Abstract
Fe65 protein interacts with the cytosolic domain of the amyloid precursor APP. Its possible involvement in gene regulation is suggested by numerous observations, including those demonstrating that it activates transcription. Here, we show that the Fe65 transcription activation domain overlaps with the WW domain of Fe65 and binds to the nucleosome assembly factor SET. This protein is required for the Fe65-mediated transactivation of a reporter gene. Two-step chromatin immunoprecipitation experiments demonstrate that a complex including Fe65/AICD/Tip60 and SET is associated with the KAI1 gene promoter. Suppression of SET levels by RNA interference shows that this protein is required for full levels of basal transcription of the KAI1 gene. These results further support the function of Fe65 and APP in gene regulation and show a new role for the SET factor.
Keywords: APP, Tip60, chromatin immunoprecipitation, KAI1, Alzheimer
Introduction
Fe65 is a multidomain protein having a WW domain in the amino-terminal half and two PTB domains (PTB1 and PTB2) in the carboxyl-terminal half. Numerous results have given support to the possible role of this protein in gene regulation. In fact, it was demonstrated that Fe65 is present in the nucleus (Minopoli et al, 2001), where it interacts with the histone acetyl transferase Tip60 (Cao & Sudhof, 2001) and was found to be associated with and to regulate the expression of the KAI1 gene promoter (Baek et al, 2002). Nuclear localization of Fe65 was also confirmed by cell immunostaining, showing the presence of Fe65 in intranuclear speckles (Muresan & Muresan, 2004; Von Rotz et al, 2004).
Fe65 interacts with the β-amyloid precursor protein APP (Fiore et al, 1995). The latter is a type I membrane protein involved in the pathogenesis of Alzheimer's disease because it is the precursor of the main constituents of Alzheimer's senile plaques. The β-amyloid peptides are generated from APP through the action of two proteases named β- and γ-secretase (Haass, 2004). The cleavage of APP by secretases generates a peptide, named AICD (APP intracellular domain), which is released from the membrane and is found in the nucleus (Gao & Pimplikar, 2001; Kimberly et al, 2001; Von Rotz et al, 2004). Intact APP functions as an extranuclear anchor for Fe65, thus preventing Fe65 nuclear translocation (Minopoli et al, 2001). The cleavage of APP probably allows Fe65 to reach the nucleus. This regulatory mechanism resembles that controlling the function of other transcription factors, such as those involving N-cadherin cleavage and the CREB-binding protein or Notch (Marambaud et al, 2003; Schweisguth, 2004). Recently, a direct involvement of Fe65 in transactivation events through its WW domain has been proposed (Cao & Sudhof, 2004). Here, we show that Fe65 is able to activate transcription depending on a small region of the protein encompassing part of its WW domain. This region binds the nucleosome assembly protein SET, and the activation of transcription mediated by Fe65 is dependent on SET. An oligomeric complex immunoprecipitated by antibodies against Fe65, APP, Tip60 or SET targets the KAI1 gene promoter, and SET is required for the basal transcription of this gene.
Results And Discussion
The transcription-activating element of Fe65
The ability of the WW domain of Fe65 to activate transcription when fused to the Gal4 DNA-binding domain (Dbd) could be due to the interaction of this domain with certain cofactors. According to this possibility, when a fusion protein, including the Gal4 Dbd and the full-length Fe65 (Gal4–Fe65f.l.; Fig 1A), was co-transfected with the G5BCAT reporter vector and with various constructs encoding fragments of Fe65, we observed that only the region including the WW domain has a marked inhibitory effect (supplementary information online). This suggests that it is titrating an endogenouss limiting factor.
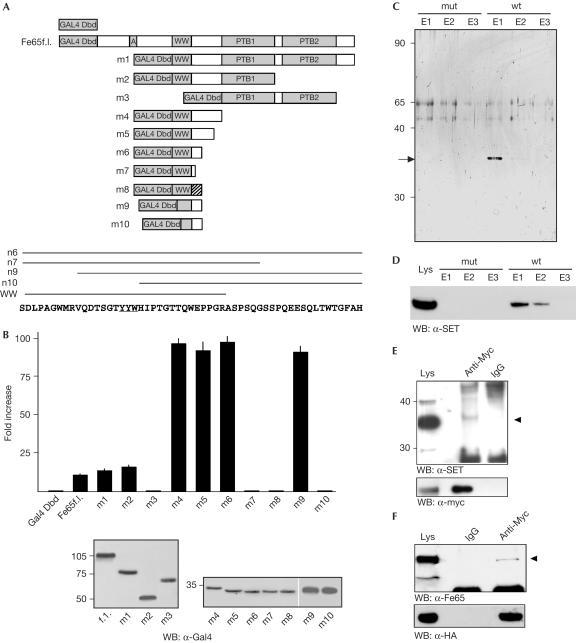
Figure 1.
SET interacts with Fe65. (A) The Gal4 Dbd was cloned upstream of the full-length Fe65 (Fe65f.l.) complementary DNA or various deletion mutants of this cDNA. Striped box in the m8 construct indicates a 19-amino-acid-long region in which the same 19 residues present downstream of the WW domain in the m6 construct are arranged in a scrambled sequence. The Fe65 sequence covered by the crucial constructs m6, m7, m9 and m10 is indicated along with that of the WW domain. (B) The deletion mutants of Fe65 fused to the Gal4 Dbd described in (A) have been co-transfected in HeLa cells with the G5BCAT vector. CAT expression is reported as fold increase compared with that observed in cells transfected with the Gal4 Dbd alone. The values are the mean+s.d. of triplicate experiments. The western blot (WB) of one representative experiment is shown below. (C) Proteins eluted from the mutant (mut) or wild-type (wt) peptide columns (fractions E1–E3) were resolved on 10% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and the gel was silver stained. The arrow indicates the 35 kDa band, which was identified as SET by mass spectrometry. (D) Proteins eluted from the mutant or wt peptide columns (fractions E1–E3) were resolved on 10% SDS–PAGE and analysed by western blot with α-SET antibody. The lysate of HEK293 cells transfected with SET expression vector was loaded as a control. (E) Proteins from HEK293 cells transfected with Myc-Fe65 were immunoprecipitated with α-Myc monoclonal antibody or with mouse IgGs. Immunoprecipitated proteins were analysed by western blot with SET antibody. (F) Proteins from HEK293 cells transfected with HA-SET were immunoprecipitated with α-HA antibody or with mouse IgGs. The western blot with Fe65 antibody of the immunoprecipitated proteins is shown. (E,F) 50 μg of the input lysate was loaded in the first lane as a migration control. The arrowheads indicate the bands of SET and Fe65.
To determine the optimal amino-acid sequence for use as bait to purify this factor by affinity chromatography, we dissected the Fe65 region responsible for the activation of transcription when fused to the Gal4 Dbd (Fig 1A). The deletion of both PTB domains of Fe65 results in an increased transactivation efficiency, in agreement with the hypothesis that they have an inhibitory role in the context of the intact molecule (Cao & Sudhof, 2004). On the contrary, the deletion mutants containing the WW domain strongly activate the transcription of the reporter gene (Fig 1B), but the WW domain alone does not act as a transcription activation domain (construct m7). In fact, at least 19 residues at the C-terminus of the WW domain (constructs m4, m5 and m6) are needed to observe a significant CAT gene expression. This 19-residue-long stretch contains some specific sequence information because it cannot be replaced by a scrambled sequence (construct m8).
The m9 protein, lacking the first nine amino acids of the N-terminus of the WW domain, activates transcription to the same extent as the m4 protein, whereas a further deletion of ten amino acids (m10 construct) is completely devoid of activity. These ten residues contain the YYW motif (underlined in Fig 1A) that is necessary for AICD–Fe65-mediated transactivation (Cao & Sudhof, 2001). Thus, it seems that the transcriptional function of Fe65 does not coincide with its WW domain. Accordingly, the minimal region of Fe65 that activates transcription (m9) does not interact with Mena, which on the contrary interacts with the Fe65 WW domain (supplementary information online; Ermekova et al, 1997).
The WW overlapping region of Fe65 interacts with SET
Thus, we have decided to use this WW overlapping region (WOR) as bait to affinity-purify the factors mediating Gal4–Fe65-induced transactivation. HEK293 cell extract was first used to challenge a peptide in which the YYW motif, necessary for transactivation, is changed to AAA, and then to challenge a second, wild-type (wt) synthetic peptide. This experimental approach allowed us to purify one protein band eluted from the second chromatography (Fig 1C). Mass spectrometry (MS) analysis of the tryptic digestion of the protein indicated that it is SET, a protein belonging to the family of nucleosome assembly proteins (von Lindern et al, 1992). Western blot analysis with an α-SET-specific antibody confirmed identity (Fig 1D).
The existence of an Fe65–SET complex is further confirmed by co-immunoprecipitation experiments. In fact, Myc-tagged Fe65 immunoprecipitates with endogenous SET (Fig 1E) and, conversely, haemagglutinin (HA)-tagged SET immunoprecipitates with endogenous Fe65 (Fig 1F).
The WW domain is a protein–protein interaction module consisting of a triple-stranded antiparallel β-sheet characterized by several conserved residues, including the two Trp that give the name to the domain (Sudol & Hunter, 2000). On the basis of the sequence of the m9 construct, it seems that the minimal module needed to interact with SET only contains the second and third strands of the β-sheet and a stretch of amino acids flanking the third strand at its N-terminus. This minimal structure does not retain the ability to interact with Mena, suggesting that the WW domain is able to interact with its partners in two ways: one requires the complete three-stranded β-sheet, whereas only two β-strands followed by a C-terminal stretch are necessary for the second type of interaction. This possibility suggests that the WW region is available for the binding of two sets of ligands or that a conformational change of this region could switch the affinity of the domain from one set of ligands to another. This hypothesis could explain the different functions proposed for Fe65, which include cytoskeleton remodelling and cell motility (Sabo et al, 2001) and gene regulation.
SET is responsible for Gal4–Fe65-mediated transactivation
To evaluate whether SET is required for transcription activation induced by Gal4–Fe65 fusion proteins, we analysed the effects of SET overexpression or suppression on the transcription activation mediated by Gal4–Fe65 proteins. SET overexpression increases by several-fold the CAT expression induced by Gal4–Fe65f.l. or by the m6 mutant, whereas it has no effect on the activation of transcription induced by the transfection of Gal4 holoprotein (Fig 2A). Furthermore, SET overexpression has no effect on transcription when the cells are transfected with Gal4–Fe65 deletion mutants that are devoid of activity, such as m3, m7, m8 and m10 (Fig 1A), indicating that SET overexpression has no nonspecific effect on transcription (Fig 2A, m10 mutant). Accordingly, these deletion mutants do not immunoprecipitate together with SET (supplementary information online).
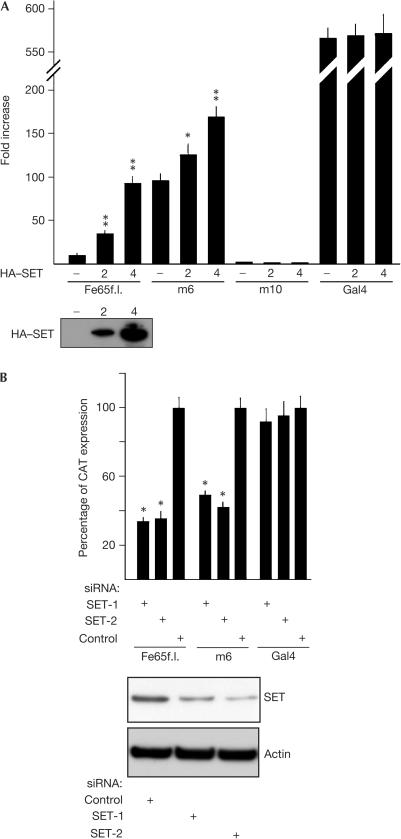
Figure 2.
SET is necessary for Fe65-dependent transcription of a reporter gene. HEK293 cells were transfected with G5BCAT vector and with either Gal4–Fe65f.l., or m6 or m10 constructs (Fig 1A) and with (A) 2 or 4 μg of a plasmid encoding the HA-SET protein or with (B) siRNA duplexes targeting SET (SET-1 and SET-2) or with control siRNA. The bars in (A) indicate the fold increase of CAT gene transcription compared with that observed in cells transfected with G5BCAT and the Gal4 Dbd alone, and those in (B) indicate the percentage of CAT gene expression measured in cells transfected with GFP-targeting siRNA as a control. SET expression was checked by western blot with α-SET antibody. Standard deviations were calculated on quadruplicate experiments (probability associated with Student's t-test: *P<0.01; **P<0.001).
Silencing of endogenous SET expression by RNA-mediated interference (RNAi) decreases the levels of SET protein to about 50% of the levels present in the cells transfected with the control double-stranded oligonucleotide (green fluorescent protein (GFP)-targeting short interfering RNA (siRNA)). This is accompanied by a significant decrease of CAT expression in cells transfected with either Gal4–Fe65f.l. or m6 construct (Fig 2B). SET RNAi has no effect on the robust transcription activation driven by the Gal4 holoprotein.
The KAI1 promoter is a target for the Fe65–SET complex
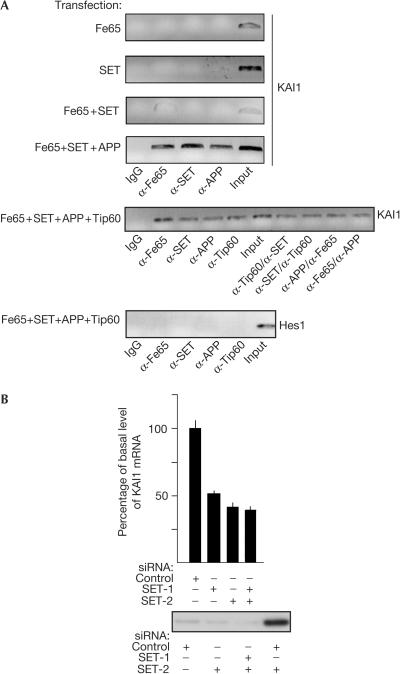
It has been demonstrated that Fe65, together with Tip60 and AICD, can be recruited on transfection to the KAI1 gene promoter (Baek et al, 2002). Therefore, we addressed the question of whether a complex containing both Fe65 and SET is similarly associated with the same KAI1 promoter. To this aim, HEK293 cells were transfected with Fe65, SET and/or APP, and Tip60 and chromatin immunoprecipitation (ChIP) assays were performed by using antibodies directed against these proteins. As shown in Fig 3A, when Fe65, SET and APP are co-transfected, a robust protein–DNA complex containing all these proteins was observed even in the absence of Tip60; hence, SET must exert a stabilizing influence similar to that of Tip60. In cells co-transfected with Fe65, SET, APP and Tip60, ChIP showed that all these proteins are present on the KAI1 gene promoter. To examine possible co-recruitment, two-step ChIPs were performed using α-Tip60/α-SET, α-SET/α-Tip60, α-APP/α-Fe65 and α-Fe65/α-APP antibody combinations. These experiments demonstrated that the four proteins can be present simultaneously on the same promoter. APP overexpression is clearly required to obtain complexes containing Fe65/SET or Fe65/SET/Tip60, supporting the hypothesis that interaction with APP renders Fe65 suitable for binding to other ligands (Cao & Sudhof, 2004). α-APP antibodies directed against C-terminal epitopes of the protein immunoprecipitate chromatin complexes; by contrast, we failed to obtain any ChIP using antibodies directed against the extracellular–intraluminal domain of APP (data not shown), suggesting that only the C-terminal domain of the protein is present in the complex.
Figure 3.
The Fe65–SET–APP complex is bound to the KAI1 promoter and SET is necessary for KAI1 gene expression. (A) ChIP assay on KAI1 promoter in HEK293 cells transfected as indicated. The antibodies used for single ChIP and the order of those used for double ChIP is reported below the experiments. 10% of the input DNA was amplified as a control. Hes1 gene ChIP is reported as a control. (B) HEK293 cells were transfected with the indicated control or SET siRNAs, and total RNA was analysed by real-time PCR to measure KAI1 mRNA. The histogram reports the mean+s.d. of triplicate experiments.
The KAI1 gene is under the control of homodimeric p50–p50 NF-κB. Interleukin-1β induces exchange of the N-CoR/TAB2/HDAC3 co-repressor complex with a Tip60-containing coactivator complex (Baek et al, 2002). The same promoter is also activated by a complex containing Tip60, Fe65 and AICD. HAT activity of Tip60 might be required for the observed transcription activation, but in some cases Tip60 may not be required. In fact, Gal4–Fe65 fusion proteins lacking the PTB1 domain, the highest affinity binding site for Tip60, are potent transcription activators (Cao & Sudhof, 2004; Fig 1B). Furthermore, PTB1 domain overexpression did not titrate any limiting factor necessary for Gal4–Fe65-dependent transactivation (supplementary information online), and Tip60 overexpression did not affect Gal4–Fe65 activity (supplementary information online). Conversely, SET is required for Gal4–Fe65-dependent transactivation, as shown by its overexpression or suppression by RNAi. Therefore, we addressed the possible role of SET in the regulation of transcription of the KAI1 gene. To this aim, we measured the level of KAI1 mRNA in cells transfected with siRNAs targeting SET mRNA. As shown in Fig 3B, the suppression of SET expression significantly decreases KAI1 mRNA levels, demonstrating that, under basal conditions, SET is quantitatively important for KAI1 gene transcription.
Although the role of SET-containing complexes in caspase-independent apoptosis has been recently shown (Fan et al, 2003), its involvement in the regulation of gene transcription is still unclear. There are several results indicating that SET might have additional activities and functions, as it interacts with p300/CBP histone acetyltransferase. The consequences of this interaction could be either an increase (Shikama et al, 2000) or inhibition (Seo et al, 2001) of the transcription of p300/CBP target genes. The results reported in this paper suggest that Fe65 is the adaptor molecule that assembles SET and Tip60 on chromatin, possibly leading to the regulation of transcription. We have also demonstrated that APP is necessary for formation of the complex and that the C-terminal tail of APP is part of the chromatin complex. The possible role of secretase-driven APP cleavage in the regulation of the observed phenomena is of significant interest, as altered APP processing could result in a change of gene expression. A link between SET functions and presenilin-dependent secretase activity is suggested by the observation that Spr-2, one of the suppressors of Sel-12 presenilin defects in Caenorhabditis elegans, is the nematode orthologue of SET (Wen et al, 2000). This relationship should be explored further.
Methods
Recombinant constructs and proteins. Vectors for Gal4 Dbd–Fe65 fusions were generated by cloning the appropriate Fe65 cDNA fragments, amplified by PCR, in-frame with the Gal4 Dbd in the pRcCMV plasmid (Invitrogen, Carlsbad, CA, USA). The sequences of PCR primers are available on request. Full-length human SET cDNA was obtained by reverse transcription–PCR using total RNA from HEK293 cells. HA-tagged SET was generated by using a reverse primer containing the HA-coding epitope sequence at the 3′ end, in the pRcCMV vector.
Cell cultures, transfections and co-precipitations. HeLa and HEK293 cells were grown at 37°C in the presence of 5% CO2 in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum (Invitrogen), 100 U/ml penicillin and 100 mg/ml streptomycin (HyClone, Logan, UT, USA). HeLa cells were transfected by calcium phosphate with 3 μg G5BCAT vector and with 3 μg of the plasmids expressing the fusion proteins. CAT expression was measured by using colorimetric CAT enzyme-linked immunoadsorbent assay (Roche Molecular Biochemicals, Mannheim, Germany). HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen) in 100 mm dishes. SET double-stranded 21-mer RNAs and control double-stranded RNA targeting GFP (Fan et al, 2003) were synthesized by Qiagen, and transfected in HEK293 with Lipofectamine 2000. Protein extracts were prepared as described (Gianni et al, 2003). For immunoprecipitations, 5 μl of antibodies was incubated with 1.5 mg of protein lysates for 2 h at 4°C, followed by a 1 h incubation with Protein A–Sepharose. Immunoprecipitates were washed three times in lysis buffer. Proteins released by boiling in SDS sample buffer were separated by Novex Bis–Tris 4–12% polyacrylamide gels (Invitrogen) and analysed by using SET (1:1,000) or Fe65 (1:2,000) antibodies.
Purification of WOR ligands. The peptides used for affinity purification were synthesized by Tufts University Core Facility (Boston, MA, USA). Both peptides carry the Strep-Tag sequence at their C-termini: wild-type peptide, WSHPQFEKGAGGVQDTSGTYYWHIPTGTTQWEPPG RASPSQGNSPQEESQLTWTGFAH; mutant peptide, the underlined sequence is changed to AAA. Peptides (100 nmol) was dissolved in 1 ml of 100 mM Tris (pH 8.0), 150 mM NaCl and 1 mM EDTA and applied on columns containing 1 ml of Strep-Tactin matrix (IBA, Göttingen, Germany; Skerra & Schmidt, 2000). Affinity chromatography was performed with HEK293 lysates in 100 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 50 mM NaF, 1 mM sodium vanadate and protease inhibitors. The lysate was precleared on a column containing Strep-Tactin matrix, then on the mutant peptide column. The flow-through was then applied on wild-type peptide column. Proteins were eluted with 50 mM ammonium bicarbonate+2.5 mM desthiobiotin and run on 10% SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Bands were excised from the gel, digested with trypsin and the peptide mixtures were analysed by MALDI-TOF mass spectrometry as previously reported (Gianni et al, 2003).
Real-time PCR. Total RNA was prepared from HEK293 cells by using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and subjected to cDNA synthesis with random hexanucleotide primers and MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA, USA) at 48°C for 1 h. The cDNA (1 μl) was then amplified for 40 cycles using SYBR Green PCR master mix (Applied Biosystems) and template-specific primers in an ABI Prism 7900 system (Applied Biosystems). Relative quantification of gene expression was performed using the comparative threshold (CT) method. Changes in mRNA expression levels were calculated following normalization to β-actin or c-Abl transcripts. The ratios obtained following normalization are expressed as fold change over calibrator samples. The primer sequences are as follows: KAI1, 5′-AGGATGCCTGGGACTACGTG and 5′-GCTCAGCGTTGTCTGTCCAGT; β-actin, 5′-TCGTGCGTGACATTAGGAG and 5′-GTCAGGCAGCTCGTAGCTCT; c-Abl, 5′-GGTATGAAGGGAGGGTGTACCA and 5′-GTGAACTAACTCAGCCAGAGTGTTGA.
ChIP. ChIP assays were performed as described (Shang & Brown, 2002). Subconfluent HEK293 cells (100-mm dishes) were fixed with 1% formaldehyde for 10 min at 20°C and then quickly rinsed with ice-cold PBS. Diluted chromatin solution was precleared in 45 μl slurry 50% Protein A–Sepharose (Sigma, St Louis, MO, USA) and 2 μg of sheared salmon sperm DNA (Invitrogen) for 2 h at 4°C. The supernatants were immunoprecipitated overnight with 2 μg of the following antibodies: α-Fe65 (I-12), α-APP (CT695, Zymed Laboratories, San Francisco, CA, USA), α-SET (Abcam, Cambridge, MA, USA) and α-Tip60 (Upstate Biotechnology). The antibodies recognizing the extracellular/intraluminal domain of APP were 6E10/4G8 (Abcam). A 2 μg portion of rabbit or goat IgG (Santa Cruz Biotechnology) was used as a control.
Protein-bound immunoprecipitated DNA was reverse crosslinked at 65°C overnight and then purified by using a PCR purification kit (Qiagen). A 2 μl portion of DNA solution was used for PCR amplification (30 cycles). Primer sequences were as follows: KAI1, 5′-GACAGGGTTTCATCCTGTTGC and 5′-GAGGATAGCCTGGCCCTAGC; Hes1, 5′-CTCAGGCGCGCGCCATTGGCC and 5′-GCTTACGTCCTTTTACTTGACTTTC. PCR products were run on a 1.0% agarose gel and analysed by ethidium bromide staining. For two-step ChIPs, the immunocomplexes were eluted by adding 100 μl of MCP beads (Pierce, New York, NY, USA) to the pelleted Sepharose beads and by shaking for 1 h at 20°C. The supernatants were collected and diluted tenfold in 1 ml final volume with dilution buffer. A 2 μg portion of second antibody was added and the immunoprecipitations were performed again as described above.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n1/extref/7400309s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
This work was funded by the VI Framework Programme of the European Commission (LSHM-CT-2003-503330), FIRB ‘Proneuro' and the Alzheimer Association.
References
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG (2002) Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell 110: 55–67 [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC (2001) A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293: 115–120 [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC (2004) Dissection of amyloid-β precursor protein-dependent transcriptional transactivation. J Biol Chem 279: 24601–24611 [DOI] [PubMed] [Google Scholar]
- Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M (1997) The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem 272: 32869–32877 [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J (2003) Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112: 659–672 [DOI] [PubMed] [Google Scholar]
- Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T (1995) The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer's amyloid precursor protein. J Biol Chem 270: 30853–30856 [DOI] [PubMed] [Google Scholar]
- Gao Y, Pimplikar SW (2001) The γ-secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc Natl Acad Sci USA 98: 14979–14984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni D et al. (2003) Platelet-derived growth factor induces the β-γ-secretase-mediated cleavage of Alzheimer's amyloid precursor protein through a Src–Rac-dependent pathway. J Biol Chem 278: 9290–9297 [DOI] [PubMed] [Google Scholar]
- Haass C (2004) Take five-BACE and the γ-secretase quartet conduct Alzheimer's amyloid β-peptide generation. EMBO J 23: 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ (2001) The intracellular domain of the β-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem 276: 40288–40292 [DOI] [PubMed] [Google Scholar]
- Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, Robakis NK (2003) A CBP binding transcriptional repressor produced by the PS1/-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell 114: 635–645 [DOI] [PubMed] [Google Scholar]
- Minopoli G, de Candia P, Bonetti A, Faraonio R, Zambrano N, Russo T (2001) The β-amyloid precursor protein functions as a cytosolic anchoring site that prevents Fe65 nuclear translocation. J Biol Chem 276: 6545–6550 [DOI] [PubMed] [Google Scholar]
- Muresan Z, Muresan V (2004) A phosphorylated, carboxy-terminal fragment of β-amyloid precursor protein localizes to the splicing factor compartment. Hum Mol Genet 13: 475–488 [DOI] [PubMed] [Google Scholar]
- Sabo SL, Ikin AF, Buxbaum JD, Greengard P (2001) The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol 153: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F (2004) Notch signaling activity. Curr Biol 14: 129–138 [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104: 119–130 [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M (2002) Molecular determinants for the tissue specificity of SERMs. Science 295: 2465–2468 [DOI] [PubMed] [Google Scholar]
- Shikama N et al. (2000) Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol Cell Biol 20: 8933–8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A, Schmidt TGM (2000) Use of the Strep-tag and streptavidin for recombinant protein purification and detection. Methods Enzymol 326: 271–304 [DOI] [PubMed] [Google Scholar]
- Sudol M, Hunter T (2000) NeW wrinkles for an old domain. Cell 103: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G (1992) Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol 12: 3346–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U (2004) The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci 117: 4435–4448 [DOI] [PubMed] [Google Scholar]
- Wen C, Levitan D, Li X, Greenwald I (2000) Spr-2, a suppressor of the egg-laying defect caused by loss of Sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily. Proc Natl Acad Sci USA 97: 14524–14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information