Abstract
Dissolved free and combined N-acetyl-d-glucosamine (NAG) is among the largest pools of amino sugars in the ocean. NAG is a main structural component in chitin and a substantial constituent of bacterial peptidoglycan and lipopolysaccharides. We studied the distribution and kinetics of NAG uptake by the phosphoenolpyruvate:NAG phosphotransferase systems (PTS) in marine bacterial isolates and natural bacterial assemblages in near-shore waters. Of 78 bacterial isolates examined, 60 took up 3H-NAG, while 18 showed no uptake. No systematic pattern in NAG uptake capability relative to phylogenetic affiliation was found, except that all isolates within Vibrionaceae took up NAG. Among 12 isolates, some showed large differences in the relationship between polymer hydrolysis (measured as chitobiase activity) and uptake of the NAG, the hydrolysis product. Pool turnover time and estimated maximum ambient concentration of dissolved NAG in samples off Scripps Pier (La Jolla, Calif.) were 5.9 ± 3.0 days (n = 10) and 5.2 ± 0.9 nM (n = 3), respectively. Carbohydrate competition experiments indicated that glucose, glucosamine, mannose, and fructose were taken up by the same system as NAG. Sensitivity to the antibiotic and NAG structural analog streptozotocin (STZ) was developed into a culture-independent approach, which demonstrated that approximately one-third of bacteria in natural marine assemblages that were synthesizing DNA took up NAG. Isolates possessing a NAG PTS system were found to be predominantly facultative anaerobes. These results suggest the hypothesis that a substantial fraction of bacteria in natural pelagic assemblages are facultative anaerobes. The adaptive value of fermentative metabolism in the pelagic environment is potentially significant, e.g., to bacteria colonizing microenvironments such as marine snow that may experience periodic O2-limitation.
In the ocean, as on land, N-acetylglucosamine (NAG) is a major component of structural polymers in bacteria, plants, and animals. Chitin, a homopolymer of NAG, is a structural material in many marine invertebrates (e.g., cuttlefish, crab, and lobster), fungi, and algae (especially some diatoms [24]). Many marine bacteria depolymerize chitin (72), with cell surface hydrolases (e.g., see reference 15 and references therein), to NAG. Bacteria, both gram-positive and gram-negative, contain NAG as a main constituent of their cell wall peptidoglycan. Peptidoglycan depolymerization to liberate NAG has not been studied; however, since peptidoglycan is a major source of the pool of dissolved nitrogenous material in the ocean (42), it is probably a large-scale process commensurate with the scale of bacterial growth dynamics in the ocean (17). Thus, NAG flux through the marine dissolved organic matter may be important for bacterial growth and carbon and nitrogen cycling.
Since NAG is potentially a good energy and nitrogen source, one might hypothesize that NAG uptake is a widespread phenotype among marine bacteria. There are only few reports on bacterial uptake of NAG in seawater. Combining in situ hybridization with microautoradiography, Cottrell and Kirchman (14) found that NAG was preferentially taken up by α-Proteobacteria and by members of the Cytophaga-Flavobacterium cluster. The mechanism of NAG transport has been studied in only a few bacteria, mainly nonmarine, e.g., Escherichia coli (e.g., see references 49 and 68), Bacillus subtilis (19, 44), Staphylococcus aureus (29), and Vibrio furnissii (7). In E. coli, NAG is transported via a NAG-specific phosphoenolpyruvate:carbohydrate phosphotransferase system (PTSNAG) and also by a system with highest affinity for mannose (PTSMan) (50, 51). These NAG-PTSs result in NAG phosphorylation during trans-membrane transit. In the cytosol, phosphorylated NAG (NAG-P) may be deacetylated and deaminated to acetate, ammonia, and fructose-6-P (7), or it may directly enter cell wall peptidoglycan biosynthesis (9). For example, Mobley et al. (44) found for B. subtilis that up to ∼90% of the NAG taken up was converted to cell wall precursors and incorporated, but it could alternatively be respired when offered as the sole carbon source. Some spirochetes (12) and E. coli (71) and B. subtilis (19) mutants that cannot synthesize NAG require it for growth. Thus, mechanistic studies of NAG transport may yield insights on the in situ physiology of marine bacteria.
PTSs occur mainly among obligate and facultative anaerobes and only rarely among strict aerobes (last reviewed in reference 56). Since oxygenated seawater constitutes the largest biome on Earth, it is of interest to know whether PTSs are rare among bacteria in oxygenated seawater (e.g., restricted to anoxic microzones). Currently, there is little information to address this question. Most marine bacteria have resisted attempts to bring them into pure cultures, which is generally considered a prerequisite for studying specific biochemical mechanisms. Consequently, we know almost nothing about the distribution of PTSs in uncultured bacteria. In fact to our knowledge, information on NAG-PTSs even in cultured marine bacteria is limited to the Vibrionaceae (7).
Our objective was to determine the distribution of NAG-PTSs among cultured marine bacteria as well as uncultured marine bacteria in coastal waters. Further, we studied the kinetic characteristics of the NAG-PTS in cultured bacteria in order to place their PTSs in the context of NAG pool turnover in the ocean.
Sensitivity to the antibiotic and NAG structural analog streptozotocin (STZ) was developed into an individual-cell, culture-independent approach to detect bacteria expressing NAG-PTS in natural marine assemblages. We further tested whether bacterial isolates expressing NAG-PTSs also expressed chitobiase (thus suggesting coupling between NAG production and uptake).
MATERIALS AND METHODS
Screening for NAG uptake and STZ sensitivity.
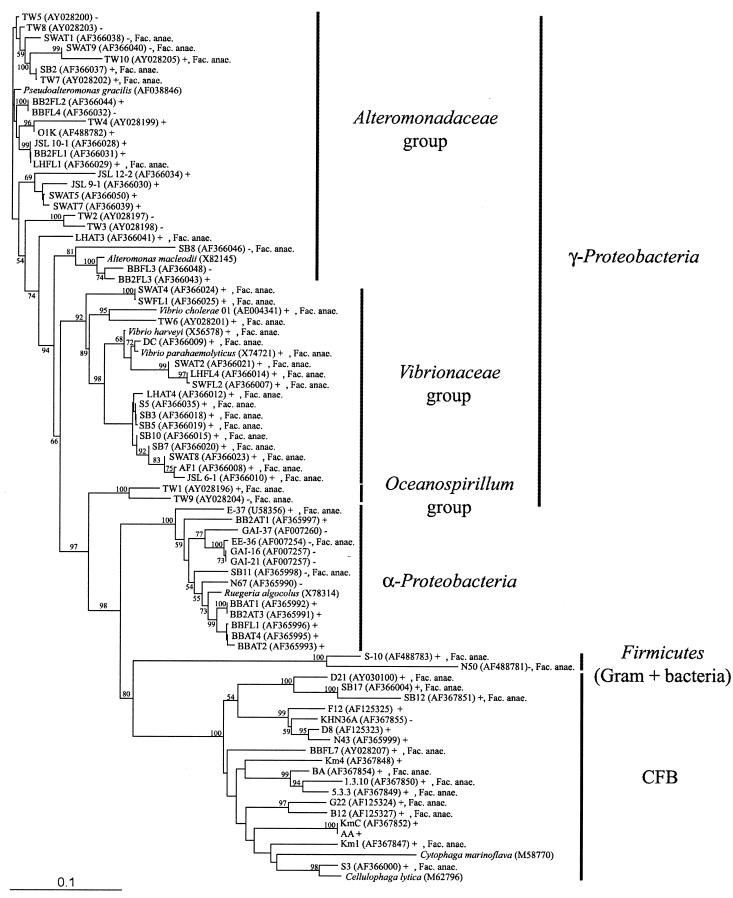
Seventy-eight marine bacterial isolates (two gram-positive isolates, 13 isolates from the α-Proteobacteria, 18 members of the Cytophaga-Flavobacterium-Bacteroides [CFB] cluster, and 45 isolates from the γ-Proteobacteria; see Fig. 1) were screened for uptake of N-acetyl-d-[1-3H]glucosamine (Amersham Pharmacia Biotech). Of these, 70 had been isolated from different marine waters by our laboratory over the last 20 years, and 5 were isolated by Moran and coworkers off the Georgia coast (22, 23). In addition, three Vibrio strains were used: Vibrio cholerae strain N16961 (40) (from Ron Taylor, Dartmouth University) and Vibrio parahaemolyticus strain BB22 and Vibrio harveyi strain BB120 (8, 9) (both from Michael Silverman, Agouron Institute).
FIG. 1.
Phylogenetic tree showing relationships of the analyzed isolates to representative bacterial 16S rRNA genes obtained from GenBank. The tree was inferred by Clustal W and the neighbor-joining method of on average approximately 400 bp. The number of bootstrap replicates supporting the branching order, from a total of 1,000 replicates, is shown at the segments. Only values of >50% are shown. Symbols and abbreviation: +, NAG uptake, complete or partial inhibition by STZ; −, no NAG uptake, no effect of STZ; Fac. anae., facultative anaerobes. GenBank accession numbers are given after the isolate names. All isolates were isolated from off Scripps Pier except for isolates with the following prefixes: SB, Santa Barbara, California coast (64); JSL, Johnson Sea Link I Research Submersible, off Florida coast; GAI, E, and EE, Georgia coast (22, 23); V. cholerae N16961 (40), V. parahaemolyticus strain BB22 (9), and V. harveyi strain BB120 (8). Isolate AA was identical to MED 11 (AF02554). The scale bar indicates base pair substitutions per nucleotide position.
Isolate names and origins are given in Fig. 1. Isolates were grown overnight in modified ZoBell 2216E broth (5 g of peptone, 1 g of yeast extract in 0.8 liter of GF/C [Whatman]-filtered seawater and 0.2 liter of MilliQ water, autoclaved at 121°C for 30 min). Cultures were then diluted in ZoBell broth and NAG (100 nM [final concentration]) was added to induce the PTS (44, 49, 68). After ∼2 h the exponentially growing cells were washed twice in artificial seawater (ASW) (66) by centrifugation (10,000 × g, 5 min, 20°C) and left in ASW for 1 to 2 h. In this screening, culture density was not quantified before the uptake assay; however, all cultures used were dense. Samples were incubated for 1 to 2 min with a mixture of 3H-labeled and unlabeled NAG (100 nM final) at 19 to 20°C and incubation terminated by filtration of 100-μl subsamples onto 0.45-μm-pore-size HA membrane filters (Millipore) followed immediately by two rinses with ice-cold sterile ASW. Formalin-fixed (2% final) controls were prepared for all experimental samples. Filters were dissolved in 1 ml of ethyl acetate and radioassayed after adding 10 ml of Ultima Gold (Fisher) liquid scintillation cocktail. Isolates that did not differ from the controls were grouped in the “no uptake” group, while isolates showing uptake (2 to 70 times as high as control) were grouped in the “uptake” group.
In order to develop the rationale for using STZ sensitivity as the basis for estimating NAG uptake in a natural bacterial assemblage, we tested whether all isolates capable of taking up NAG were sensitive to STZ. This compound is an N-methyl-N-nitrosourea derivative of N-acetyl-d-glucosamine produced by Streptomyces achromogenes (28). Its mode of action is at the level of DNA structure and/or synthesis (52) and may be due to the intracellular conversion of accumulated STZ to diazomethane, a strong alkylating agent affecting primarily replicating DNA (38). STZ (Sigma) was dissolved in MilliQ water (5 mg ml−1, pH ≈ 5.3), sterile filtered, and stored at −20°C for <1 to 2 weeks before use. To test for STZ sensitivity, isolates (in triplicate) were grown overnight in ZoBell broth with or without STZ (50 μg ml−1). Isolates with no visible growth were scored as highly sensitive. Isolates showing growth were further tested by measuring the inhibition of short-term [3H]thymidine incorporation by STZ. This was because the observed growth (overnight) could have been due to the presence of STZ-resistant mutants in the inoculum (27) and/or the lack of sufficient STZ activity due to the rapid STZ degradation at neutral pH (our observations and reference 65). The isolates were grown overnight in a seawater medium (Bacto Peptone, 0.010 g; Casamino Acids, 0.058 g; glucose, 0.125 g; NH4Cl, 0.30 mM; NaH2PO4, 0.25 mM; ferric citrate, 0.20 μM; EDTA, 0.20 mM; seawater, 1.0 liter; G. F. Steward and F. Azam, unpublished data), induced, and washed as described above. Then they were incubated with and without STZ (50 μg ml−1) for 30 min before aliquots for [3H]thymidine incorporation measurements were taken (below).
Concentration dependence of NAG uptake.
Uptake velocity for NAG was further studied for 10 of the 78 isolates (above) representing three phylogenetic groups. Following growth and induction, as described, they were counted microscopically and triplicate samples of 1 × 108 to 5 × 108 cells ml−1 were incubated with 3H-NAG at seven different concentrations (1 to 100 nM) for 1 min before filtration and radioassay (above). Samples for subsequent cell counts were fixed at several time points during the course of the experiments to correct for any growth.
Chitobiase activity.
We sought to elucidate a possible relationship between chitin hydrolysis (measured as chitobiase activity) and NAG uptake in marine isolates. A 5% (wt/vol) slurry of chitin (90% purified crustacean shells and 10% γ-chitin; both from Calbiochem, La Jolla, Calif.) in MilliQ water was autoclaved and mixed with ZoBell broth containing NAG (100 nM) to a final chitin concentration of ∼1%. Twelve isolates were grown overnight in this broth to ensure induction of chitobiase and PTS. Then, fresh medium was added to obtain exponential growth. After ∼2 h, cells were washed once by centrifugation and aliquots fixed for cell counts. For chitobiase activity measurements, the cultures were diluted 10-fold and aliquoted into a 96-well white microtiter plate (Falcon). Five replicate samples and three controls (medium without cells) for each isolate were incubated at room temperature in the dark with the fluorogenic substrate 4-methylumbelliferyl N-acetyl-β-d-glucosaminide (100 nM; Sigma). Fluorescence was measured using an HTS 7000 series BioAssay microtiter plate fluorometer (excitation wavelength = 360 nm; emission wavelength = 465 nm; Perkin-Elmer, Norwalk, Conn.). Fluorometer measurements were calibrated with 4-methylumbelliferone. The NAG uptake was measured in triplicate aliquots of undiluted cultures in 1-min incubations with 3H-NAG (100 nM) as above.
PTS activity in isolates.
PTS activity was measured as phosphoenolpyruvate (PEP)-dependent phosphorylation of 3H-NAG (35; modified by M. H. Saier, Jr. [personal communication]). Cells were grown overnight and treated as prior to the screening for NAG uptake. Two milliliters of culture was lysed in a cooled French press and kept on ice. The lysed culture (100 μl) was mixed with 100 μl of assay mix yielding the following final concentrations: 25 mM KF, 12.5 mM MgCl2, 2.5 mM dl-dithiothreitol (Sigma), 25 mM phosphate buffer (pH 7.4), and 1.0 μM 3H-NAG and with and without a phosphate donor (5 mM PEP, pH 6 to 8; Sigma). The mixture was incubated for 20 min at 19 to 20°C and then applied to columns (0.7 by ∼7 to 8 cm) of AG 1-X8 resin, 20-50 mesh (Bio-Rad), which had been washed with 30 ml of 1 M NaCl followed by 60 ml of MilliQ water. After sample addition, the column was washed with 70 ml MilliQ water and phosphorylated NAG was eluded from the column with 20 ml 1 M LiCl. One milliliter of eluates from assays with and without PEP was radioassayed in 10 ml of Ultima Gold and compared. Isolates incapable of NAG uptake and mixes with no added cell extracts served as negative controls.
Mixed acid fermentation among the isolates.
The presence of facultative anaerobic isolates capable of mixed acid fermentation of mannitol or glucose was determined on a seawater-agar medium containing the pH indicator phenol red (10 g of peptone, 1 g of beef extract, 10 g of mannitol or glucose, 0.025 g of phenol red, and 15 g of agar in 1 liter of 80:20 seawater-MilliQ water). As anoxic conditions developed inside single plate colonies, mixed acid fermentation could be seen as a pronounced plate color change (red to bright yellow). One to two isolates were used per plate.
Field studies:
Inhibition of bacterial [3H]thymidine and single-cell bromodeoxyuridine (BrdU) incorporation by STZ. Screening the isolates, we established that bacteria taking up NAG were killed or severely inhibited by STZ while bacteria with no NAG uptake were unaffected. By comparing [3H]thymidine and BrdU incorporation in seawater samples with and without added STZ, we would get a means of estimating the proportion of bacterial production due to those bacteria expressing NAG-PTS. On 24 and 29 May and 1 June 2001, we measured bacterial abundance and the effect of STZ on [3H]thymidine incorporation in samples obtained from the upper 0.5-m depth off Scripps Pier (32°53′N, 117°15′W). Incubations were initiated within 10 min of sampling at in situ temperature ±1°C. On May 24 and June 1, the effect on BrdU incorporation was also measured. Triplicate samples were incubated with and without freshly made STZ (100 μg ml−1) for 1 h before the addition of [3H]thymidine (20 nM, see below) or BrdU (20 μm). Triplicate samples (100 ml) were incubated with BrdU (Boehringer-Mannheim) for 5 h in the dark on a shaker. BrdU-incorporating cells were quantified by the method of Hamasaki and Azam (26; unpublished data). Cells were concentrated by centrifugation (15,000 × g, 10 min, 4°C), fixed in 2% paraformaldehyde, further concentrated by microcentrifugation (15,000 × g, 10 min, 4°C), resuspended in 50:50 100% ethanol and phosphate-buffered saline (PBS) buffer (135 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.0), and stored at 4°C. Concentrates were spotted onto glass slides and ethanol-dehydrated. Endogenous peroxidase was quenched with 3% H2O2 in PBS for 10 min, followed by two washes in PBS, fixation with 3 to 4% paraformaldehyde in PBS for 30 min and two washes in PBS. The cells were permeabilized with 0.01 N HCl for 5 min, treated with pepsin (2 mg ml−1) for 3 h at 37°C, washed 3 times with PBS, and treated with 0.1% (wt/vol) lysozyme (3 mg ml−1) in TE buffer (100 mM Tris-Cl, 50 mM EDTA, pH 8.0) for 15 min followed by three washes with PBS. Nucleic acids were denatured overnight with 1:100 nuclease in incubation buffer (BrdU labeling and detection kit III; Boehringer Mannheim) at 4°C and washed twice with PBS. BrdU was probed with a peroxidase (POD)-anti-BrdU antibody 1:500 in PBS containing 0.1% bovine serum albumin for 3 h in a humidity chamber followed by four washes with PBS. Signal amplification was obtained by adding biotinyl tyramide (1:50) in amplification diluent (TSA-INDIRECT [ISH] kit; Renaissance) for 10 min and three washes with TNT buffer (10 mM Tris-Cl, 150 mM NaCl, 0.05% Tween 20, pH 8.0) followed by incubation with streptavidin-fluorescein (1:500) in TNT buffer containing 0.5% bovine serum albumin for 30 min. After three washes in TNT buffer, the cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg ml−1), washed with PBS, and mounted with antifading solution (46). All incubations were at room temperature, except where indicated. Twenty fields for each of the triplicate samples were counted (corresponding to >300 DAPI- stained cells and >100 fluorescein-stained cells per sample). A sample incubated with nonradioactive thymidine instead of BrdU and an exponentially growing culture incubated with BrdU served as negative and positive controls, respectively.
NAG pool turnover and ambient concentration off Scripps Pier.
The turnover time of NAG (f/t = fraction of label taken up per time; turnover time = t/f) was estimated in samples taken off Scripps Pier 12 times during the period from 24 April to 4 June 2001. Sampling time was consistently during late afternoon. Bacterial abundance and [3H]leucine incorporation (to estimate bacterial production; below) were also measured at most samplings. Triplicate samples and duplicate formalin-killed controls were incubated with 3H-NAG (3 nM) for 1 h at in situ temperature ±1°C. Incubations were filtered and radioassayed as described above.
On three occasions (24 April, 21 and 23 May) the upper limit of the ambient concentration of NAG was estimated as Sn + Km (Sn is the ambient concentration, Km is a constant similar to the Michaelis constant, Km) by Wright-Hobbie linearization (70) of the Michaelis-Menten equation. Triplicate samples and duplicate formalin-killed controls were incubated with 3H-NAG (1, 3, 10, and 30 nM [final concentrations]) for 1 h. Incubations were filtered and radioassayed as described above.
Carbohydrate competition experiment.
The effect of 8 carbohydrates on the uptake of NAG was determined in a sample obtained off Scripps Pier on 26 May, 2001. Triplicate 5-ml aliquots with a mixture of labeled and unlabeled NAG (100 nM, specific activity 1 Ci mmol−1) and carbohydrates at 1, 10, or 1,000 times the concentration of the added NAG were incubated for 1 h at in situ temperature ±1°C. For each concentration, duplicate formalin-fixed samples served as controls. Six replicate samples and four formalin-fixed samples were used to determine the NAG uptake with no competing carbohydrates added. The sterile-filtered carbohydrate stock-solutions were made in distilled water at sufficiently high concentrations that the sample dilution was always ≤1%. Incubations were filtered and radioassayed as described above.
Bacterial abundance and production.
Formalin fixed (2% final) samples were stained with DAPI (1 μg ml−1) for 10 min and filtered onto black 0.2-μm-pore-size polycarbonate filters (Poretics). Within a few hours of sampling >200 bacteria filter−1 (or >20 fields filter−1) were counted at ×1,250 using epifluorescence microscopy (Olympus BH-2). To estimate variability among filters, triplicate sets of filters were made for a seawater sample and for a culture sample. Variability was on average 5.1%.
Bacterial production was measured by [3H]leucine (33) or [3H]thymidine incorporation (21), as modified for microcentrifugation by Smith and Azam (63). Triplicate 1.7-ml aliquots were incubated with l-[4, 5-3H]leucine or with [methyl-3H]thymidine (20 nM final concentration; DuPont NEN) in sterile 2.0-ml capacity polypropylene tubes for ca. 1 h at in situ temperature ±1°C. Samples with 5% trichloroacetic acid added prior to the addition of isotope served as blanks. Bacterial carbon production was calculated from leucine incorporation as in Simon and Azam (62). Thymidine incorporation was converted to carbon production using 1.8 × 1018 cells mol of thymidine incorporated−1 (20) and a carbon-to-cell ratio of 20 fg of C bacterium−1 (36).
RESULTS
NAG uptake in isolates.
Of the 78 isolates, 60 took up NAG while 18 did not show any measurable uptake (Fig. 1; Table 1).
TABLE 1.
Distribution of NAG uptake and facultative anaerobes in isolatesc
| Phylogeny (total no. in group) | Parameter | Uptake groupa (+) | No-uptake groupa (−) |
|---|---|---|---|
| Firmicutes (2) | NAG uptake | 1 | 1 |
| % Fac. anae.c | 100 | 100 | |
| CFB (18) | NAG uptake | 17 | 1 |
| % Fac. anae. | 65 | 0 | |
| α-Proteobacteria (13) | NAG uptake | 7 | 6 |
| % Fac. anae. | 14 | 33 | |
| Oceanospirillumb (2) | NAG uptake | 1 | 1 |
| % Fac. anae. | 100 | 100 | |
| Vibrionaceaeb (19) | NAG uptake | 19 | 0 |
| % Fac. anae. | 100 | ||
| Alteromonadaceaeb (24) | NAG uptake | 15 | 9 |
| % Fac. anae. | 33 | 33 | |
| Total (78) | NAG uptake | 60 | 18 |
| % Fac. anae. | 63 | 39 |
Groups defined on the basis of NAG uptake; see Materials and Methods.
γ-Proteobacteria.
For individual isolates, see Fig. 1. Fac. anae., facultative anaerobes.
STZ sensitivity in isolates.
Of the 60 isolates showing NAG uptake, 41 isolates had no visible growth overnight when STZ was present. For the remaining 19 isolates showing NAG uptake, STZ reduced thymidine incorporation by, on average, 58% ± 28%. Tests of a few of the isolates, which did not grow overnight in the presence of STZ, showed ∼100% inhibition of thymidine incorporation by STZ. All 18 isolates that did not take up NAG showed unchanged thymidine incorporation and were unaffected by STZ.
Facultative anaerobes among isolates.
Of the 60 isolates that took up NAG and were completely or partly inhibited by STZ, 38 (63%) were facultative anaerobes. Seven (39%) of the 18 isolates that were unaffected by STZ were also facultative anaerobes (Fig. 1; Table 1). Isolates affiliated with different phylogenetic groups varied in their ability to take up NAG or to ferment glucose or mannitol. Notably, all isolates within the Vibrionaceae group took up NAG, their growth was completely inhibited by STZ, and they were all facultative anaerobes (Fig. 1; Table 1).
NAG uptake, chitobiase activity, and PTS in selected isolates.
NAG uptake was not significantly correlated to chitobiase activity (r2 = 0.12); however, isolates with low NAG uptake rates consistently showed low chitobiase activities (Table 2). Phosphorylation of NAG during uptake was tested for eight isolates known to take up NAG and for two isolates not taking up NAG. All eight converted NAG to NAG-P while no phosphorylation was found for the two negative controls (Table 2).
TABLE 2.
PTS, NAG uptake, and chitobiase activity for bacterial isolates
| Isolatea | PTSb | NAG uptake (amol cell−1 h−1) | Chitobiase activity (amol cell−1 h−1) | Chitobiase/NAG uptake |
|---|---|---|---|---|
| BB2AT1 (α) | NDc | 7.3 | 14 | 2 |
| BBFL1 (α) | + | 23.8 | 4,355 | 183 |
| Km1 (CFB) | + | 12.0 | 2,553 | 213 |
| F3 (CFB) | ND | (0.03) | 29 | 967 |
| TW6 (γ) | + | 52.5 | 4,134 | 79 |
| LHAT4 (γ) | + | 23.4 | 6,950 | 297 |
| S-5 (γ) | + | 19.8 | 10,375 | 524 |
| SB3 (γ) | + | 14.0 | 15,849 | 1,132 |
| V. cholerae (γ) | + | 15.3 | 13,581 | 888 |
| JSL 12-2 (γ) | + | 0.5 | 487 | 974 |
| LHFL1 (γ) | − | 0.0 | 231 | |
| SWAT1 (γ) | − | (0.04) | 0 |
α, α-Proteobacteria; γ, γ-Proteobacteria; CFB, Cytophaga-Flavobacterium-Bacteroides.
PTS assays were performed in experiments separate from the NAG uptake at chitobiase activity experiment. Standard deviations were always below 10%.
ND, not determined.
Concentration-dependence of NAG uptake.
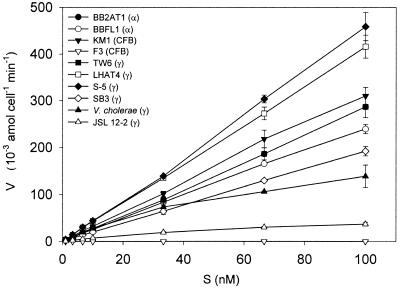
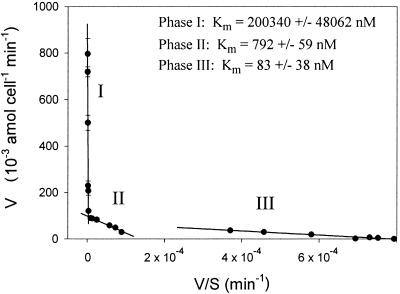
Uptake of NAG was highly variable for 10 analyzed isolates (Fig. 2). Little or no evidence for saturation kinetics precluded accurate determination of transport constants. Concentration dependence of uptake for a single isolate (JSL 12-2) was measured over a broad substrate range (1 nM to 1 mM). An Eadie-Hoftsee plot of the data indicates multiphasic uptake kinetics (Fig. 3).
FIG. 2.
Dependence of uptake rates on NAG concentration for 10 isolates. Error bars indicate SD.
FIG. 3.
Eadie-Hoftsee plot of NAG uptake data for isolate JSL 12-2 (γ-Proteobacteria) indicating the presence of multiphasic uptake systems. The half-saturation constants (Km ± standard error) were calculated by Lineweaver-Burk plots of the uptake data. The substrate range was 1 nM to 1 mM NAG. Error bars indicate standard deviations.
NAG uptake in natural assemblages off Scripps Pier; substrate specificity.
Inhibition of uptake by other carbohydrates was variable but indicated that glucosamine, mannose, glucose, and fructose affected NAG uptake (Table 3). However, only the inhibition by glucose, which inhibited the uptake of 3H-NAG to the same degree as unlabeled NAG, was consistent with competitive inhibition.
TABLE 3.
Inhibition of NAG uptake by various carbohydrates in samples from off Scripps Piera
| Carbohydrate | Concn (mM) | % of control (mean ± SD) |
|---|---|---|
| Control | No addition | 100 ± 8 |
| N-acetyl-glucosamine | 0.1 | 57 ± 29 |
| 1 | 0 ± 0 | |
| 100 | 0 ± 0 | |
| Glucosamine | 0.1 | 71 ± 3 |
| 1 | 66 ± 29 | |
| 100 | 59 ± 27 | |
| Mannose | 0.1 | 89 ± 7 |
| 1 | 89 ± 2 | |
| 100 | 45 ± 19 | |
| Glucose | 0.1 | 55 ± 10 |
| 1 | 0 ± 0 | |
| 100 | 0 ± 0 | |
| Fructose | 0.1 | 57 ± 23 |
| 1 | 82 ± 27 | |
| 100 | 45 ± 13 | |
| Mannitol | 0.1 | 85 ± 19 |
| 1 | 77 ± 17 | |
| 100 | 81 ± 13 | |
| Sucrose | 0.1 | 90 ± 7 |
| 1 | 90 ± 23 | |
| 100 | 68 ± 9 | |
| Lactose | 0.1 | 74 ± 6 |
| 1 | 97 ± 11 | |
| 100 | 76 ± 12 |
3H-NAG concentration was 0.1 mM. n = 3; control, n = 6.
NAG pool size and turnover.
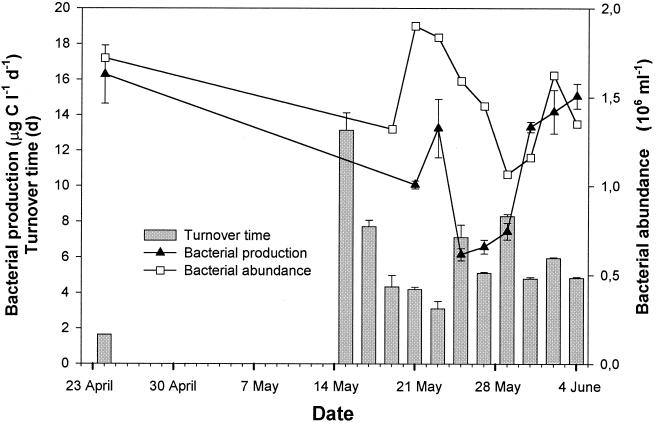
The NAG turnover time ranged from 1.6 to 13.1 days (average 5.9 ± 3.0 days; Fig. 4). The turnover time was negatively correlated to bacterial production (r2 = 0.46; n = 9) but only weakly negatively correlated to bacterial abundance (r2 = 0.30; n = 10). The ambient NAG concentration was unknown; however, an upper limit (Km + Sn) was estimated to average 5.2 ± 0.9 nM in three in situ uptake experiments.
FIG. 4.
Bacterial production and abundance, and turnover time of NAG (f/t = fraction of label taken up per time; turnover time = t/f) in samples from off Scripps Pier. The NAG concentration was 3 nM. Error bars indicate standard deviations.
STZ inhibition of bacterial growth in natural assemblages.
The effect of STZ on thymidine incorporation was examined on three occasions, yielding an average reduction of 28.4% (Table 4). The effect of STZ on BrdU incorporation was examined at the single-cell level. The BrdU-antibody-labeled cells as a percentage of DAPI stained cells in control samples were 31% ± 4% and 46% ± 1%. On both occasions, additions of STZ led to a decrease of 43% (Table 4).
TABLE 4.
Effects of STZ on thymidine and BrdU incorporation in samples from off Scripps Piera
| Date | Bacterial abundance (106 ml−1) | Thymidine incorporation
|
BrdU incorporation
|
||||
|---|---|---|---|---|---|---|---|
| μg of C liter−1 day−1
|
Reduction (%) | % of DAPI-stained cells
|
Reduction (%) | ||||
| Without STZ | With STZ | Without STZ | With STZ | ||||
| 24 May | 2.06 | 22.8 ± 1.2 | 15.7 ± 0.6 | 31 | 31 ± 4 | 18 ± 8 | 43 |
| 29 May | 1.11 | 16.5 ± 1.0 | 11.6 ± 0.9 | 30 | NDb | ND | ND |
| 1 June | 1.71 | 15.1 ± 0.8 | 11.5 ± 0.9 | 24 | 46 ± 1 | 26 ± 3 | 43 |
Values are means ± standard deviations.
ND, not determined.
DISCUSSION
Importance of studying transport systems in natural assemblages.
Transmembrane transport is a true measure of organic matter flux into marine bacteria. The substrate concentration gradients across the cell membrane being on the order of, e.g., 106 for amino acids (62), pelagic bacteria would be expected to have highly efficient, energy-dependent, concentrative transport systems. However, our knowledge of specific transport systems employed by the bacteria in the sea is rudimentary and largely extrapolated from research on nonmarine enteric bacteria or from limited work on marine isolates. What transport mechanisms are dominantly used by the diverse as-yet-uncultured phylotypes in the natural assemblages has been an intractable question. Yet this knowledge is essential for a mechanistic understanding of the adaptive strategies of bacteria in the ocean and how the ecosystem expression of bacterial adaptive strategies influences the oceanic carbon cycle. The present study demonstrates that concerted studies of isolates and natural assemblages can yield insights into the expression of specific transport phenotypes (the PTS in the present study) by the bacteria in marine ecosystems.
Broad phylogenetic distribution of NAG-PTS.
Studies to date have shown that NAG uptake in bacteria is consistently via a PTS. So, the ability to take up NAG might be equated with the presence of a PTS for NAG transport. However, this generalization has not been tested in marine bacteria. Therefore, we first tested this relationship, in 8 NAG-uptake-positive isolates (out of a total of 78 isolates that we examined for NAG uptake). All 8 isolates were positive for PEP-dependent phosphorylation of NAG in cell extracts, a standard criterion for the presence of the expression of a PTS. We then assumed that all isolates that took up NAG also expressed a NAG-PTS.
Of the 78 isolates examined, 77% took up NAG. They belonged to all major bacterial phyla represented (γ- and α-Proteobacteria, gram-positive bacteria, and members of the Cytophaga-Flavobacterium-Bacteroides cluster). Likewise, isolates negative for NAG uptake also belonged to all major phylogenetic groups within Bacteria. This finding is consistent with the latest compilation of bacteria for which the presence or absence of the PTS has been documented (56), which shows that there is no obvious pattern in PTS distribution among the major bacterial phyla. (However, PTSs are found preferentially among obligate and facultative anaerobes and only rarely among strict aerobes [16, 55, 56, 57, 59; discussed further below].) If our isolates were representative of the bacteria in the ocean, then a large majority of bacteria would be expected to have the genetic capability to express NAG-PTSs. This led us to ask whether indeed the expression of NAG-PTSs is widespread among the natural assemblages (using STZ sensitivity as an indicator; see below).
STZ sensitivity as an indicator of NAG-PTS-expressing cells.
STZ and NAG compete for the same transport systems (2, 30, 38, 39), both are phosphorylated during uptake, and (in E. coli) both are accumulated to similar concentration gradients (2). Once inside the cell, STZ-P kills the cell by degrading its DNA. Bacteria with mutations in the PTS operon (2, 30, 37) or those unable to synthesize or energize the PTS (39) are unaffected. Our test of the effect of STZ in marine bacteria led to the conclusion that STZ kills or severely inhibits bacteria expressing NAG uptake while bacteria negative for NAG uptake are unaffected. On the basis of this finding, we reasoned that STZ sensitivity of bacterial growth would be a valid indicator of NAG-PTS-expressing bacteria in the natural assemblages. Thus, we were able to address the question of what fraction of the growing (BrdU-positive) bacteria is expressing NAG-PTSs? A second, technically simpler yet biogeochemically interesting, question that we addressed was, what fraction of total bacterial production is due to those bacteria that express NAG-PTS?
Indeed, a substantial proportion of DNA-synthesizing bacteria in samples from Scripps Pier expressed a NAG-PTS. STZ inhibited [3H]thymidine incorporation by 28.4% ± 3.8% and BrdU incorporation at the single-cell level by 43% (Table 4). These percentages are lower than for our isolates where 77% took up NAG. There may be a bias in favor of NAG-PTS-positive isolates and/or we may have underestimated the BrdU-positive cells if those with low rates of incorporation had gone undetected.
While we did not test it, the STZ sensitivity approach might reasonably be combined with species-specific oligonucleotide probes to study PTSs in specific bacterial species. Finally, since STZ degrades the target bacteria DNA, one might be able to detect, and quantify in an assemblage, the STZ-sensitive bacteria simply by a modified DNA staining protocol (e.g., see reference 73).
Uptake kinetics.
The kinetic characteristics of bacterial transport systems reflect adaptations to nutrient-poor environments (Fig. 2). Our analysis of the concentration dependence of NAG influx indicated large differences in uptake affinities and capacities between bacteria. We used a low concentration range (1 to 100 nM). In this range most isolates showed no tendency towards uptake saturation indicating Km values much higher than 100 nM. The near-linear relationship between NAG concentration (S) and uptake velocity (V) indicates that these isolates would be highly responsive to elevated NAG levels in their environment. We recognize that linear S versus V plots could also indicate that the uptake was by simple Fickian diffusion across the membrane. However, based on the survey of 8 isolates, all of which utilized a PTS in NAG uptake, it seems likely that most bacteria taking up NAG use PTS rather than simple diffusive transport. Analysis of S versus V data over a broader range of S showed more complex kinetic patterns but with a tendency towards saturation. As exemplified in Fig. 3, some bacteria showed evidence of multiple or multiphasic uptake systems. Such kinetic response would enable bacteria to achieve enhancement of V over a broad range of environmental concentrations. This type of kinetic pattern has previously been described for d-glucose uptake by a marine isolate (45). Multiphasic transport kinetics might be a strategy for optimizing uptake along NAG concentration gradients. Such gradients are likely to exist since NAG is mainly produced by the action of hydrolases at particulate loci in seawater (as would be the case of any monomer released at a particulate locus [4]). Multiphasic kinetics would thus enable the bacteria interacting with a NAG source particle (e.g., a chitinous particle) to achieve a tight hydrolysis-uptake coupling (discussed further below).
Hydrolysis-uptake coupling.
Some isolates showed large differences in the relationship between polymer hydrolysis (measured as chitobiase activity) and uptake of NAG, the hydrolysis product. Most had a high ratio of chitobiase/uptake (from 183 to 1,132); however, there were some interesting exceptions. BB2AT1 and TW6 had high NAG uptake but low chitobiase activity while LHFL1 had undetectable NAG uptake but significant chitobiase activity (Table 1). We speculate that in natural bacterial assemblages some bacteria specialize in polymer hydrolysis but are less efficient in taking up hydrolysis products. Other species have low or no enzyme production and highly efficient monomer uptake and might depend on inefficient hydrolysis-uptake systems in other bacteria. Such cross-feeding has recently been demonstrated for a model community consisting of proteolytic and nonproteolytic Pseudomonas fluorescens (69). Another example is from a survey of V. parahaemolyticus-related isolates associated with copepods by Kaneko and Colwell (31). These authors defined two main clusters of isolates: group I-A was chitinolytic and could be important in the initial colonization of copepods; group II-A did not possess a chitinase but could utilize NAG and, thus, could be important in the remineralization of copepod detritus.
NAG pool turnover.
The NAG pool turnover time ranged from 1.6 to 13.1 days (average, 5.9 ± 3.0 days; Fig. 4). This range is longer than for d-glucose turnover in seawater in previous studies (6, 53). Since we measured NAG turnover rate only at one place and only a few times we cannot say whether NAG pool turnover is generally slower than d-glucose turnover. Further, induction of transport systems as well as utilization of NAG during the 1 -h incubation could potentially have affected the measured uptake.
There are no reports of NAG concentration in seawater. However, our measurements of (Km + Sn) provided an upper limit of 5.2 ± 0.9 nM on NAG concentration in our samples. The widespread expression of NAG-PTSs, and particularly the presence of high-affinity transport (above), would be consistent with (indeed predict) a tight control of NAG pool concentration to nanomolar levels in seawater. This situation is similar to bacterial control of excellent substrates (e.g., amino acids) at nanomolar levels and within narrow limits.
Substrate specificity.
In natural assemblages, total or partial inhibition of NAG uptake in seawater by glucose, glucosamine, mannose, and fructose suggested a common transport system(s). However, except for glucose, the measured inhibitive effects were not consistent with competitive inhibition. Potentially, the results have been biased by, e.g., induction of transport systems or substrate-induced growth during the incubation. Nevertheless, the effects indicate that PTSNAG, which is specific for NAG, is not the sole uptake system for NAG in natural assemblages. Glucose and NAG had roughly equal uptake affinities (unlabeled NAG and glucose inhibited 3H-NAG uptake equally). The use of a single system to transport multiple structurally similar substrates could be economical but may affect the cell's ability to coordinate substrate fluxes. Optimizations may vary with the microenvironmental concentrations of the substrates. Further insights might be gained by studying substrate competition at a range of concentrations, but this was not done in our study.
PTS in an “aerobic” ocean.
PTS is energetically more favorable than ATP-driven active transport systems since it accomplishes substrate phosphorylation as well as transmembrane translocation at the expense of a single high-energy molecule (56, 58). This is especially important to organisms in oxygen-depleted environments carrying out glycolysis, which has a low energy yield. For instance, in E. coli, a facultative anaerobe, the glucose-PTS has an intermediate affinity for its substrate, and under glucose-limited growth an alternative, ATP-dependent high-affinity transport system (Mgl) is induced (18). However, Mgl is not expressed under anaerobiosis and the PTS becomes the main mechanism (41).
In the ocean, >90% of the bacteria are free-living in the water column (25, 67) and, thus, thrive in an aerobic and nutrient-limited environment. If we extrapolate from the experimental evidence from E. coli and from the energetics of PTS in aerobic and anaerobic environments, then PTS would not be expected to be a prevalent system among pelagic bacteria. As an explanation of the widespread distribution of PTS indicated by our study, we suggest that fermentative metabolism and a high capacity uptake system (as a PTS; 41) is an adaptation to growth on particles, where low-oxygen and nutrient-rich conditions may prevail. This proposed scenario fits well with speculations on the accumulation of bacteria with low-affinity high-capacity uptake systems in substrate production microzones (6) and with previous suggestions of “particle-specialist bacteria” in the sea (3, 5, 54, 61). Indeed, in the pelagic marine environment, where organic matter is produced largely as particles, bacterial adaptations to interact with particles to optimize substrate uptake might include the ability to survive in low (or no) oxygen microenvironments. However, it should be noted that the traditional view that some marine aggregates contain low-oxygen or anaerobic microzones (e.g., see references 1, 10, 32, and 60) has recently been challenged by studies indicating that remineralization of aggregates may not be oxygen limited (47) and that anoxia in marine aggregates may be a transient phenomenon (48).
So, are PTS-positive bacteria in the sea facultative anaerobes? This generalization was tested for our 78 isolates. Indeed, significantly more facultative anaerobes were found among the isolates which took up NAG (63%) than among isolates with no NAG uptake (39%). It should be noted that these are conservative estimates since we tested only for mixed acid fermentation and not alcohol fermentation and only for organisms capable of fermenting glucose or mannitol. Notably, all isolates within the Vibrionaceae group of γ-Proteobacteria took up NAG and were all facultative anaerobes. Similarly, Meadow et al. (43) found a glucose-PTS and a fructose-PTS in 15 Vibrio species grown in seawater. A PTS has also been documented in V. furnissii (7) and V. parahaemolyticus (34). This seemingly ubiquitous presence of PTSs in Vibrionaceae could be an important physiological feature, which may help explain the prevalence of this group in various environments including the oceans (13).
Implications for studying bacterial ecology.
Our study highlights that “biochemical interrogations” of bacteria in natural marine assemblages can yield insights into their ecology and indeed into the nature of their microenvironments. For instance, this approach has led to the hypothesis that anaerobic microzones are significant in the ecology of a substantial fraction of bacteria in the coastal ocean. Further, we emphasize that iterative approaches based on building conceptual bridges between studies of pure cultures and natural assemblages can be quite powerful. The use of STZ in this mode here illustrates the point. Such an approach has the potential to contribute to the eventual goal of understanding the mechanistic bases of bacterial ecosystem activities in the ocean.
Acknowledgments
We are grateful to Koji Hamasaki for performing the single-cell BrdU procedure and to Milton H. Saier, Jr., for providing the PTS protocol and for numerous pieces of advice. We thank Forest Rohwer for help with construction of the phylogenetic tree and Richard A. Long, Kay D. Bidle, and Laura B. Fandino for providing isolates and sequences, and for general support. We thank Morten Søndergaard for suggestions on ways to improve the manuscript.
This work was supported by an NIAID grant (RO1 AI46600) to F.A. and by a Ph.D. stipend from the University of Copenhagen to L.R.
REFERENCES
- 1.Alldredge, A. L., and Y. Cohen. 1987. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science 235:689-691. [DOI] [PubMed] [Google Scholar]
- 2.Ammer, J., M. Brennenstuhl, P. Schindler, J.-V. Höltje, and H. Zähner. 1979. Phosphorylation of streptozotocin during uptake via the phosphoenolpyruvate:sugar phosphotransferase system in Escherichia coli. Antimicrob. Agents Chemother. 16:801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 4.Azam, F., and J. W. Ammerman. 1984. Cycling of organic matter by bacterioplankton in pelagic marine ecosystems: microenvironmental considerations, p. 345-360. In M. J. R. Fasham (ed.), Flows of energy and materials in marine ecosystems. Plenum Press, New York, N.Y.
- 5.Azam, F., and B. C. Cho. 1987. Bacterial utilization of organic matter in the sea, p. 261-281. In M. M. Fletcher, J. G. Jones, and T. R. G. Gray (ed.), Ecology of microbial communities. Cambridge University Press, Cambridge, United Kingdom.
- 6.Azam, F., and R. Hodson. 1981. Multiphasic kinetics for D-glucose uptake by assemblages of natural marine bacteria. Mar. Ecol. Prog. Ser. 6:213-222. [Google Scholar]
- 7.Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria. J. Biol. Chem. 266:24276-24286. [PubMed] [Google Scholar]
- 8.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belas, R., M. Simon, and M. Silverman. 1986. Regulation of lateral gene transcription in Vibrio parahaemolyticus. J. Bacteriol. 167:210-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi, M., D. Marty, J.-L. Teyssié, and S. W. Fowler. 1992. Strictly aerobic and anaerobic bacteria associated with sinking particulate matter and zooplankton fecal pellets. Mar. Ecol. Prog. Ser. 88:55-60. [Google Scholar]
- 11.Brock, T. D., M. T. Madigan, J. M. Martinko, and J. Parker. 1994. Biology of microorganisms. Prentice-Hall International Inc., Englewood Cliffs, N.J.
- 12.Canale-Parola, E. 1977. Physiology and evolution of spirochetes. Bacteriol. Rev. 41:181-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colwell, R. 1984. Vibrios in the environment. John Wiley & Sons Inc., New York, N.Y.
- 14.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cottrell, M. T., D. N. Wood, L. Yu, and D. L. Kirchman. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the α- and γ-subclasses of the Proteobacteria. Appl. Environ. Microbiol. 66:1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dills, S. S., A. Apperson, M. R. Schmidt, and M. H. Saier, Jr. 1980. Carbohydrate transport in bacteria. Microbiol. Rev. 44:385-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducklow, H. W. 2000. Bacterial production and biomass in the oceans, p. 85-120. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss Inc., New York, N.Y.
- 18.Ferenci, T. 1996. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol. Rev. 18:301-317. [DOI] [PubMed] [Google Scholar]
- 19.Freese, E. B., R. M. Cole, W. Klofat, and E. Freese. 1970. Growth, sporulation, and enzyme defects of glucosamine mutants of Bacillus subtilis. J. Bacteriol. 101:1046-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuhrman, J. A., J. W. Ammerman, and F. Azam. 1980. Bacterioplankton in the coastal euphotic zone: distribution, activity and possible relationships with phytoplankton. Mar. Biol. 60:201-207. [Google Scholar]
- 21.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 22.González, J. M., W. B. Whitman, R. E. Hodson, and M. A. Moran. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gooday, G. W. 1990. The ecology of chitin degradation, p. 387-430. In K. C. Marshall (ed.), Advances in microbial ecology. Plenum Press, New York, N.Y.
- 25.Griffith, P., F.-K. Shiah, K. Gloersen, H. W. Ducklow, and M. Fletcher. 1994. Activity and distribution of attached bacteria in Chesapeake Bay. Mar. Ecol. Prog. Ser. 108:1-10. [Google Scholar]
- 26.Hamasaki, K., and F. Azam. 2001. Application of bromodeoxyuridine (BrdU) incorporation to estimate the growth rates of individual bacterial cells in natural seawater, p. 02.006. In 9th International Symposium on Microbial Ecology. International Society for Microbial Ecology, Amsterdam, The Netherlands.
- 27.Hanka, L. J., and W. T. Sokolski. 1960. Bacterial resistance to streptozotocin. Antibiot. Annu. 1959-1960:255-261. [PubMed] [Google Scholar]
- 28.Herr, R. R., T. E. Eble, M. E. Bergy, and H. K. Jahnke. 1960. Isolation and characterization of streptozotocin. Antibiot. Annu. 1959-1960:236-240. [PubMed] [Google Scholar]
- 29.Imada, A., Y. Nozaki, F. Kawashima, and M. Yoneda. 1977. Regulation of glucosamine utilization in Staphylococcus aureus and Escherichia coli. J. Gen. Microbiol. 100:329-337. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson, G. R., F. Poy, and J. W. Lengeler. 1990. Inhibition of Streptococcus mutans by the antibiotic streptozotocin: mechanisms of uptake and the selection of carbohydrate-negative mutants. Infect. Immun. 58:543-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko, T., and R. R. Colwell. 1978. The annual cycle of Vibrio parahaemolyticus in Chesapeake Bay. Microb. Ecol. 4:135-155. [DOI] [PubMed] [Google Scholar]
- 32.Karl, D. M., and B. D. Tilbrook. 1994. Production and transport of methane in oceanic particulate organic matter. Nature 368:732-734. [Google Scholar]
- 33.Kirchman, D. L., E. K'nees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubota, Y., S. Iuchi, and S. Tanaka. 1982. Evidence for the existence of a trehalose-specific enzyme II of the phosphoenolpyruvate:sugar phosphotranferase system in Vibrio parahaemolyticus. FEMS Microbiol. Lett. 13:5-7. [Google Scholar]
- 35.Kundig, W., and S. Roseman. 1966. A phosphoenolpyruvate-hexose phosphotransferase system from Escherichia coli. Methods Enzymol. 9:396-403. [Google Scholar]
- 36.Lee, S. H., and J. A. Fuhrman. 1987. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lengeler, J. W. 1979. Streptozotocin—an antibiotic superior to penicillin in the selection of rare bacterial mutations. FEMS Microbiol. Lett. 5:417-419. [Google Scholar]
- 38.Lengeler, J. W. 1980. Analysis of the physiological effects of the antibiotic streptozotocin on Escherichia coli K12 and other sensitive bacteria. Arch. Microbiol. 128:196-203. [DOI] [PubMed] [Google Scholar]
- 39.Lengeler, J. W. 1980. Characterisation of mutants of Escherichia coli K12, selected by resistance to streptozotocin. Mol. Gen. Genet. 179:49-54. [DOI] [PubMed] [Google Scholar]
- 40.Levine, M. M., R. E. Black, M. L. Clements, D. R. Nalin, L. Cisneros, and R. A. Finkelstein. 1981. Volunteer studies in development of vaccines against cholera and enterotoxigenic Escherichia coli: a review, p. 443-459. In T. Holme, J. Holmgren, M. H. Merson, and R. Möbley (ed.), Acute enteric infections in children. New prospects for treatment and prevention. Elsevier/North Holland Publishing Co., Amsterdam, The Netherlands.
- 41.Manché, K., L. Notley-McRobb, and T. Ferenci. 1999. Mutational adaptation of Escherichia coli to glucose limitation involves distinct evolutionary pathways in aerobic and oxygen-limited environments. Genetics 153:5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy, M. D., J. I. Hedges, and R. Benner. 1998. Major bacterial contribution to marine dissolved organic nitrogen. Science 281:231-234. [DOI] [PubMed] [Google Scholar]
- 43.Meadow, N. D., R. Revuelta, V. N. Chen, R. R. Colwell, and S. Roseman. 1987. Phosphoenolpyruvate:glycose phosphotransferase system in species of Vibrio, a widely distributed marine bacterial genus. J. Bacteriol. 169:4893-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mobley, H. L. T., R. J. Doyle, U. N. Streips, and S. O. Langemeier. 1982. Transport and incorporation of N-acetyl-d-glucosamine in Bacillus subtilis. J. Bacteriol. 150:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissen, H., P. Nissen, and F. Azam. 1984. Multiphasic uptake of D-glucose by an oligotrophic marine bacterium. Mar. Ecol. Prog. Ser. 16:155-160. [Google Scholar]
- 46.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 47.Ploug, H. 2001. Small-scale oxygen fluxes and remineralization in sinking aggregates. Limnol. Oceanogr. 46:1624-1631. [Google Scholar]
- 48.Ploug, H., M. Kühl, B. Buchholz-Cleven, and B. B. Jørgensen. 1997. Anoxic aggregates—an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 13:285-294. [Google Scholar]
- 49.Plumbridge, J. A. 1990. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J. Bacteriol. 172:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postma, P. W., and J. W. Lengeler. 1985. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 49:232-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reusser, F. 1971. Mode of action of streptozotocin. J. Bacteriol. 105:580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rich, J. H., H. W. Ducklow, and D. L. Kirchman. 1996. Concentrations and uptake of neutral monosaccharides along 140°W in the equatorial Pacific: contribution of glucose to heterotrophic bacterial activity and the DOM flux. Limnol. Oceanogr. 41:595-604. [Google Scholar]
- 54.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romano, A. H., S. J. Eberhard, S. L. Dingle, and T. D. McDowell. 1970. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in bacteria. J. Bacteriol. 104:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romano, A. H., and M. H. Saier, Jr. 1992. Evolution of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Section I. Physiologic and organismic considerations, p. 143-170. In R. D. Mortlock (ed.), The evolution of metabolic function. CRC Press, Boca Raton, Fla.
- 57.Romano, A. H., J. D. Trifone, and M. Brustolon. 1979. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in fermentative bacteria. J. Bacteriol. 139:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roseman, S. 1969. The transport of carbohydrates by a bacterial phosphotransferase system. J. Gen. Physiol. 54:138-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saier, J. R., M. H. 1977. Bacterial phosphoenolpyruvate:sugar phosphotransferase systems: structural, functional, and evolutionary interrelationships. Bacteriol. Rev. 41:856-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shanks, A. L., and M. L. Reeder. 1993. Reducing microzones and sulfide production in marine snow. Mar. Ecol. Prog. Ser. 96:43-47. [Google Scholar]
- 61.Sieburth, J. M. 1979. Sea microbes. Oxford University Press, New York, N.Y.
- 62.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 63.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 64.Smith, D. C., G. F. Steward, R. A. Long, and F. Azam. 1995. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep-Sea Res. II 42:75-97. [Google Scholar]
- 65.Sokolski, W. T., J. J. Vavra, and L. J. Hanka. 1960. Assay methods and antibacterial studies on streptozotocin. Antibiot. Annu. 1959-1960:241-246. [PubMed] [Google Scholar]
- 66.Trick, C. G. 1989. Hydroxamate-siderophore production and utilization by marine eubacteria. Curr. Microbiol. 18:375-378. [Google Scholar]
- 67.Turley, C. M., and P. J. Mackie. 1994. Biogeochemical significance of attached and free-living bacteria and the flux of particles in the NE Atlantic Ocean. Mar. Ecol. Prog. Ser. 115:191-203. [Google Scholar]
- 68.White, R. J. 1970. The role of the phosphoenolpyruvate phosphotransferase system in the transport of N-acetyl-D-glucosamine by Escherichia coli. Biochem. J. 118:89-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Worm, J., L. E. Jensen, T. S. Hansen, M. Søndergaard, and O. Nybroe. 2000. Interactions between proteolytic and non-proteolytic Pseudomonas fluorescens affect protein degradation in a model community. FEMS Microbiol. Ecol. 32:103-109. [DOI] [PubMed] [Google Scholar]
- 70.Wright, R. T., and J. E. Hobbie. 1966. Use of glucose and acetate by bacteria and algae in aquatic systems. Ecology 47:447-464. [Google Scholar]
- 71.Wu, H. C., and T. C. Wu. 1971. Isolation and characterization of a glucosamine-requiring mutant of Escherichia coli K-12 defective in glucosamine-6-phosphate synthetase. J. Bacteriol. 105:455-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zobell, C. E., and S. C. Rittenberg. 1938. The occurrence and characteristics of chitinoclastic bacteria in the sea. J. Bacteriol. 35:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zweifel, U. L., and Å. Hagström. 1995. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts). Appl. Environ. Microbiol. 61:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]