Abstract
The process of disulphide bond formation in the endoplasmic reticulum of eukaryotic cells was one of the first mechanisms of catalysed protein folding to be discovered. Protein disulphide isomerase (PDI) is now known to catalyse all of the reactions that are involved in native disulphide bond formation, but despite more than 40 years of study, its mechanism of action is still not fully understood. This review discusses recent advances in our understanding of the human PDI family of enzymes and focuses on their functional properties, substrate interactions and some recently identified family members.
Keywords: disulphide bond formation, endoplasmic reticulum, protein disulphide isomerase, protein folding, thiol-disulphide oxidoreductase
Introduction
Native disulphide bond formation is a complex process. Disulphide bonds must not only be formed (oxidation), but also incorrect bonds must be broken (reduction) or rearranged (isomerization). During the past couple of years there have been several significant advances in our understanding of these processes—for instance, concerning the role of the endoplasmic reticulum (ER) oxidase Ero1 and the source of oxidizing equivalents (for a review, see Tu & Weissman, 2004). Although there has been a strong focus on oxidation reactions, isomerization rather than oxidation is rate limiting during the in vitro refolding of many disulphide-containing proteins. As isomerization reactions are thought to be catalysed only by members of the protein disulphide isomerase (PDI) family, a better knowledge of their mechanisms of action is crucial to our understanding of native disulphide bond formation.
The enzyme PDI is a multi-domain, multi-functional member of the thioredoxin superfamily (for reviews, see Freedman et al, 2002; Ferrari & Söling, 1999). PDI can catalyse thiol-disulphide oxidation, reduction and isomerization, the last of which occurs directly through intramolecular disulphide rearrangement or through cycles of reduction and oxidation (Schwaller et al, 2003). PDI comprises two thioredoxin-like catalytic domains, a and a′, which are separated by two non-catalytic domains, b and b′. The catalytic domains contain a characteristic CXXC active-site motif, with the two amino acids that lie between the cysteine residues having a major role in determining the redox potential of the enzyme and hence its function as a thiol-disulphide reductase, oxidase or isomerase. Despite attempts for more than 30 years to crystallize PDI, the structure of this protein, or of any other catalytically active eukaryotic PDI-family member, has not yet been determined. The structures of the human PDI a and b domains have been solved by nuclear magnetic resonance (NMR), and both have a thioredoxin-fold (Kemmink et al, 1996, 1997). Given the homology between a and a′, and b and b′, it is probable that PDI consists of four domains each with a thioredoxin-fold, plus a short acidic carboxy-terminal extension; however, it is unknown how these domains are orientated with respect to each other. Recently, a 19-amino-acid linker region was identified between the b′ and a′ domains (Pirneskoski et al, 2004), which would potentially allow more flexibility between these domains than between the other domains. However, the physiological relevance of this finding is unknown.
The PDI family
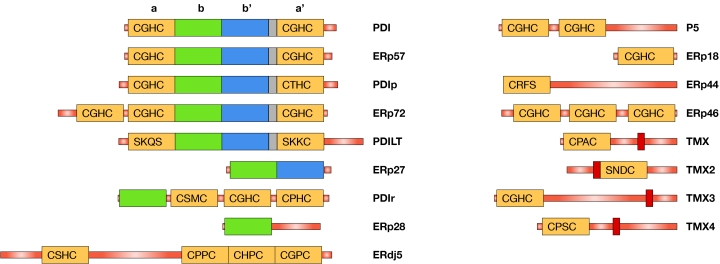
During the past two years, several new human PDI-family members have been reported: ERp18 (Alanen et al, 2003; Knoblach et al, 2003), ERp44 (Anelli et al, 2002, 2003), ERp46 (Knoblach et al, 2003; Sullivan et al, 2003), ERdj5 (Cunnea et al, 2003; Hosoda et al, 2003), thioredoxin-related transmembrane protein 2 (TMX2; Meng et al, 2003) and PDILT (van Lith et al, 2004). When added to the previously known family members PDI, PDIp, ERp57, ERp72, P5, PDIr, ERp28 (Freedman et al, 2002; Ferrari & Söling, 1999) and TMX (Matsuo et al, 2001), there are now 14 human PDI-family members in the ER, with a wide range of domain architectures and active-site chemistries (Fig 1; Table 1). For the new PDI-family members, the ability to catalyse thiol-disulphide exchange reactions has only been shown for ERp18. In addition to these 14 proteins, three unpublished PDI-family members (ERp27, TMX3 and TMX4), the ER localization of which has been confirmed (L.W.R. and L.E., unpublished data), can be found in public databases (for accession numbers, see Table 1).
Figure 1.
Schematic overview of the human protein disulphide isomerase family. Thioredoxin-like domains are represented by rectangles with the active-site sequence added for catalytic domains (yellow). The catalytically inactive b′ domain is shown in blue and other catalytically inactive domains are in green. The linker region between b′ and a′ domains is coloured in grey, and transmembrane regions are shown in red. Signal sequences are not shown. Note that the catalytic domain of ERp18 contains a putative 23-amino-acid insert between β3 and α3 (Alanen et al, 2003) that is not shown in the figure.
Table 1.
Sequence features of members of the human protein disulphide isomerase family
| Name* | SwissProt accession | Length | ER retention | N-glycosylation sites (putative) | a-like domains | Charge pair sequence | Conserved arginine |
|---|---|---|---|---|---|---|---|
| PDI | P07237 | 508 | KDEL | 0 | 2 | E47–K81, E391–K424 | Yes, yes |
| ERp57 | P30101 | 505 | QEDL | 0 | 2 | E51–K82, E400–K433 | Yes, yes |
| PDIp | Q13087 | 525 | KEEL | 3 | 2 | E65–K99, K412–E445 | Yes, yes |
| ERp72 | P13667 | 645 | KEEL | 0 | 3 | E85–K119, E200–K234, E549–K582 | Yes, yes, yes |
| ERp65 | Q8N807 | 584 | KEEL | 8 | 2 | L66–K100, M411–K444 | No, no |
| ERp27 | Q96DN0 | 273 | KVEL | 1 | 0 | – | – |
| PDIr | Q14554 | 519 | KEEL | 0 | 3 | M176–N209, M299–A334, M420–A453 | No, yes, no |
| ERp28 | P30040 | 261 | KEEL | 1 | 0 | – | – |
| ERdj5 | Q8IXB1 | 793 | KDEL | 1 | 4 | N152–A183, D474–T505, D582–S613, D694–K725 | Yes, no, yes, yes |
| P5 | Q15084 | 440 | KDEL | 0 | 2 | E49–A80, E184–A219 | Yes, yes |
| ERp18 | O95881 | 172 | EDEL | 1 | 1 | I60–N93 | No |
| ERp44 | Q9BS26 | 406 | RDEL | 0 | 1 | N52–R89 | Yes |
| ERp46 | Q8NBS9 | 432 | KDEL | 0 | 3 | M83–K118, K211–K244, K344–E378 | Yes, yes, yes |
| TMX | Q9H3N1 | 280 | Unknown | 0 | 1 | E50–K82 | Yes |
| TMX2† | Q9Y320 | 296 | KKDK | 2 | 1 | E161–K193 | ?‡ |
| TMX3 | Q96JJ7+ | 454 | KKKD | 2 | 1 | D47–K81 | Yes |
| TMX4 | Q9H1E5 | 349 | Unknown | 1 | 1 | K58–K90 | No |
*This table lists the 17 human protein disulphide isomerase (PDI)-family members the endoplasmic reticulum (ER) location of which has been confirmed. In addition, Q96MT2 is probably an ER-located human PDI-family member.
†The version of TMX2 included here represents the consensus version from human cDNA databases and is consistent with homologous proteins found in a wide range of species. A longer version of TMX2 (Q8NBP9, 372 amino acids, of which the first 281 are identical to the sequence listed here) has been published (Meng et al, 2003).
‡The alignment of TMX2 with other family members in this region is ambiguous. It is possible that the conserved arginine is present in the sequence.
+TMX3 starts at M33 of Q96JJ.
Substrate interactions
To function as catalysts of protein thiol-disulphide exchange, PDI-family members must be able to interact with their substrates. In PDI, the b′ domain provides the primary peptide- or non-native protein-binding site, but other domains also contribute to binding (Klappa et al, 1998). Detailed in vitro enzymology on linear combinations of PDI domains has shown that the isolated a and a′ domains can catalyse thiol-disulphide exchange reactions in peptide and protein substrates, but that a combination of a catalytic domain and the b′ domain is required for simple isomerization reactions. Moreover, all of the thioredoxin-like domains of PDI are required for isomerization reactions that involve substantial changes in structure in the substrate (Darby et al, 1998). The implications from these studies are that the a and a′ domains contain as-yet-unidentified, low-affinity binding sites for non-native proteins, whereas the b′ domain contains a higher affinity binding site by which PDI holds substrates during isomerization reactions. The b domain of PDI has neither been implicated in substrate binding (Klappa et al, 1998) nor does the addition of the b domain add to the catalytic ability of PDI domain constructs (Darby et al, 1998), which suggests that this domain may have a structural role in PDI rather than a direct catalytic role.
Using structural models of the b′ domains of PDI and PDIp in combination with the known specificity of substrate binding by PDIp (Ruddock et al, 2000), the substrate-binding site in the b′ domain of PDI was recently mapped to a small hydrophobic binding pocket located where the active site is found in catalytic thioredoxin -like domains (Pirneskoski et al, 2004). Mutations designed to partially fill or occlude this pocket resulted in a significant reduction in substrate binding by PDI.
Whereas PDI interacts directly with and also folds non-native proteins, ERp57 is known to act in vitro and in vivo on glycosylated substrates through its interaction with the ER-resident lectins calnexin and calreticulin (Zapun et al, 1998; Oliver et al, 1997). The primary interaction site on calreticulin has been localized to the highly acidic tip of the P-domain (Frickel et al, 2002), which forms an elongated hairpin loop (Ellgaard et al, 2001). The regions of ERp57 that are important for this interaction are the b′ domain and, to a lesser extent, the C-terminal positively charged region (Russell et al, 2004; Silvennoinen et al, 2004; Urade et al, 2004; Pollock et al, 2004). The binding site for calreticulin in the b′ domain of ERp57 has been mapped to residues that are equivalent to those that form the substrate-binding site in PDI (Russell et al 2004).
The picture that emerges from these studies—supported by a recent study on the Wind protein (Barnewitz et al, 2004), which is the Drosophila homologue of ERp28—is that PDI-family members use a conserved binding pocket located in a non-catalytic thioredoxin-like domain for high-affinity binding of substrates or substrate-interacting cofactors. This, in turn, is likely to be important for the proteins' ability to catalyse isomerization reactions. In most cases described so far, this binding site is located in b′-like domains, which are found in PDI, PDIp, ERp57, ERp72, ERp27 and PDILT. An alignment of these domains (Fig 2) reveals that whereas PDIp, ERp27 and PDILT are more PDI-like and are therefore probably able to bind non-native proteins, ERp72 is more ERp57-like, and many of the residues that are implicated in ERp57–calreticulin interactions are conserved in ERp72. However, there is no experimental evidence yet to suggest an ERp72–calreticulin interaction.
Figure 2.
Multiple sequence alignment of the b′-like domains of the human protein disulphide isomerase family. The alignment was constructed from a large number of single and multiple alignments and adjusted by hand, taking secondary structure predictions into account. Amino acids identical or similar to each other in four or more proteins are highlighted. Residues in which mutation inhibits the interaction of ERp57 with calreticulin (Russell et al, 2004) are indicated with an asterisk below the alignment, whereas those in which mutation inhibits the interaction of protein disulphide isomerase (PDI) with the peptide substrate Δ-somatostatin (Pirneskoski et al, 2004) are indicated with an asterisk above the alignment. At six of the seven sites implicated in ERp57–calreticulin interactions, ERp57 and ERp72 share identity or similarity with each other but not with the PDI/PDIp/PDILT/ERp27 cluster.
The specificity of substrate binding by the b′ domains of the human PDI-family is poorly understood. The binding site in ERp57 is specialized for interacting with the tip of the P-domain of calreticulin and calnexin (Frickel et al, 2002; Russell et al, 2004; Pollock et al, 2004), whereas the binding of peptides to PDIp is dependent on the presence in the substrate of a single tyrosine or tryptophan residue with no adjacent negative charge (Ruddock et al, 2000). In the structural model of the PDI b′ domain (Pirneskoski et al, 2004), the hydrophobic binding pocket is large enough to accommodate only one or two hydrophobic amino-acid side chains, which is consistent with the single amino-acid specificity of PDIp. In addition, several unsatisfied backbone and side-chain hydrogen bonds were found in the immediate vicinity of the binding pocket. Therefore, this binding site seems well suited for interaction with non-native substrates that contain exposed hydrophobic groups and unsatisfied backbone hydrogen bonds; that is, proteins that might require the assistance of a protein-folding catalyst such as PDI.
Predicted functional properties
Although the name of the family implies that all members have a role in protein disulphide isomerization, only a subset are able to catalyse this reaction efficiently, whereas others are probably not directly involved in native disulphide bond formation.
There are four prominent determinants of the enzymatic activity of PDI-family members: the active-site sequence; the presence or absence of additional residues that also modulate the pKa of the active-site cysteines; the presence or absence of a glutamic acid–lysine charge pair that is involved in proton transfer reactions; and, for the ability to catalyse isomerization reactions, a high-affinity substrate-binding site in a non-catalytic domain (discussed above). The sequences of individual PDI proteins therefore allow predictions to be made about the types of reaction that they perform.
The most common active-site motif in human PDI-family members is CXHC, which is found in efficient thiol-disulphide oxidants of the ER and bacterial periplasm. This motif is present in PDI, PDIp, ERp57, ERp72, P5, ERp46, TMX3, one of the catalytic domains of ERdj5 and two domains of PDIr (Fig 1). By contrast, three of the active sites of ERdj5 contain a CXPC motif that is found in the thioredoxins, which are thiol-disulphide reductants. The surface-exposed amino-terminal cysteine of the CXXC motif of PDI is essential for any thiol-disulphide reaction and is missing from ERp27, ERp28, PDILT and TMX2. ERp44 lacks the C-terminal active-site cysteine that is required for many thiol-disulphide exchange reactions to proceed efficiently and whose presence makes mixed disulphide intermediates with substrates very transient. The N-terminal active-site cysteine of ERp44 thus forms more stable mixed disulphides, and by this mechanism it mediates the ER retention of proteins, including Ero1 and nascent secretory proteins (Anelli et al, 2002, 2003). The function of the non-thioredoxin-like C-terminal two-thirds of ERp44 is unknown, as are the features that determine its substrate-binding specificity.
Recently, a conserved arginine that is present in many members of the PDI family has been reported to modulate the pKa of the active-site cysteine residues by moving into and out of the active-site locale (Lappi et al, 2004). This motion has been implicated in the timing mechanism that allows a single catalyst to act as an efficient isomerase and oxidase of protein substrates, and to allow for the release of non-productive folding substrates. This arginine is important for the catalysis of oxidation by PDI, ERp57, ERp72 and P5 and is also conserved in most of the other PDI-family member a-like domains (Table 1).
In addition to a CXXC active site and a modulation of the pKa values of the active-site cysteines, efficient completion of the catalytic cycle for oxidation or reduction requires numerous proton transfer reactions both within the catalyst and to and from the substrate (Lappi et al, 2004). In the thioredoxins, a buried, charged glutamic acid–lysine pair that is located under the CXXC active site has been shown to be important for the catalytic activity of thioredoxin (Dyson et al, 1997) and for the oxidative activity of PDI and ERp57 (L.W.R., unpublished data). The glutamic acid—the presumed proton acceptor—of this charged pair is conserved in many of the PDI-family members (Table 1).
The presence of a CXHC active site in combination with the three other determinants of enzymatic activity suggests that PDI, PDIp, ERp57 and ERp72 are involved in disulphide bond oxidation and isomerization, which has been confirmed in vitro for PDI, ERp57 and ERp72 (see, for example, Darby et al, 1998; Frickel et al, 2004; Rupp et al, 1994). By contrast, ERp27, ERp28, PDILT and TMX2, which lack the CXXC active-site motif, are probably not involved directly in native disulphide bond formation. P5, TMX3 and ERp46, which all lack a b′-like domain but retain the other features and CXHC active sites, would be expected to be efficient oxidases, and ERdj5, which contains CXPC active sites, an efficient reductase. The unusual active site of TMX makes it difficult to predict its functional role, but the protein contains the other features that are required for the efficient catalysis of thiol-disulphide exchange reactions. The remaining family members, PDIr, ERp18 and TMX4, lack the glutamic-acid proton acceptor and therefore would be expected to be relatively inefficient catalysts of these reactions. This latter prediction is confirmed by in vitro data showing that ERp18 has only 15% of the oxidase activity of the a domain of PDI (Alanen et al, 2003), and PDIr has only 2% of the isomerase activity of PDI (Horibe et al, 2004) and 6% of its oxidase activity (L.W.R., unpublished data). It has been reported that the low isomerase activity of PDIr might be due to it acting on only a subset of proteins, including α1-antitrypsin (Horibe et al, 2004); however, it is noteworthy that a mutant PDIr with all of its active-site cysteines mutated to serine retained 57% of the activity of wild-type PDIr in the reactivation of α1-antitrypsin. This finding implies a reaction mechanism that is independent of thiol-disulphide exchange.
Future directions
Despite the large body of work on the activities of PDI-family members in vitro and in vivo there are still numerous unanswered questions regarding the physiological functions of the individual proteins and their mechanisms of action, especially within the complex environment of the ER.
The intransigence of catalytically active PDI-family members to crystallization is a major obstacle towards our understanding of them, as is the lack of assays with endogenous substrates for which the rate-limiting steps are well defined. Although some data on the relative activities of PDI-family members are appearing in the literature, a systematic comparison of the different activities that each family member from a single organism has towards a range of substrates is required. It will also be interesting to compare the set of PDI-family members present in different organisms, as this might teach us something about the co-evolution of these proteins and the need for individual organisms to catalyse specific processes.
The known presence of nearly 20 members of the PDI family in humans implies a complexity to the system that we are far from understanding. In particular, it will be necessary to determine whether the different enzymes either have overlapping or separate and distinct substrate specificities. Unfortunately, no gene knock-down studies of PDI-family members in mammalian tissue culture cells have been published. Whether this reflects that no effects have been observed or that it is too difficult at present to evaluate the effects of such experiments is not clear. Other obvious questions to address include a systematic evaluation of the transcriptional regulation of all of these proteins, the cellular mechanisms for regulation of their redox state and the physiological relevance of some unusual locations that have been reported for PDI-family members (for a review, see Turano et al 2002).
There is also a need to understand how PDI-family members interact with each other and with other ER-resident folding catalysts and chaperones. The presence of a J-domain in ERdj5 suggests that this protein cooperates with ER chaperones of the heat-shock protein 70 (Hsp70) family (Cunnea et al, 2003; Hosoda et al, 2003). There is already evidence for chaperone organization within the ER, and PDI, ERp72, P5 have been reported to form a complex with several ER chaperones and folding factors (Meunier et al, 2002). However, much more work is needed to determine the dynamics of such complexes, how they are formed, and what their effects are on the folding process. Other interaction partners that act as modulators also need to be identified.
PDI has been studied for more than 40 years, but the list of what we do not know about this enzyme is still long. With the discovery of novel proteins that belong to the PDI family, the field faces exciting challenges in the years to come.


Acknowledgments
The authors thank E. Frickel, R. Freedman, M. Molinari and members of the Ellgaard and Ruddock groups for critical reading of the manuscript. The authors also thank the Swiss National Research Foundation, ETH Zurich, Biocenter Oulu, Academy of Finland, University of Oulu and the Sigrid Juselius Foundation for financial support.
References
- Alanen HI, Williamson RA, Howard MJ, Lappi AK, Jäntti HP, Rautio SM, Kellokumpu S, Ruddock LW (2003) Functional characterisation of ERp18, a new endoplasmic reticulum located thioredoxin superfamily member. J Biol Chem 278: 28912–28920 [DOI] [PubMed] [Google Scholar]
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R (2002) ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J 21: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Alessio M, Bachi A, Bergamelli L, Bertoli G, Camerini S, Mezghrani A, Ruffato E, Simmen T, Sitia R (2003) Thiol-mediated protein retention in the endoplasmic reticulum: the role of Erp44. EMBO J 22: 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnewitz K, Guo C, Sevvana M, Ma Q, Sheldrick GM, Söling H-D, Ferrari DM (2004) Mapping of a substrate-binding site in the protein disulfide isomerase-related chaperone Wind based on protein function and crystal structure. J Biol Chem 279: 39829–39837 [DOI] [PubMed] [Google Scholar]
- Cunnea PM et al. (2003) ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J Biol Chem 278: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Darby NJ, Penka E, Vincentelli R (1998) The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J Mol Biol 276: 239–247 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Jeng MF, Tennant LL, Slaby I, Lindell M, Cui DS, Kuprin S, Holmgren A (1997) Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry 36: 2622–2636 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Riek R, Herrmann T, Güntert P, Braun D, Helenius A, Wuthrich K (2001) NMR structure of the calreticulin P-domain. Proc Natl Acad Sci USA 98: 3133–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari DM, Söling HD (1999) The protein disulphide-isomerase family: unraveling a string of folds. Biochem J 339: 1–10 [PMC free article] [PubMed] [Google Scholar]
- Freedman RB, Klappa P, Ruddock LW (2002) Catalytic and binding domains in the mechanism and specificty of protein disulphide isomerases: a theme with variations. EMBO Rep 3: 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel E-M, Riek R, Jelesarov I, Helenius A, Wüthrich K, Ellgaard L (2002) TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci USA 99: 1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel E-M et al. (2004) ERp57 is a multifunctional thiol-disulfide oxidoreductase. J Biol Chem 279: 18277–18287 [DOI] [PubMed] [Google Scholar]
- Horibe T, Gomi M, Iguchi D, Ito H, Kitamura Y, Masuoka T, Tsujimoto I, Kimura T, Kikuchi M (2004) Different contributions of the three CXXC motifs of human protein-disulfide isomerase-related protein to isomerase activity and oxidative refolding. J Biol Chem 279: 4604–4611 [DOI] [PubMed] [Google Scholar]
- Hosoda A, Kimata Y, Tsuru A, Kohno K (2003) JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J Biol Chem 278: 2669–2676 [DOI] [PubMed] [Google Scholar]
- Kemmink J, Darby ND, Dijkstra K, Nilges M, Creighton TE (1996) Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy. Biochemistry 35: 7684–7691 [DOI] [PubMed] [Google Scholar]
- Kemmink J, Darby ND, Dijkstra K, Nilges M, Creighton TE (1997) The folding catalyst protein disulfide isomerase is constructed of active and inactive thioredoxin modules. Curr Biol 7: 239–245 [DOI] [PubMed] [Google Scholar]
- Klappa P, Ruddock LW, Darby NJ, Freedman RB (1998) The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J 17: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Keller BO, Groenendyk J, Aldred S, Zheng J, Lemire BD, Li L, Michalak M (2003) ERp19 and ERp46, new members of the thioredoxin family of endoplasmic reticulum proteins. Mol Cell Proteomics 2: 1104–1119 [DOI] [PubMed] [Google Scholar]
- Lappi AK, Lensink M, Alanen HI, Salo KEH, Lobell M, Juffer A, Ruddock LW (2004). A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J Mol Biol 335: 283–295 [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Akiyama N, Nakamura H, Yodoi J, Noda M, Kizaka-Kondoh S (2001) Identification of a novel thioredoxin-related transmembrane protein. J Biol Chem 276: 10032–10038 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang C, Chen J, Peng S, Cao Y, Ying K, Xie Y, Mao Y (2003) Cloning and identification of a novel cDNA coding thioredoxin-related transmembrane protein 2. Biochem Genet 41: 99–106 [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood Y-K, Chung KT, Hendershot LM (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, van der Wal FJ, Bulleid NJ, High S (1997) Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science 275: 86–88 [DOI] [PubMed] [Google Scholar]
- Pirneskoski A, Klappa P, Lobell M, Williamson RA, Byrne L, Alanen HI, Salo KE, Kivirikko KI, Freedman RB, Ruddock LW (2004) Molecular characterisation of the principal substrate binding site of the ubiquitous folding catalyst protein disulphide isomerase. J Biol Chem 279: 10374–10381 [DOI] [PubMed] [Google Scholar]
- Pollock S, Kozlov G, Pelletier M-F, Trempe J-F, Jansen G, Sitnikov D, Bergeron JJ, Gehring K, Ekiel I, Thomas DY (2004) Specific interaction of ERp57 and calnexin determined by NMR spectroscopy and an ER two-hybrid system. EMBO J 23: 1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddock LW, Freedman RB, Klappa P (2000) Specificity in substrate binding by protein folding catalysts: tyrosine and tryptophan residues are the recognition motifs for the binding of peptides to the pancreas-specific protein disulfide isomerase PDIp. Protein Sci 9: 758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp K, Birnbach U, Lundström J, Van Nguyen P, Söling H-D (1994) Effects of CaBP2, the rat analog of ERp72, and of CaBP1 on the refolding of denatured proteins: comparison with protein disulfide isomerase. J Biol Chem 269: 2501–2507 [PubMed] [Google Scholar]
- Russell SJ, Ruddock LW, Salo KEH, Oliver JD, Roebuck QP, Llewellyn DH, Roderick HL, Koivunen P, Myllyharju J, High S (2004) The primary substrate binding site in the b′ domain of ERp57 is adapted for ER lectin association. J Biol Chem 279: 18861–18869 [DOI] [PubMed] [Google Scholar]
- Schwaller M, Wilkinson B, Gilbert HF (2003) Reduction–reoxidation cycles contribute to catalysis of disulfide isomerisation by protein-disulfide isomerase. J Biol Chem 278: 7154–7159 [DOI] [PubMed] [Google Scholar]
- Silvennoinen L, Myllyharju J, Ruoppolo M, Orrù S, Caterino M, Kivirikko KI, Koivunen P (2004) Identification and characterisation of structural domains of human Erp57. Association with calreticulin requires several domains. J Biol Chem 279: 13607–13615 [DOI] [PubMed] [Google Scholar]
- Sullivan DC, Huminiecki L, Moore JW, Boyle JJ, Poulsom R, Creamer D, Barker J, Bicknell R (2003) EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J Biol Chem 278: 47079–47088 [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano C, Coppari S, Altieri F, Ferraro A (2002) Proteins of the PDI family: unpredicted non-ER locations and functions. J Cell Physiol 193: 154–163 [DOI] [PubMed] [Google Scholar]
- Urade R, Okudo H, Kato H, Moriyama T, Arakaki Y (2004) ER-60 domains responsible for interaction with calnexin and calreticulin. Biochemistry 43: 8858–8868 [DOI] [PubMed] [Google Scholar]
- van Lith M, Hartigan N, Hatch J, Benham AM (2004) PDILT: a divergent testis-specific PDI with a non-classical SXXC motif that engages in disulfide dependent interactions in the endoplasmic reticulum. J Biol Chem [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJM, Thomas DY (1998) Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with Erp57. J Biol Chem 273: 6009–6012 [DOI] [PubMed] [Google Scholar]