Abstract
The signalling functions of Rho-family GTPases are based on the formation of distinctive protein–protein complexes. Invaluable insights into the structure–function relationships of the Rho GTPases have been obtained through the resolution of several of their structures in complex with regulators and downstream effectors. In this review, we use these complexes to compare the binding and specificity-determining sites of the Rho GTPases. Although the properties that characterize these sites are diverse, some fundamental conserved principles that govern their intermolecular interactions have emerged. Notably, all of the interacting partners of the Rho GTPases, irrespective of their function, bind to a common set of conserved amino acids that are clustered on the surface of the switch regions. This conserved region and its specific structural characteristics exemplify the convergence of the Rho GTPases on a consensus binding site.
Keywords: effector, GAP, GDI, GEF, Rho GTPase, switch region
Introduction
The small guanine nucleotide-binding proteins of the Rho family (Rho GTPases) act as molecular switches by cycling between a GDP-bound inactive state and a GTP-bound active state. Their activity is controlled by three groups of regulatory proteins: guanine nucleotide-dissociation inhibitors (GDIs), which interact only with the GDP-bound state and sequester the GTPase from the membrane into the cytoplasm; guanine nucleotide-exchange factors (GEFs), which bind independently of the nucleotide-bound state and accelerate the exchange reaction of bound GDP for GTP; and GTPase-activating proteins (GAPs), which interact only with the GTP-bound state and stimulate the slow intrinsic GTP-hydrolysis reaction (Vetter & Wittinghofer, 2001). In addition to these cellular regulators, bacterial pathogens produce toxins that specifically target Rho GTPases by mimicking GAPs and GEFs (Boquet & Lemichez, 2003). The formation of the active GTP-bound state of the GTPase is accompanied by a conformational change in two regions (known as switch I and II), which provide a platform for the selective interaction with functionally diverse proteins (the so-called downstream effectors) that initiate a network of cytoplasmic and nuclear signalling cascades (Bishop & Hall, 2000; Burridge & Wennerberg, 2004).
Rho GTPases are important regulators of cellular processes ranging from fundamental events (for example, the establishment of cell polarity) to highly specialized activities (for example, the contraction of vascular smooth-muscle cells; Etienne-Manneville & Hall, 2002). However, it remains conceptually unclear how these downstream signalling events and the Rho GTPases themselves are spatially regulated. This question becomes even more complicated when the large number of possible interactions between the 24 GTPases and the four GDIs, 66 GEFs, 83 GAPs and 101 effectors is considered (Burridge & Wennerberg, 2004; Wennerberg & Der, 2004). A detailed picture of the molecularswitch function of the Rho GTPases, and their interactions with regulators and effectors, has emerged from structures that have been solved using X-ray crystallography or nuclear magnetic resonance (NMR) methods (Corbett & Alber, 2001; Vetter & Wittinghofer, 2001).
To further understand how different interacting partners recognize Rho GTPases, we have focused on the interacting surface of the Rho proteins by analysing 26 GTPase structures in complex with their regulators and effectors. Our analysis shows that GDIs, GEFs, GAPs and effectors, although structurally diverse, all share a consensus binding site that overlaps with the switch I and II regions (encompassing amino acids 29–42 and 62–68, respectively) that undergo nucleotide-dependent conformational changes (Wei et al, 1997; Ihara et al, 1998). It is important to note that the amino-acid numbering used in this review is based on the RhoA sequence.
Focal points of bimolecular interactions
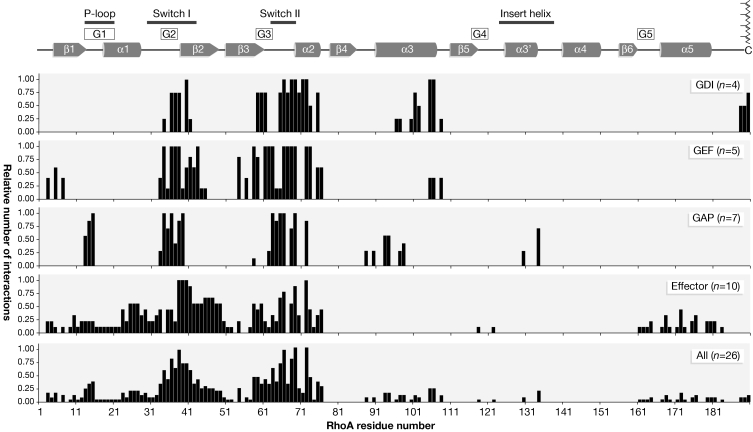
The large number of structures that are available for RhoA, Rac1 and Cdc42, which are the best-studied members of the Rho family, in complex with various regulators and effectors (Table 1), provides a unique opportunity to study the common characteristics of the interactions between Rho GTPases and their binding partners. We calculated the relative number of interactions of all residues of the Rho GTPases with GDIs, GEFs, GAPs and effectors, and these data are plotted as histograms in Figure 1. Most binding partners contact residues in the switch regions and in the α2-helix of the GTPase, which becomes more evident when the contacts of all binding partners are taken into account (Fig 1). In addition, different binding partners contact other regions that are specific to their function: all regulatory proteins bind the α3-helix, GDIs also recruit the prenylated carboxy (C)-terminus and GAPs seem to be the only proteins that contact the insert helix. By contrast, effector proteins exhibit various interaction patterns depending on the effector type (see below) and predominantly bind to the amino (N)-terminal third of the GTPase or the region around the α5-helix. To discover which residues of the Rho GTPases are mostly involved in protein–protein interactions, we compiled an alignment of the intermolecular contacts in the switch regions (Fig 2). A notable finding is that, with the exception of the effectors p67phox (67-kD subunit of the phagocyte NADPH oxidase; Lapouge et al, 2000) and the first binding site of the protein kinase Nα (PKNα; Maesaki et al, 1999), all Rho GTPase-binding partners, irrespective of their structural and functional diversity, interact with common elements of the switch regions and the α2-helix (Fig 2). The most prominent contact residues are two leucines (Leu69 and Leu72) of the α2-helix, which both make crucial hydrophobic contacts with nearly all interacting proteins. In addition, 10 other residues (Tyr34, Pro36, Thr37, Val38, Phe39, Glu40, Gln63, Asp65, Tyr66 and Arg68; colour coded in Fig 2) of the switch I and II regions are involved in most interactions, irrespective of the conformational state of the GTPases or the function of the interacting partner. To illustrate the parts of the surface of the Rho GTPases that are involved in protein–protein interactions, the solvent-accessible surface of RhoA was coloured according to the number of interactions in which each residue participates (Fig 3). A remarkable conclusion is that the contacting residues mentioned above are clustered at the surface of the GTPase irrespective of whether it is bound to GDP or GTP. Previous structural analyses of complexes of small GTPases from diverse families (Ras, Rho, Ran, Arf and Rab) showed that almost all of the surface residues are involved in at least one interaction (Corbett & Alber, 2001). By contrast, our analysis indicates that a large area on the surface of the Rho GTPases is not involved in any interaction that has been described so far (Fig 3).
Table 1.
Structures ofRho-GTPase complexes
| Complex* | PDB ID‡ | Res (Å)§ | References |
|---|---|---|---|
| GDIs | |||
| RhoA·GDP·GDI-1 | 1CC0 | 5.0 | Longenecker et al, 1999 |
| Rac1·GDP·GDI-1 | 1HH4 | 2.7 | Grizot et al, 2001 |
| Rac2·GDP·GDI-2 | 1DS6 | 2.3 | Scheffzek et al, 2000 |
| Cdc42·GDP·GDI-1 | 1DOA | 2.6 | Hoffman et al, 2000 |
| GEFs | |||
| RhoA·Dbs | 1LB1 | 2.8 | Snyder et al, 2002 |
| Rac1·Tiam1 | 1FOE | 2.8 | Wothylacke et al, 2000 |
| Cdc42·Dbs | 1KZ7 | 2.4 | Rossman et al, 2002 |
| Cdc42·ITSN | 1KI1 | 2.3 | Snyder et al, 2002 |
| Cdc42·SopE¶ | 1GZS | 2.3 | Buchwald et al, 2002 |
| GAPs | |||
| RhoA·GDP·AlF4−·GAP∥ | 1TX4 | 1.6 | Rittinger et al, 1997b |
| RhoA·GDP·MgF3−·GAP∥ | 1OW3 | 1.8 | Graham et al, 2002 |
| Rac1·GDP·AlF3·ExoS¶ | 1HE1 | 2.0 | Würtele et al, 2001 |
| Rac1·GDP·AlF3·SptP¶ | 1G4U | 2.3 | Stebbins & Galán, 2000 |
| Cdc42·GppNHp·GAP∥ | 1AM4 | 2.7 | Rittinger et al, 1997a |
| Cdc42·GDP·AlF3·GAP∥ | 1GRN | 2.1 | Nassar et al, 1998 |
| Cdc42·GDP·AlF3·GAP∥,# | 2NGR | 1.9 | Nassar et al, 1998 |
| Effectors | |||
| RhoA·GppNHp·ROCK | 1S1C | 2.6 | Dvorsky et al, 2004 |
| RhoA·GTPγS·PKNα | 1CXZ | 2.2 | Maesaki et al, 1999 |
| Rac1·GTP·p67phox** | 1E96 | 2.4 | Lapouge et al, 2000 |
| Rac1·GppNHp·arfaptin | 1I4T | 2.6 | Tarricone et al, 2001 |
| Rac1·GDP·arfaptin | 1I4D | 2.5 | Tarricone et al, 2001 |
| Cdc42·GppNHp·ACK**,‡‡ | 1CF4 | NMR | Mott et al, 1999 |
| Cdc42·GppCH2p·WASP‡‡ | 1CEE | NMR | Abdul-Manan et al, 1999 |
| Cdc42·GppNHp·PAK**,‡‡ | 1E0A | NMR | Morreale et al, 2000 |
| Cdc42·GppCH2p·PAK‡‡ | 1EES | NMR | Gizachew et al, 2000 |
| Cdc42·GppNHp·Par6**,‡‡ | 1NF3 | 2.1 | Garrard et al, 2003 |
*GTPase–GDI complexes are in the GDP-bound form,GTPase–GEF complexes are nucleotide free,GTPase–GAP complexes are in the GppNHp-bound ground state and in the GDP-aluminium fluoride (AlF4−/AlF3) or GDP-magnesium fluoride (MgF3−)-bound transition states,GTPase–effector complexes are in GTPγS-, GppNHp- or GppCH2p- (GTP analogues) bound states.
‡Protein Data Bank identification number.
§The numbers indicate the resolution of the crystal structures and nuclear magnetic resonance (NMR) represents those structures that have been solved in solution.
∥The catalytic domain of p50GAP has been used.
¶SopE, ExoS and SptP are regulators from bacterial pathogens.
#GAP mutant (R305A) has been used.
**Constitutive active mutants (Gln61 to Leu) of the respective GTPase have been used.
‡‡Cdc42/Rac interactive binding (CRIB)-containing effector proteins.ACK, activated Cdc42-associated tyrosine kinase; Dbs,Dbl's big sister; GAP,GTPase-activating protein; GDI, guanine nucleotide-dissociation inhibitor; GEF, guanine nucleotide-exchange factor; ITSN, intersectin; PAK, p21-activated kinase; PKN, protein kinase N; ROCK, Rho kinase;WASP,Wiskott–Aldrich syndrome protein.
Figure 1.
Structural motifs and intermolecular contact sites of Rho GTPases. Structural elements (α-helices are represented as cylinders and β-strands as arrows), the guanine nucleotide-binding peptide loops (G1–G5), the conserved sequence motifs and the isoprenylation site at the C-terminus of RhoA are highlighted on the top according to Ihara et al (1998). The frequencies of intermolecular contacts of interacting partners with Rho GTPases are shown as histograms. The relative numbers of GTPase interactions with the individual guanine nucleotide-dissociation inhibitors (GDIs), guanine nucleotide-exchange factors (GEFs), GTPase-activating proteins (GAPs) and effectors, and all interacting partners are plotted as a function of the residue numbers of RhoA. The relative numbers of interactions for the corresponding residues of Rho GTPases are calculated as the numbers of contacts (defined as ≤4 Å) divided by the maximal number of contacts. 'n' indicates the number of analysed complexes.
Figure 2.
Frequency alignment of residues of Rho GTPases that are involved in intermolecular interactions. Aligned sequences of the switch I, β3, switch II and α2 regions of RhoA, Rac1 and Cdc42 are shown together with the frequency of involvement of the residues in the interaction with regulators and effectors using 26 complexes (Table 1). The residues that are most frequently involved in the interfaces are colour coded. The symbol '+' indicates an interaction with the binding partners, whereas the symbol '−' represents no intermolecular contact. GAPRA refers to a GTPase-activating protein (GAP) in which the arginine finger (Arg85) is replaced by an alanine. It is important to note that protein kinase Nα (PKNα) binds RhoA at two sites: contact site 1 (1cs; outside of the switch regions) and contact site 2 (2cs; inside the switch regions). Moreover, the former, along with p67phox, does not make contact with any of the colour-coded residues. ACK, activated Cdc42-associated tyrosine kinase; Dbs, Dbl's big sister; GDI, guanine nucleotide-dissociation inhibitor; ITSN, intersectin; PAK, p21-activated kinase; ROCK, Rho kinase; WASP, Wiskott–Aldrich syndrome protein.
Figure 3.
Switch regions as the focal points of bimolecular interactions. Residues of RhoA that mediate the interactions with guanine nucleotide-dissociation inhibitors (GDIs), guanine nucleotide-exchange factors (GEFs), GTPase-activating proteins (GAPs) and effectors are coloured from yellow (corresponding to one contact) to red (corresponding to a maximal number of 24 contacts from the 26 analysed complexes). Grey indicates the residues that are not involved in any interactions. The GDP-bound state and the GTP-γS-bound state of the RhoA structures are shown on the upper and lower panels, respectively, in ribbon (A, D) and surface (B, E) representation in the same orientation. (B) and (E) are also rotated 180° around the indicated axis with respect to one another (C,F). The labelled residues that interact most frequently with their partner proteins are shown in a ball-and-stick configuration in (A) and (D). The position of some structural elements (helices α2 and α3 or β2/β3 strands) and sequence motifs (switch I and II or the insert helix) are shown in (A) and (D).
To address the structural characteristics of this focal point of the Rho GTPases in more detail, we concentrated on the involvement of these 12 residues in interactions with regulatory and effector proteins.
Interaction with regulatory proteins
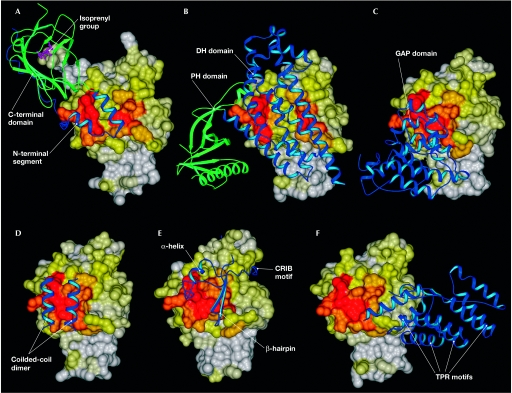
In stimulated cells, isoprenylated Rho GTPases are associated specifically with the cellular membrane, which is essential for their biological activity. In resting cells, however, GDIs extract them from the membrane and create a cytosolic pool of inactivated Rho GTPases (Oloffson, 1999). GDI structures in complex with GDP-bound RhoA, Cdc42, Rac2 and Rac1 (Longenecker et al, 1999; Hoffman et al, 2000; Scheffzek et al, 2000; Grizot et al, 2001, respectively) have shown that GDI proteins comprise two domains and show a conserved binding mode to Rho GTPases (Figs 1,4A). Their short, N-terminal helix–loop–helix domain binds to the switch I, β3, switch II and α2 regions, whereas the C-terminal immunoglobulin-like domain binds to the switch II and α3 regions, and to the covalently bound isoprenyl group that is attached to the C-terminus of the GTPase. The extensive contacts between the GDI and the GTPase result in a well-ordered conformation of the switch regions, which is probably responsible for the ability of the GDI to inhibit GDP dissociation and GTP hydrolysis (Hoffman et al, 2000; Scheffzek et al, 2000).
Figure 4.
Complexes of regulators and effectors with their respective GTPases. (A) Cdc42·GDP·GDI. (B) RhoA·Dbs. (C) RhoA·GDP·AlF4−·GAP. (D) RhoA·GppNHp·ROCK. (E) Cdc42·GppCH2p·WASP. (F) Rac1(Q61L)·GTP·p67phox. The surfaces of the GTPases are coloured as in Fig 3. The guanine nucleotide-dissociation inhibitor (GDI), guanine nucleotide-exchange factor (GEF), GTPase-activating protein (GAP) and effector domains are shown as ribbons. CRIB, Cdc42/Rac-interacting binding; Dbs, Dbl's big sister; DH, Dbl homology; PH, pleckstrin homology; ROCK, Rho kinase; TPR, tetraricopeptide repeat; WASP, Wiskott–Aldrich syndrome protein.
Compared with the GEF, GAP and effector interfaces (see below), there are only a few residues on the GTPase that determine the specificity of the GTPase–GDI interaction (Fig 2). Among other theories, contact with the invariant Thr37 of switch I, which is essential for the coordination of the Mg2+ ion and, therefore, nucleotide binding, provides a plausible explanation for the ability of the GDI protein to inhibit nucleotide exchange on the GTPase. Another residue is Arg68, which, together with the conserved Tyr66, Leu69 and Leu72 residues, contributes to the stability of the N-terminal segment of the GDI. Arg68 also contacts the C-terminal domain of the GDI (Fig 4A). Replacement of the equivalent arginine residue by alanine or glutamic acid has been shown to compromise bridging contacts, which disrupts the Cdc42–GDI interaction (Lin et al, 2003; Gandhi et al, 2004).
GEFs for Rho GTPases belong mostly to the Dbl family, and contain the catalytically active Dbl homology (DH) domain followed by an adjacent pleckstrin homology (PH) domain (Erickson & Cerione, 2004). Crystal structures of different DH–PH tandem domains in complex with nucleotide-free Rho GTPases (Worthylake et al, 2000; Rossman et al, 2002; Snyder et al, 2002) have shown that the DH domain, which is composed of a flattened elongated α-helical bundle, binds switch I, β2/β3, switch II, α2 and α3 of the GTPase (Figs 1,4B).
The main feature of the nucleotide-exchange reaction that is carried out by GEFs is, in contrast to GDIs, the disruption of the favourable interactions that are necessary for Mg2+-ion coordination and tight nucleotide binding (Worthylake et al, 2000; Buchwald et al, 2002; Rossman et al, 2002; Snyder et al, 2002). However, the DH domain does not directly impinge on the nucleotide-binding site; rather, it induces conformational rearrangements of the switch regions that promote nucleotide ejection—an action that has been described as a 'push-and-pull' mechanism (Vetter & Wittinghofer, 2001). During the 'push' phase, the DH domain interacts with switch I and anchors it in a new position by strong electrostatic contacts of a highly conserved glutamic acid—for example, Glu639 in Dbl's big sister (Dbs)—with Thr37. The coordination of the Mg2+ ion is therefore disrupted while the switch II region is 'pulled' towards the nucleotide-binding site by the extensive interactions between Tyr66, Arg68, Leu69 and Leu72, and the DH domain (Fig 4B). A similar mechanism has been put forward for bacterial protein toxins, such as the Salmonella cytotoxin SopE, although this toxin shares neither detectable sequence nor structural homology with the Dbl family (Buchwald et al, 2002).
In contrast to the Dbl family members, T-lymphoma invasion and metastasis-inducing protein 1 (Tiam1) and intersectin, the PH domain of Dbs directly contacts the switch II Arg68 (Fig 4B; Rossman et al, 2002; Snyder et al, 2002), which is similar to the C-terminal domain of GDIs, even though the basic actions of these two regulators are reciprocal (Vetter & Wittinghofer, 2001; Erickson & Cerione, 2004). Whereas these contacts are an integral element of the GDI mechanism, the role of the tandem PH domains of the Rho GTPasespecific GEFs remains unclear.
Hydrolysis of the bound GTP is the timing mechanism that returns Rho GTPases to their GDP-bound inactive state and thereby completes the GTPase cycle. The intrinsic GTP-hydrolysis reaction is slow, but can be stimulated by several orders of magnitude through interaction of the GTPase with GAPs. The Rho-specific GAPs are defined by the presence of a conserved catalytic domain (Moon & Zheng, 2003), which is sufficient for GTPase binding and the stimulation of their GTP-hydrolysis reaction. So far, several GAP structures in complex with the GTPase in the ground state (Rittinger et al, 1997a) or the transition state (Rittinger et al, 1997b; Nassar et al, 1998; Graham et al, 2002) have provided insights into the structural characteristics and the regulatory mechanism of the GAPs. The GAP domain, which contains a bundle of nine α-helices packed together in an anti-parallel arrangement, interacts with the P-loop, the switch I and II regions, and the nucleotide itself (Figs 1,4C).
The basic mechanism of GTPase stimulation relies on the stabilization of the highly mobile switch regions and the transition state of the GTP-hydrolysis reaction by supplying a catalytic arginine to the active site (Gamblin & Smerdon, 1998; Scheffzek et al, 1998; Kosloff & Selinger, 2001; Vetter & Wittinghofer, 2001). Therefore, GAPs position the catalytically crucial Gln63 in an appropriate conformation towards a nucleophilic water molecule, which hydrolyses GTP and neutralizes developing negative charges on the leaving group during the phosphoryl-transfer reaction. Replacement of this so-called 'arginine finger' by an alanine greatly diminishes the catalytic capacity of the respective GAPs, which are still able to bind the GTPase with high affinity (Ahmadian et al, 1997; Leonard et al, 1998; Graham et al, 1999). Most remarkably, the same mechanistic strategy has been shown for bacterial GAPs, such as the Salmonella typhimurium virulence factor SptP and the Pseudomonas aeruginosa cytotoxin ExoS, even though they do not share any sequence or structural similarity with eukaryotic Rho GAP domains (Stebbins & Galán, 2000; Würtele et al, 2001).
The effector-binding site
The ability of the Rho GTPases to control a wide range of intracellular signalling pathways is attributed to their association with their cellular targets: the effector proteins. Unlike regulators that interact with Rho GTPases to modulate their switch function, effectors require the GTPase to be in a specific conformational arrangement to accomplish their own intrinsic function.
The crystal structures of the GTPase-binding domains (GBDs) of PKN and Rho kinase (ROCK) in complex with RhoA have shown that the domains, as predicted from their primary structure, form α-helical coiled-coils that are arranged in an anti-parallel and parallel fashion, respectively (Maesaki et al, 1999; Dvorsky et al, 2004). A 13-residue left-handed coiled-coil at the C-terminal part of the ROCK–GBD, which is designated as the minimal Rho-interacting motif, binds exclusively to the switch and α2 regions of RhoA (Fig 4D). Conversely, the RhoA–PKN complex points to two possible contact sites on RhoA (Maesaki et al, 1999): contact site 1 consists of the α1, β2/β3 and α5 regions of RhoA (Fig 1), whereas contact site 2 overlaps remarkably well with the ROCK-binding site (Figs 2,4D). Complex structures of Cdc42 with effector proteins containing a Cdc42/Rac interactive binding (CRIB) motif, which have been determined mostly by NMR spectroscopy owing to their high flexibility (Table 1; Abdul-Manan et al, 1999; Mott et al, 1999; Gizachew et al, 2000; Morreale et al, 2000; Garrard et al, 2003), have shown that the GBD of this class of effectors makes extensive contact with the surface of Rho GTPases (Fig 4E). It binds with its β-hairpin and C-terminal α-helix to the α1, switch I and II regions, and wraps around the α5 and β2 regions of the GTPase with its extended N-terminus, which encompasses the CRIB motif (Fig 4E).
Two other effectors, arfaptin and p67phox, show unexpected features of their structures and contact sites on the GTPase (Lapouge et al, 2000; Tarricone et al, 2001). Arfaptin adopts an elongated crescentshaped dimer of three helix coiled-coils that makes contact with the switch I and II, and α2 regions of Rac1, regardless of its nucleotide-bound state (Tarricone et al, 2001), and structurally mimics the DH domain of Tiam1 (Cherfils, 2001). p67phox comprises an α-helical domain that consists of four so-called tetratricopeptide repeat (TPR) motifs (D'Andrea & Regan, 2003) that bind α1, the N-terminal residues of switch I, and the G3 and G5 loops, but not the switch II region or the principal parts of switch I (Figs 1,2,4F; Lapouge et al, 2000). It has also been proposed that the switch regions might be the contact sites for a third protein that is associated with the Rac1·GTP·p67phox complex (Diebold & Bokoch, 2001; Hoffman & Cerione, 2001).
The GTPase–effector complex structures mentioned so far have not clarified the mechanism of effector activation, which requires the disruption of intramolecular autoinhibition and the exposure of their functional domains. A common feature of effector complexes is that, with the exception of p67phox, they all make intensive contacts with the switch/α2 regions of Rho GTPases (Fig 2), which indicates that this site probably serves as the platform for the GTP-dependent recognition of effectors. The two invariant leucines (Leu69 and Leu72), which form crucial hydrophobic contacts with almost all effector domains, have been put forward as essential elements for the Cdc42/Rac-mediated activation of CRIB-containing effectors (Morreale et al, 2000). A different activation mechanism has been implicated for the Rhospecific effectors PKN and ROCK, which use other domains to bind cooperatively to sites outside of the switch regions of RhoA (Blumenstein & Ahmadian, 2004).
Plasticity and specificity
The high plasticity of the consensus binding site explains how a single GTPase can interact with various regulatory proteins and effectors. Although the Rho GTPases exist in two main states, a comparison of the three-dimensional arrangement of the respective amino-acid side chains explains their ability to adopt a range of interfaces together with their interacting partner. In principle, GDIs and GEFs recognize the inactive GDP-bound state of the GTPase, and cause considerable induced fit on binding, as the pattern of their consensus binding site shows clear differences (Figs 3B,4A,4B). Conversely, the formation of tight complexes of GAPs and effectors with the active GTP-bound state of the GTPase induces only moderate conformational changes in the respective interacting interface (Figs 3E,4C–F).
Another important issue is the specificity of such a bimolecular interaction, which cannot be explained simply by minor amino-acid deviations in the consensus binding site or on the basis of the presented structural comparison of Rho GTPases. Rather, it requires complex biophysical phenomena, such as molecular recognition and overall dynamics, in the context of the interacting molecules. Replacement of the amino acids outside the contact regions of Rho GTPases has been shown to affect their specificity and affinity towards their binding partners (Haeusler et al, 2003; Heo & Meyer, 2003). The characteristic features of the respective interacting partner that cannot be mimicked by another protein are the result of the binding of concerned moieties, and are therefore specific for each class of protein complexes. GDIs provide their N-terminal segment, which recognizes and binds the switch regions to initiate GTPase displacement from the membrane by the function of their C-terminal domain. By contrast, the DH domain of GEFs binds the β2/β3 region and thereby initiates conformational changes of the switch regions, which are required for nucleotide exchange. The catalytic domain of GAPs directly binds the switch regions and supplies an arginine finger to the active site to switch off the signal transduction. By contrast, the GBDs of the effectors recognize the switch regions of active Rho GTPases and induce the rearrangement of their autoinhibitory conformation, which leads to their activation. To attain the functional state, however, effectors require further contact regions beyond the consensus binding site to accomplish signal transduction.
Conclusions
The molecular-switch function of the small GTPases, which has evolved to provide a rapid but transient transmission of signals in cells, is modulated by four classes of binding partners: GDIs, GEFs, GAPs and effectors. In this review, we have shown that the switch apparatus of the Rho GTPases contains a distinct consensus binding site that is the main determinant of the bimolecular interaction. Owing to its internal plasticity and the function of the binding partner, this site adopts various conformations that are complementary to, and specific for, the surfaces of individual interacting partners. Numerous functionally and structurally distinct proteins are therefore able to recognize a single GTPase.


Acknowledgments
We appreciate the comments and discussions provided by our group members: L. Hemsath, D. Fiegen, L.C. Haeusler, L. Blumenstein and A. Eberth. We apologize for not being able to cite the work of many colleagues owing to space constraints. Work in the authors' laboratory is supported, in part, by a European Community Marie Curie Fellowship, the Volkswagen-Stiftung, the Deutsche Forschungsgemeinschaft, the Max Planck Society, the Verband der Chemischen Industrie and the Bundesministerium für Bildung und Forschung.
References
- Abdul-Manan N et al. (1999) Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott–Aldrich syndrome' protein. Nature 399: 379–383 [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAPstimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 [DOI] [PubMed] [Google Scholar]
- Bishop AL, Hall A (2000) Rho GTPases and their effector proteins. Biochem J 348: 241–255 [PMC free article] [PubMed] [Google Scholar]
- Blumenstein L, Ahmadian MR (2004) Models of the cooperative mechanism for Rho-effector recognition: implications for RhoA-mediated effector activation. J Biol Chem Oct 8 [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Boquet P, Lemichez E (2003) Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol 13: 238–246 [DOI] [PubMed] [Google Scholar]
- Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K (2002) Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J 21: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Wennerberg K (2004). Rho and Rac take center stage. Cell 116: 167–179 [DOI] [PubMed] [Google Scholar]
- Cherfils J (2001) Structural mimicry of DH domains by Arfaptin suggests a model for the recognition of Rac–GDP by its guanine nucleotide exchange factors. FEBS Lett 507: 280–284 [DOI] [PubMed] [Google Scholar]
- Corbett KD, Alber T (2001) The many faces of Ras: recognition of small GTP-binding proteins. Trends Biochem Sci 26: 710–716 [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28: 655–562 [DOI] [PubMed] [Google Scholar]
- Diebold BA, Bokoch GM (2001). Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nat Immunol 2: 211–215 [DOI] [PubMed] [Google Scholar]
- Dvorsky R, Blumenstein L, Vetter IR, Ahmadian MR (2004) Structural insights into the interaction of ROCKI with the switch regions of RhoA. J Biol Chem 279: 7098–7104 [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cerione RA (2004) Structural elements, mechanism, and evolutionary convergence of rho protein–guanine nucleotide exchange factor complexes. Biochemistry 43: 837–842 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Gamblin SJ, Smerdon SJ (1998) GTPase-activating proteins and their complexes. Curr Opin Struct Biol 8: 195–201 [DOI] [PubMed] [Google Scholar]
- Gandhi PN, Gibson RM, Tong X, Miyoshi J, Takai Y, Konieczkowski M, Sedor JR, Wilson-Delfosse AL (2004) An activating mutant of Rac1 that fails to interact with Rho GDP-dissociation inhibitor stimulates membrane ruffling in mammalian cells. Biochem J 378: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrard SM, Capaldo CT, Gao L, Rosen MK, Macara IG, Tomchick DR (2003). Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J 22: 1125–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizachew D, Guo W, Chohan KK, Sutcliffe MJ, Oswald RE (2000). Structure of the complex of Cdc42Hs with a peptide derived from p-21 activated kinase. Biochemistry 39: 3963–3971 [DOI] [PubMed] [Google Scholar]
- Graham DL, Eccleston JF, Lowe PN (1999) The conserved arginine in rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with Rho.GDP and aluminium fluoride. Biochemistry 38: 985–991 [DOI] [PubMed] [Google Scholar]
- Graham DL et al. (2002) MgF3− as a transition state analog of phosphoryl transfer. Chem Biol 9: 375–381 [DOI] [PubMed] [Google Scholar]
- Grizot S, Faure J, Fieschi F, Vignais PV, Dagher MC, Pebay-Peyroula E (2001) Crystal structure of the Rac1–RhoGDI complex involved in NADPH oxidase activation. Biochemistry 40: 10007–10013 [DOI] [PubMed] [Google Scholar]
- Haeusler LC, Blumenstein L, Stege P, Dvorsky R, Ahmadian MR (2003) Comparative functional analysis of the Rac-isoforms. FEBS Lett 555: 556–560 [DOI] [PubMed] [Google Scholar]
- Heo WD, Meyer T (2003) Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell 113: 315–328 [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione RA (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100: 345–356 [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Cerione RA (2001). Rac inserts its way into the immune response. Nat Immunol 2: 194–196 [DOI] [PubMed] [Google Scholar]
- Ihara K et al. (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J Biol Chem 273: 9656–9666 [DOI] [PubMed] [Google Scholar]
- Kosloff M, Selinger Z (2001) Substrate assisted catalysis: application to G proteins. Trends Biochem Sci 26: 161–166 [DOI] [PubMed] [Google Scholar]
- Lapouge K, Smith SJ, Walker PA, Gamblin SJ, Smerdon SJ, Rittinger K (2000) Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol Cell 6: 899–907 [DOI] [PubMed] [Google Scholar]
- Leonard DA, Lin R, Cerione RA, Manor D (1998) Biochemical studies of the mechanism of action of the Cdc42 GTPase-activating protein. J Biol Chem 273: 16210–16215 [DOI] [PubMed] [Google Scholar]
- Lin Q, Fuji RN, Yang W, Cerione RA (2003) RhoGDI is required for Cdc42-mediated cellular transformation. Curr Biol 13: 1469–1479 [DOI] [PubMed] [Google Scholar]
- Longenecker KL et al. (1999) How RhoGDI binds Rho. Acta Crystallogr D Biol Crystallogr 55: 1503–1515 [DOI] [PubMed] [Google Scholar]
- Maesaki R, Ihara K, Shimizu T, Kuroda S, Kaibuchi K, Hakoshima T (1999) The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1. Mol Cell 4: 793–803 [DOI] [PubMed] [Google Scholar]
- Moon SY, Zheng Y (2003) Rho GTPase-activating proteins in cell regulation. Trends Cell Biol 13: 13–22 [DOI] [PubMed] [Google Scholar]
- Morreale A et al. (2000) Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat Struct Biol 7: 384–388 [DOI] [PubMed] [Google Scholar]
- Mott HR et al. (1999). Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature 399: 384–388 [DOI] [PubMed] [Google Scholar]
- Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA (1998) Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol 5: 1047–1052 [DOI] [PubMed] [Google Scholar]
- Olofsson B (1999) Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal 11: 545–554 [DOI] [PubMed] [Google Scholar]
- Rittinger K et al. (1997a) Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature 388: 693–697 [DOI] [PubMed] [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ (1997b) Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transitionstate analogue. Nature 389: 758–762 [DOI] [PubMed] [Google Scholar]
- Rossman KL, Worthylake DK, Snyder JT, Siderovski DP, Campbell SL, Sondek J (2002) A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J 21: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Wittinghofer A (1998) GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci 23: 257–262 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P (2000) The Rac–RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat Struct Biol 7: 122–126 [DOI] [PubMed] [Google Scholar]
- Snyder JT et al. (2002) Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol 9: 468–475 [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Galán JE (2000) Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol Cell 6: 1449–1460 [DOI] [PubMed] [Google Scholar]
- Tarricone C et al. (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411: 215–219 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Wei Y et al. (1997) Crystal structure of RhoA-GDP and its functional implications. Nat Struct Biol 4: 699–703 [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Der CJ (2004) Rho-family GTPases: it's not only Rac and Rho (and I like it). J Cell Sci 117: 1301–1312 [DOI] [PubMed] [Google Scholar]
- Worthylake DK, Rossman KL, Sondek J (2000). Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408: 682–688 [DOI] [PubMed] [Google Scholar]
- Würtele M et al. (2001) How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nat Struct Biol 8: 23–26 [DOI] [PubMed] [Google Scholar]