Google Scholar]
- 3.Boufadel, M. C., P. Reeser, M. T. Suidan, B. A. Wrenn, J. Cheng, X. Du, T. H. L. Huang, and A. D. Venosa. 1999. Optimal nitrate concentration for the biodegradation of n-heptadecane in a variably-saturated sand column. Environ. Technol. 20:191-199. [Google Scholar]
- 4.Bragg, J. R., R. C. Prince, E. J. Harner, and R. M. Atlas. 1994. Effectiveness of bioremediation for the Exxon-Valdez oil-spill. Nature 368:413-418. [Google Scholar]
- 5.Bruns, A., and L. Berthe-Corti. 1999. Fundibacter jadensis gen. nov., sp. nov., a new slightly halophilic bacterium, isolated from intertidal sediment. Int. J. Syst. Bacteriol. 49:441-448. [DOI] [PubMed] [Google Scholar]
- 6.Button, D. K., B. R. Robertson, P. W. Lepp, and T. M. Schmidt. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64:4467-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. J., J. R. Stephen, A. P. Richter, A. D. Venosa, J. Bruggemann, S. J. Macnaughton, G. A. Kowalchuk, J. R. Haines, E. Kline, and D. C. White. 2000. Phylogenetic analysis of aerobic freshwater and marine enrichment cultures efficient in hydrocarbon degradation: effect of profiling method. J. Microbiol. Methods 40:19-31. [DOI] [PubMed] [Google Scholar]
- 8.Chung, W. K., and G. M. King. 2001. Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl. Environ. Microbiol. 67:5585-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochran, P. K., C. A. Kellogg, and J. H. Paul. 1998. Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants. Mar. Ecol. Prog. Ser. 164:125-133. [Google Scholar]
- 10.Curtis, T. P., and N. G. Craine. 1998. The comparison of the diversity of activated sludge plants. Water Sci. Technol. 37:71-78. [Google Scholar]
- 11.Dutta, T. K., and S. Harayama. 2001. Biodegradation of n-alkylcycloalkanes and n-alkylbenzenes via new pathways in Alcanivorax sp. strain MBIC 4326. Appl. Environ. Microbiol. 67:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1988. Isolation and complete nucleotide determination of entire genes. Characterisation of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelhardt, M. A., K. Daly, R. P. J. Swannell, and I. M. Head. 2001. Isolation and characterization of a novel hydrocarbon-degrading, Gram-positive bacterium, isolated from intertidal beach sediment, and description of Planococcus alkanoclasticus sp. nov. J. Appl. Microbiol. 90:237-247. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1989. PHYLIP—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 15.Fernandez, A., S. Y. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floodgate, G. D. 1995. Some environmental aspects of marine hydrocarbon bacteriology. Aquat. Microb. Ecol. 9:3-11. [Google Scholar]
- 17.Gauthier, M. J., B. Lafay, R. Christen, L. Fernandez, M. Acquaviva, P. Bonin, and J.-C. Bertrand. 1992. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42:568-576. [DOI] [PubMed] [Google Scholar]
- 18.Geiselbrecht, A. D., R. P. Herwig, J. W. Deming, and J. T. Staley. 1996. Enumeration and phylogenetic analysis of polycyclic aromatic hydrocarbon-degrading marine bacteria from Puget Sound sediments. Appl. Environ. Microbiol. 62:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman, M. J., R. C. Prince, R. M. Garrett, K. K. Garrett, R. E. Bare, K. R. O'Neil, M. R. Sowlay, S. M. Hinton, K. Lee, G. A. Sergy, E. H. Owens, and C. C. Guenette. 2000. Microbial diversity in oiled and un-oiled shoreline sediments in the Norwegian Arctic, p. 775-787. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Microbial biosystems: new frontiers. Proceedings of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbiology, Kentville, Nova Scotia, Canada.
- 20.Harayama, S., H. Kishira, Y. Kasai, and K. Shutsubo. 1999. Petroleum biodegradation in marine environments. J. Mol. Microbiol. Biotechnol. 1:63-70. [PubMed] [Google Scholar]
- 21.Head, I. M., and R. P. J. Swannell. 1999. Bioremediation of petroleum hydrocarbon contaminants in marine habitats. Curr. Opin. Biotechnol. 10:234-239. [DOI] [PubMed] [Google Scholar]
- 22.Hedlund, B. P., A. D. Geiselbrecht, T. J. Bair, and J. T. Staley. 1999. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium. Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 65:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, S. C., and J. H. Paul. 1996. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142:27-38. [Google Scholar]
- 24.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y.
- 25.Kasai, Y., H. Kishira, K. Syutsubo, and S. Harayama. 2001. Molecular detection of marine bacterial populations on beaches contaminated by the Nakhodka tanker oil-spill accident. Environ. Microbiol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 26.Kolber, Z. S., F. G. Plumley, A. S. Lang, J. T. Beatty, R. E. Blankenship, C. L. Van Dover, C. Vetriani, M. Koblizek, C. Rathgeber, and P. G. Falkowski. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492-2495. [DOI] [PubMed] [Google Scholar]
- 27.Leahy, J. G., and R. R. Colwell. 1990. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, K., and E. M. Levy. 1992. Microbial degradation of petroleum in an intertidal beach environment—in situ sediment enclosures studies, p. 140-155. In Marine ecosystem enclosed experiments. Proceedings of a symposium. International Development Centre, Ottawa, Ontario, Canada.
- 29.Lee, K., G. H. Tremblay, and E. M. Levy. 1993. Bioremediation: application of slow-release fertilizers on low-energy shorelines, p. 449-454. In Proceedings of the 1993 Oil Spill Conference. American Petroleum Institute, Washington, D.C.
- 30.MacNaughton, S. J., J. R. Stephen, A. D. Venosa, G. A. Davis, Y. J. Chang, and D. C. White. 1999. Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinhassi, J., F. Azam, J. Hemphala, R. A. Long, J. Martinez, U. L. Zweifel, and A. Hagstrom. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 33.Prince, R. C. 1997. Bioremediation of marine oil spills. Trends Biotechnol. 15:158-160. [Google Scholar]
- 34.Röling, W. F. M., B. M. van Breukelen, M. Braster, M. T. Goeltom, J. Groen, and H. W. van Verseveld. 2000. Analysis of microbial communities in a landfill leachate polluted aquifer using a new method for anaerobic physiological profiling and 16S rDNA based fingerprinting. Microb. Ecol. 40:177-188. [DOI] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Shiba, T., Y. Shioi, K.-I. Takamiya, D. C. Sutton, and C. R. Wilkinson. 1991. Distribution and physiology of aerobic bacteria containing bacteriochlorophyll a on the east and west coasts of Australia. Appl. Environ. Microbiol. 57:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, V. H., D. W. Graham, and D. D. Cleland. 1998. Application of resource-ratio theory to hydrocarbon biodegradation. Environ. Sci. Technol. 32:3386-3395. [Google Scholar]
- 38.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco.
- 39.Swannell, R. P. J., B. C. Croft, A. L. Grant, and K. Lee. 1995. Evaluation of bioremediation agents in beach microcosms. Spill Sci. Technol. Bull. 2:151-159. [Google Scholar]
- 40.Swannell, R. P. J., K. Lee, and M. McDonagh. 1996. Field evaluations of marine oil spill bioremediation. Microbiol. Rev. 60:342-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swannell, R. P. J., D. Mitchell, G. Lethbridge, D. Jones, D. Heath, M. Hagley, M. Jones, S. Petch, R. Milne, R. Croxford, and K. Lee. 1999. A field demonstration of the efficacy of bioremediation to treat oiled shorelines following the Sea Empress incident. Environ. Technol. 20:863-873. [Google Scholar]
- 42.Swannell, R. P. J., D. J. Mitchell, J. C. Waterhouse, I. P. Miskin, I. M. Head, S. Petch, D. M. Jones, A. Willis, K. Lee, and J. E. Lepo. 2000. Impact of bioremediation treatments on the biodegradation of buried oil and predominant bacterial populations, p. 759-765. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Microbial biosystems: new frontiers. Proceedings of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbiology, Kentville, Nova Scotia, Canada.
- 43.Syutsubo, K., H. Kishira, and S. Harayama. 2001. Development of specific oligonucleotide probes for the identification and in situ detection of hydrocarbon-degrading Alcanivorax strains. Environ. Microbiol. 3:371-379. [DOI] [PubMed] [Google Scholar]
- 44.Tilman, G. D. 1982. Resource competition and community structure. Princeton University Press, Princeton, N.J.
- 45.Van de Peer, Y., and R. de Wachter. 1994. Treecon for windows—a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 46.Venosa, A. D., M. T. Suidan, B. A. Wrenn, K. L. Strohmeier, J. R. Haines, B. L. Eberhart, D. King, and E. Holder. 1996. Bioremediation of an experimental oil spill on the shoreline of Delaware Bay. Environ. Sci. Technol. 30:1764-1775. [Google Scholar]
- 47.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 48.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. B. Moore, W. R. Abraham, H. Lunsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]
- 49.Yurkov, V. V., and J. T. Beatty. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695-724. [DOI] [PMC free article] [PubMed] [Google Scholar]