Abstract
NMR structures of recombinant prion proteins from various species expressed in Escherichia coli have been solved during the past years, but the fundamental question of the relevancy of these data relative to the naturally occurring forms of the prion protein has not been directly addressed. Here, we present a comparison of the cellular form of the bovine prion protein isolated and purified from healthy calf brains without use of detergents, so that it contains the two carbohydrate moieties and the part of the GPI anchor that is maintained after enzymatic cleavage of the glycerolipid moiety, with the recombinant bovine prion protein expressed in E. coli. We show by circular dichroism and 1H-NMR spectroscopy that the three-dimensional structure and the thermal stability of the natural glycoprotein and the recombinant polypeptide are essentially identical. This result indicates possible functional roles of the glycosylation of prion proteins in healthy organisms, and provides a platform and validation for future work on the structural biology of prion proteins, which will have to rely primarily on the use of recombinant polypeptides.
Keywords: cellular prion protein, circular dichroism spectroscopy, NMR, prion protein structure, transmissible spongiform encephalopathy
Introduction
The development of transmissible spongiform encephalopathies (TSEs) has been shown to be linked to the presence of the host-encoded prion protein PrP (Büeler et al, 1993). Furthermore, a conformational transition from cellular PrP in the healthy organism, PrPC, to a disease-related, presumably infectious scrapie form, PrPSc, has been proposed as the critical event in TSE pathogenesis (Alper et al, 1967; Griffith, 1967; Prusiner, 1991). Keen interest is thus focused on the three-dimensional (3D) structures of the two, or possibly several, forms of the prion protein that seem to be key factors for rationalizing TSE pathogenesis. Because only very low yields of PrPC are obtained through purification from natural sources, and PrPSc is highly insoluble in aqueous solvents, no atomic resolution X-ray crystal or NMR solution structures are as yet available either for natural PrPC or for PrPSc. Optical spectroscopy measurements showed a high α-helix content for PrPC purified from hamster brain, and a high content of βsheet structure for PrPSc (Caughey et al, 1991; Pan et al, 1992; Pergami et al, 1999). Further studies of PrPSc using X-ray diffraction provided additional support for its amyloid nature, and, on the basis of electron microscopy data, a left-handed β-helix structure was proposed for PrPSc (Nguyen et al, 1995; Wille et al, 2002).
In view of the limited accessibility of prion proteins from natural sources, structural studies have so far been focused on recombinant prion proteins expressed in Escherichia coli (Riek et al, 1996, 1997; Donne et al, 1997; López García et al, 2000; Zahn et al, 2000). Although the intact disulphide bond of wild-type PrP was present in these preparations, the recombinant proteins did not include the other post-translational modifications of natural PrP, namely the attachment of two glycosyl moieties and a carboxy-terminal GPI anchor (Bolton et al, 1985; Stahl et al, 1990). To validate the relevancy of the data obtained with recombinant PrP, this paper describes the isolation of sufficient quantities of natural PrPC from healthy calf brains for in vitro physico-chemical studies, and presents a detailed structural comparison with recombinant PrP on the basis of circular dichroism (CD) and 1H-NMR spectroscopy.
Results And Discussion
Isolation and purification of natural bPrPC
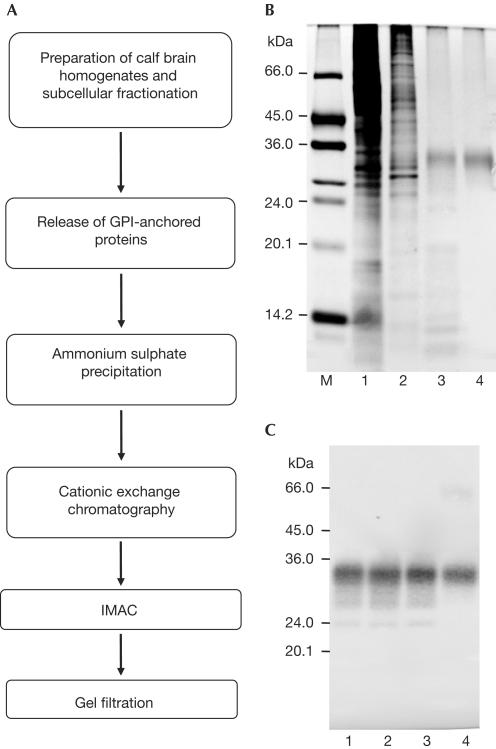
The basis for the present structural investigations of the natural, full-length PrPC was the development of an efficient method for the isolation from brain tissue and purification of intact, natively folded PrPC with all its post-translational modifications (Fig 1A). The first step included the preparation of brain homogenates and subcellular fractionation by a series of centrifugation steps, which yielded a membrane fraction. In contrast to previously published protocols for the purification of natural PrPC (Pan et al, 1992; Pergami et al, 1996), in which the protein was separated from the membrane with the use of detergents, we treated the membrane fraction with the phosphatidylinositolspecific phospholipase C (PI-PLC) to release the GPI-anchored proteins (Stahl et al, 1990). PI-PLC cuts the glycerolipid portion of the GPI anchor, which remains in the membrane fraction, whereas the soluble part of the GPI anchor remains attached to the protein. This step enabled the isolation and purification of the protein without the use of detergents or denaturants, which could have modified the structure of the protein (Pergami et al, 1999).
Figure 1.
Isolation and purification of bPrPC from healthy calf brains. (A) Purification scheme. (B,C) Analysis of natural bPrPC after the principal purification steps. Samples were analysed by an SDS–12% (w/v) PAGE followed by silver staining (B), and by immunostaining after western blotting (C). Lane M: molecular mass standard; lane 1: soluble fraction after isolation of natural bPrPC from the cell membranes and ammonium sulphate precipitation; lane 2: pooled fractions after cationic exchange chromatography on SP-Sepharose; lane 3: pooled fractions after IMAC; lane 4: purified natural bPrPC after gel filtration on Superose12.
Soluble fractions of natural bPrPC were pooled, concentrated by ammonium sulphate precipitation, resolubilized and purified by conventional chromatography techniques, such as cationic exchange chromatography, Co2+-immobilized metal affinity chromatography (IMAC) and gel filtration on Superose12 (Fig 1). After the final purification step, the diglycosylated form of the natural bPrPC was recovered as the main product.
The amount and purity of natural bPrPC obtained after the last two purification steps were estimated by scanning the corresponding silver-stained gels on a densitometer and determination of the total amount of proteins. On average, 25±5 μg of bPrPC with a purity of ∼60% was obtained from one calf brain after purification on Co2+-IMAC and concentrating. The average amount of bPrPC obtained from one calf brain in the final sample was estimated to be 15±5 μg, as determined by its absorbance at 280 nm, with a purity of >93%. Matrix-associated laser desorption ionization (MALDI) mass spectrometry of this preparation showed a mass distribution with a peak at 31,962 Da, and Edman sequencing showed that the purified protein contained the correct amino terminus for the mature bovine prion protein after cleavage of the N-terminal signal sequence. This preparation was then used for the biophysical and structural comparisons of natural bPrPC with recombinant full-length bPrP(23–230) (Figs 2, 3). All measurements in this study were performed at pH 4.5, as pH values between 4.5 and 5.2 had been used for the previous 3D NMR structure determinations of recombinant bPrP(23–230) (López García et al, 2000) and prion proteins from other species (Riek et al, 1996, 1997; Donne et al, 1997; Zahn et al, 2000).
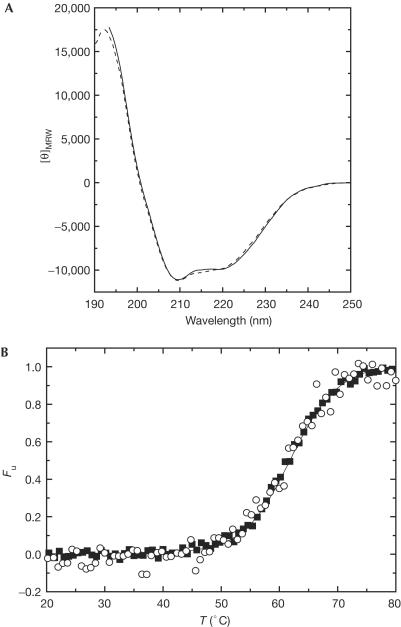
Figure 2.
Regular polypeptide secondary structure and thermal stability of recombinant bPrP(23–230) and natural bPrPC. (A) Far-UV CD spectra of recombinant bPrP(23–230) (dashed line) and natural bPrPC (solid line) at 20°C. [θ]MRW, mean residue ellipticity (deg/cm2/dmol). (B) Normalized thermal unfolding transitions of recombinant bPrP(23–230) (filled squares) and natural bPrPC (open circles) monitored by CD at 222 nm and a heating rate of 1°C/min. Fu, fraction of unfolded protein.
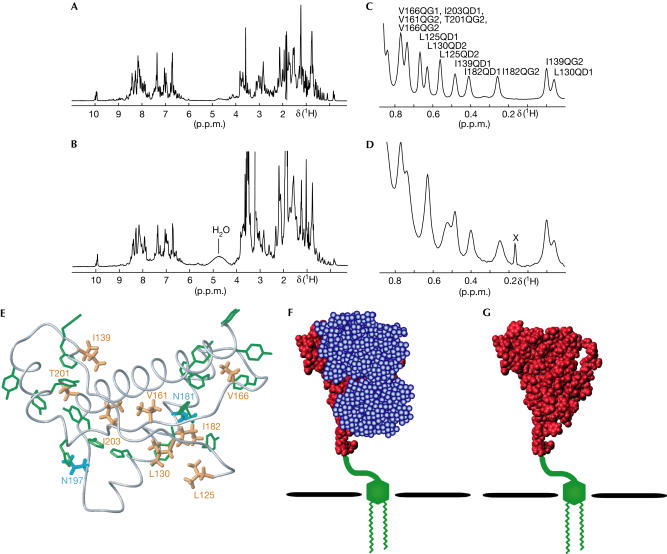
Figure 3.
Tertiary structure characterization with 1H-NMR at 900 MHz. (A) 1D 1H-NMR spectrum of recombinant bPrP(23–230). (B) 1D 1H-NMR spectrum of natural bPrPC. (C,D) Expanded plots of the region 0.9 to −0.1 p.p.m. of the 1H-NMR spectra of (A) and (B), respectively. In (C), the peaks are labelled with the previously obtained resonance assignments (Lopez Garcia et al, 2000). In (D), a spurious peak is marked with the label X. (E) Location of the methyl groups with high fieldshifted resonance lines in the NMR structure of recombinant bPrP(23–230) (Lopez Garcia et al, 2000). The polypeptide backbone of the C-terminal domain of bPrP(23–230) is represented by a grey spline function through the α-carbon positions. The side chains with high fieldshifted methyl 1H-NMR lines are shown in orange colour, and the aromatic residues that cause the high field shifts of these methyl groups are shown in green. The two N-glycosylation sites are coloured in cyan. (F,G) Space-filling models of the C-terminal globular domain of residues 125–230 on the basis of the NMR structure of recombinant bPrP(23–230). (F) bPrPC with the two glycosylation moieties in blue, the GPI anchor in green and the polypeptide chain in red. (G) bPrPC without the two glycosylation moieties.
Secondary structure and stability of natural bPrPC
Figure 2A compares the far-UV circular dichroism (CD) spectra of natural bPrPC and recombinant bPrP(23–230), which has the full length of the mature protein. The two spectra are nearly identical, and both show the typical shape for α-helical secondary structure, with two minima at 208 and 222 nm. In the wavelength range considered, there are no significant contributions to the CD spectrum either from carbohydrate moieties or from the GPI anchor, and hence Fig 2A shows that there could be at most small local differences between the secondary structures in the two proteins.
As several previous studies led to the hypothesis that the glycosylation of PrPC could modulate the conversion of PrPC into PrPSc by conferring increased stability on the structure of PrPC and thereby reducing the efficiency of conversion (Taraboulos et al, 1990; Kocisko et al, 1994; Lehmann & Harris, 1997; Zuegg & Gready, 2000), we further measured thermal unfolding transitions using the far-UV CD signal at 222 nm (Fig 2B). The thermal unfolding transitions of natural bPrPC and recombinant bPrP(23–230) have closely similar sigmoidal shapes, representing cooperative unfolding with apparent melting temperatures of 60.6°C for natural bPrPC and 61.0°C for recombinant bPrP(23–230). This demonstrates that glycosylation of natural bPrPC has no significant effect on the thermal stability of the PrPC polypeptide fold at the slightly acidic pH of 4.5 used here, which is similar to the pH value in endosomes, where the transformation to PrPSc presumably takes place (Borchelt et al, 1992; Arnold et al, 1995).
Tertiary structure of natural bPrPC
Homonuclear 1H-NMR spectroscopy was used for a comparison of the tertiary structures of recombinant bPrP(23–230) and natural bPrPC, as bPrPC isolated from calf brains is available only with natural isotope distribution. One-dimensional (1D) 1H-NMR spectra of the two proteins (Fig 3A,B) were recorded on a 900 MHz spectrometer at pH 4.5 in a mixed solvent of 90% H2O/10% D2O. Overall, the two spectra have many features in common, and obvious differences can readily be rationalized. Thus, the increased linewidths in the spectrum of natural bPrPC (Fig 3B) when compared with the unglycosylated recombinant prion protein (Fig 3A) result from its higher molecular mass, and the additional intense lines between 1 and 4 p.p.m. in Fig 3B are due to the glycans in the natural bPrPC. The visual impression of extensive similarities between the two spectra of Fig 3A,B is substantiated by closer examination of the wellseparated peripheral chemical shift regions. The N-terminal flexible tail in recombinant bPrP(23–230) contributes indole N–H resonance lines of eight tryptophan residues near 10.2 p.p.m. (López García et al, 2000; Fig 3A). A corresponding line pattern is visible in the spectrum of natural bPrPC (Fig 3B), showing that the flexible N-terminal tail is also present in natural bPrPC.
In the region between 0.9 and −0.1 p.p.m., the 1H-NMR spectrum of recombinant bPrP(23–230) (Fig 3C) shows resonance lines that have been assigned to ring-currentshifted methyl groups in the globular C-terminal domain (López García et al, 2000). These methyl groups are distributed throughout the core of the domain (Fig 3E), and even subtle local structural rearrangements in the C-terminal domain would result in significant shifts of some of these methyl resonance lines (Wüthrich, 1986). Comparison of the ring-currentshifted methyl resonance lines in recombinant bPrP(23–230) (Fig 3C) and natural bPrPC (Fig 3D) shows a 1:1 coincidence if one allows for small shifts for L125 and L130 between the two proteins. Furthermore, line broadening observed for residue I182 could be attributed to direct steric influence of the oligosaccharide moiety at the adjacent Asn 181. We also recorded two-dimensional (2D) homonuclear correlation and nuclear Overhauser enhancement (NOE) spectra of the natural bPrPC sample of Fig 3B,D. However, because of the low protein concentration and the correspondingly poor signal-to-noise ratio, no additional information could be derived from the 2D 1H-NMR spectra.
Conclusions
In summary, the CD data on the regular secondary structures as well as the comparison of ring-current-shifted methyl groups in recombinant bPrP(23–230) and in natural bPrPC show that the 3D structure of the C-terminal protein domain of residues 125–230 is maintained in the two proteins, and that the post-translational modifications hardly affect the 3D structure and the thermal stability of the cellular prion protein. In view of the data in Fig 2, it seems difficult to maintain the previously mentioned hypothesis that under-glycosylation of PrPC could destabilize the cellular prion protein so as to facilitate the conversion into PrPSc (Taraboulos et al, 1990; Kocisko et al, 1994; Lehmann & Harris, 1997; Zuegg & Gready, 2000). Other possible functions of the glycosylation might thus be more important. For example, because of the dynamics and plasticity of the oligosaccharides near the N-glycosidic linkages, these sugar moieties can be expected to protect extensive regions of the protein surface from intermolecular contacts (Fig 3F,G). Furthermore, as our results exclude the possibility that principal conformational changes of PrPC are induced by the post-translational modifications, it could be that the structure of PrPSc is actually more sensitive to the post-translational modifications. This would be in line with the finding that differently glycosylated forms of PrPSc might be used as biological markers for the classification of different prion strains (Collinge et al, 1996). Considering that continued work on the structural biology of prion diseases will necessarily have to rely primarily on experiments with recombinant prion proteins, the present data on the effects of the post-translational modifications on structure and stability of PrPC present an important foundation for the validation and interpretation of new results on PrP structure and function in health and disease.
Methods
Expression and purification of Bacillus cereus PI-PLC in E. coli B. cereus PI-PLC was expressed and purified as previously described (Ryan et al, 1996).
Brain tissues Calf brains (200–400 g each) obtained from the slaughterhouse were stored at −80°C until use.
Isolation and purification of natural bPrPC Brain homogenates (20–30%, w/v) were prepared in buffer A (0.32 M sucrose, 20 mM Tris–HCl and 5 mM EDTA at pH 7.5) using an Ultra Turrax T18 tissue homogenizer. The homogenates were centrifuged at 3,000g for 10 min, homogenized again in buffer A and centrifuged at 3,000g for 10 min. To obtain a membrane fraction, the supernatants were combined and centrifuged at 100,000g for 45 min. The membrane fraction was suspended in 150 ml of 20 mM Tris–HCl with 5 mM EDTA at pH 7.5, and incubated for 2 h at 37°C with 10–20 U/ml recombinant B. cereus PI-PLC to release the GPI-anchored membrane proteins. The fraction was then diluted with one volume of 20 mM Tris–HCl with 5 mM EDTA at pH 7.5, and centrifuged at 100,000g for 45 min to separate the PI-PLC-released proteins from the insoluble membrane fraction. The pellet was resuspended once to repeat the separation procedure. The supernatants of PI-PLC-released proteins from three calf brains were pooled and subjected to 45% ammonium sulphate precipitation. The pellet was resuspended in 100 ml of buffer B (10 mM 3-(N-morpholino)-propanesulphonic acid (MOPS), 1 mM phenylmethylsulphonyl fluoride (PMSF), 10 μg/ml leupeptin and 10 μg/ml aprotinin at pH 7.5). This suspension was centrifuged and then applied to an SP-Sepharose cation exchange column (10 ml) that had been equilibrated with buffer B, from which it was eluted with a linear gradient (150 ml) from 200 to 800 mM NaCl in buffer B. Fractions containing natural bPrPC were combined and applied to three coupled 1 ml HiTrap Co2+-immobilized metal affinity columns (Amersham Biosciences, Dusendorf, Germany) equilibrated with buffer C (10 mM MOPS, 150 mM NaCl, 20 mM imidazole, 1 mM PMSF, 10 μg/ml leupeptin and 10 μg/ml aprotinin at pH 7.5). The protein was eluted with a linear gradient (90 ml) from 20 to 120 mM imidazole in buffer C. Fractions containing natural bPrPC of 30 calf brains were combined, concentrated using Centricon YM-3 (Millipore, Bedford, MA, USA), injected in 150 μl portions to a tandem Superose12 column (24 ml each; Amersham) and eluted with 50 mM sodium phosphate at pH 7.0. In this purification step, the diglycosylated form of bPrPC was mainly recovered, whereas un- and monoglycosylated bPrPC seemed to remain on the column. Homogenous bPrPC fractions were pooled, concentrated using Centricon-3 and washed with distilled water to remove the buffer.
On average, 10–20 μg of bPrPC was obtained from each calf brain. The purity of the natural bPrPC sample was >93%, as judged by densitometry of a silver-stained SDS–12% (w/v) polyacrylamide gel. The correct N-terminal sequence of the bPrPC preparation was verified by Edmann sequencing, and a mass distribution with a principal peak of 31,962 Da was determined by MALDI mass spectrometry. Digestion of natural bPrPC with peptide-N-glycosidase F (PNGaseF) resulted in a single band that migrated with a higher molecular mass than the recombinant protein on an SDS–12% (w/v) polyacrylamide gel (data not shown). This size corresponds to the molecular mass of unglycosylated bPrPC containing the soluble part of the GPI anchor, thus confirming the aforementioned composition of our final product.
Expression and purification of recombinant full-length bPrP(23–230) The recombinant full-length bPrP(23–230) was purified as described elsewhere (Zahn et al, 1997).
Protein concentrations The total protein content was determined by the method of Bradford (BioQuant, Merck, Darmstadt, Germany) using bovine serum albumin as standard (Bradford, 1976). Protein concentrations in the final samples and in the samples for the CD and NMR measurements were measured using a molar extinction coefficient ɛ280=61,000 M−1 cm−1 for recombinant bPrP(23–230) and natural bPrPC (Gill & von Hippel, 1989).
Circular dichroism measurements Far-UV CD spectra and thermal transitions were recorded on a JASCO-710 spectropolarimeter at protein concentrations of 3.7–6.4 μM in 20 mM sodium acetate, pH 4.5, in a 0.1 cm cuvette. The far-UV CD spectra were performed at 20°C. For the thermal transitions, the samples were heated from 20 to 85°C at a constant heating rate of 1°C/min in a 0.1 cm cuvette, and the mean residue ellipticity was recorded at 222 nm. The data were normalized and corrected for the pre- and post-transitional baselines.
NMR measurements 1H-NMR spectra of natural bPrPC and recombinant bPrP(23–230) were recorded on a Bruker AVANCE DRX 900 spectrometer at a 1H-frequency of 900 MHz with protein concentrations of 14.3 μM for the natural bPrPC and 116 μM for the recombinant bPrP(23–230) in a mixed solvent of 90% H2O/10% D2O, pH 4.5, at 20°C.
Acknowledgments
We thank Ms Y. Auchli for technical assistance, Dr R. Brunisholz from the Protein Service Laboratory for N-terminal protein sequencing and MALDI mass spectrometry, and Dr M. Ryan and Dr O.H. Griffith for providing us with the expression system for B. cereus PI-PLC. This work was supported by the Schweizerischer Nationalfonds and the ETH Zürich through the National Center of Competence in Research (NCCR) Structural Biology.
References
- Alper T, Cramp WA, Haig DA, Clarke MC (1967) Does the agent of scrapie replicate without nucleic acid? Nature 214: 764–766 [DOI] [PubMed] [Google Scholar]
- Arnold JE, Tipler C, Laszlo L, Hope J, Landon M, Mayer RJ (1995) The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. J Pathol 176: 403–411 [DOI] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bolton DC, Meyer RK, Prusiner SB (1985) Scrapie PrP 27–30 is a sialoglycoprotein. J Virol 53: 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Taraboulos A, Prusiner SB (1992) Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J Biol Chem 267: 16188–16199 [PubMed] [Google Scholar]
- Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C (1993) Mice devoid of PrP are resistant to scrapie. Cell 73: 1339–1347 [DOI] [PubMed] [Google Scholar]
- Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS (1991) Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry 30: 7672–7680 [DOI] [PubMed] [Google Scholar]
- Collinge J, Sidle KC, Meads J, Ironside J, Hill AF (1996) Molecular analysis of prion strain variation and the aetiology of ‘new variant' CJD. Nature 383: 685–690 [DOI] [PubMed] [Google Scholar]
- Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc Natl Acad Sci USA 94: 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Griffith JS (1967) Self-replication and scrapie. Nature 215: 1043–1044 [DOI] [PubMed] [Google Scholar]
- Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, Caughey B (1994) Cell-free formation of protease-resistant prion protein. Nature 370: 471–474 [DOI] [PubMed] [Google Scholar]
- Lehmann S, Harris DA (1997) Blockade of glycosylation promotes acquisition of scrapie-like properties by the prion protein in cultured cells. J Biol Chem 272: 21479–21487 [DOI] [PubMed] [Google Scholar]
- López García F, Zahn R, Riek R, Wüthrich K (2000) NMR structure of the bovine prion protein. Proc Natl Acad Sci USA 97: 8334–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JT, Inouye H, Baldwin MA, Fletterick RJ, Cohen FE, Prusiner SB, Kirschner DA (1995) X-ray diffraction of scrapie prion rods and PrP peptides. J Mol Biol 252: 412–422 [DOI] [PubMed] [Google Scholar]
- Pan KM, Stahl N, Prusiner SB (1992) Purification and properties of the cellular prion protein from Syrian hamster brain. Protein Sci 1: 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergami P, Jaffe H, Safar J (1996) Semipreparative chromatographic method to purify the normal cellular isoform of the prion protein in nondenatured form. Anal Biochem 236: 63–73 [DOI] [PubMed] [Google Scholar]
- Pergami P, Bramanti E, Ascoli GA (1999) Structural dependence of the cellular isoform of prion protein on solvent: spectroscopic characterization of an intermediate conformation. Biochem Biophys Res Commun 264: 972–978 [DOI] [PubMed] [Google Scholar]
- Prusiner SB (1991) Molecular biology of prion diseases. Science 252: 1515–1522 [DOI] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K (1996) NMR structure of the mouse prion protein domain PrP(121–231). Nature 382: 180–182 [DOI] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K (1997) NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett 413: 282–288 [DOI] [PubMed] [Google Scholar]
- Ryan M, Smith MP, Vinod TK, Lau WL, Keana JF, Griffith OH (1996) Synthesis, structure–activity relationships, and the effect of polyethylene glycol on inhibitors of phosphatidylinositolspecific phospholipase C from Bacillus cereus. J Med Chem 39: 4366–4376 [DOI] [PubMed] [Google Scholar]
- Stahl N, Borchelt DR, Prusiner SB (1990) Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositolspecific phospholipase C. Biochemistry 29: 5405–5412 [DOI] [PubMed] [Google Scholar]
- Taraboulos A, Rogers M, Borchelt DR, McKinley MP, Scott M, Serban D, Prusiner SB (1990) Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc Natl Acad Sci USA 87: 8262–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, Cohen FE, Agard DA, Prusiner SB (2002) Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci USA 99: 3563–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K (1986) NMR of Proteins and Nucleic Acids, pp 30–31. New York, NY: Wiley [Google Scholar]
- Zahn R, von Schroetter C, Wüthrich K (1997) Human prion proteins expressed in Escherichia coli and purified by high-affinity column refolding. FEBS Lett 417: 400–404 [DOI] [PubMed] [Google Scholar]
- Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, Lopez Garcia F, Billeter M, Calzolai L, Wider G, Wüthrich K (2000) NMR solution structure of the human prion protein. Proc Natl Acad Sci USA 97: 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuegg J, Gready JE (2000) Molecular dynamics simulation of human prion protein including both N-linked oligosaccharides and the GPI anchor. Glycobiology 10: 959–974 [DOI] [PubMed] [Google Scholar]