Abstract
Restarting stalled replication forks partly depends on the break-induced recombination pathway, in which a DNA double-stranded break (DSB) is created on the stalled replication fork to initiate the downstream recombination cascades. Single-stranded DNA gaps accumulating on stalled replication forks are potential targets for endonucleases to generate DSBs. However, it is unclear how this process is executed and which nucleases are involved in eukaryotic cells. Here, we identify a novel gap endonuclease (GEN) activity of human flap endonuclease 1 (FEN-1), critical in resolving stalled replication fork. In response to replication arrest, FEN-1 interacts specifically with Werner syndrome protein for efficient fork cleavage. Replication protein A facilitates FEN-1 interaction with DNA bubble structures. Human FEN-1, but not the GEN-deficient mutant, E178A, was shown to rescue the defect in resistance to UV and camptothecin in a yeast FEN-1 null mutant.
Keywords: DNA replication fork, flap endonuclease 1, Wemer syndrome protein
Introduction
The DNA replisome frequently encounters DNA lesions, stable DNA secondary structures, and DNA-bound protein complexes, such as transcription factors (Cox, 2001). In each of these situations, the replisome disassembles and the DNA replication fork collapses. Mishandling or failure to repair the damaged replication fork can lead to mutagenesis, tumorigenesis or cellular lethality. Previous studies have shown that break-induced recombination (BIR) has a vital role in repairing stalled replication forks (Cox, 2001; Michel, 2000). In BIR, conversion of stalled replication forks into recombination substrates, DNA double-stranded breaks (DSBs), is the initial and critical step. It has been shown that extensive accumulation of single-stranded DNA (ssDNA) regions occurs in response to replication fork arrest (Sogo et al, 2002). These ssDNA regions were thought to be the sites cleaved by structure-specific endonuclease to trigger downstream BIR (Michel, 2000). However, it is unclear how this process is executed and which nucleases are involved in eukaryotic cells. Here, we provide in vitro and in vivo data, which demonstrate a novel gap endonuclease (GEN) activity of the flap endonuclease 1 (FEN-1) complexes and its possible role in processing stalled DNA replication forks.
Results And Discussion
Replication fork cleavage is a new property of FEN-1
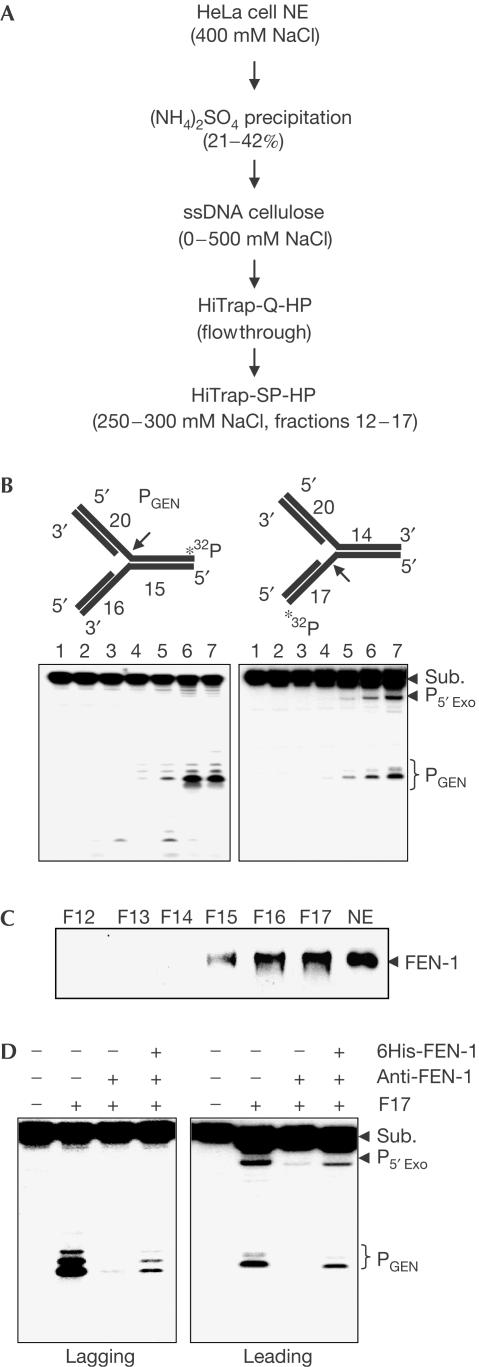
To identify the human GEN activity capable of cleaving replication forks, we fractionated HeLa cell nuclear extracts (Fig 1A) and partially purified a GEN activity that specifically cleaves DNA replication-fork-like structures at the ssDNA region on either the lagging (Fig 1B, left panel) or the leading (Fig 1B, right panel) strand template. However, GEN cleavage of the lagging strand was about threefold more efficient than that of the leading template strand. We also noted a 5′ blunt end exonuclease activity in the GEN fraction (Fig 1B). SDS–polyacrylamide gel electrophoresis (SDS–PAGE) of this partially purified GEN fraction showed a prominent band migrating at about 43 kDa, which was subsequently identified as FEN-1 by protein microsequencing (Mass Spectrometry Laboratory, City of Hope, National Medical Center) and further confirmed by western blotting (Fig 1C). Depletion of FEN-1 from the GEN sample with anti-FEN-1 protein A–agarose resulted in loss of the GEN activity, which was recovered by addition of recombinant human FEN-1 (Fig 1D). We detect no XPF, XPG or exonuclease 1 in western analysis of fractions containing GEN activity.
Figure 1.
Identification of GEN from HeLa cell nuclear extracts. (A) Purification scheme for the GEN activity. The specified fraction in each step contained the GEN activity. (B) GEN cleavage of 5′ (left) or 3′ (right) DNA double-stranded flap substrates (oligos 1, 2, 4, 5) by HiTrap-SP-HP fractions 12–17. A 500 ng portion of each fraction was assayed and the reaction was analysed with sequencing PAGE. Lanes 2–7 in both panels correspond to fractions 12–17, respectively. (C) Detection of FEN-1 protein in GEN fractions by western blotting with purified anti-FEN-1 antibody. (D) FEN-1 accounts for the identified GEN activity. FEN-1 was depleted from F17 with anti-FEN-1-bound protein A-agarose. Nuclease activities were assayed with 500 ng F17, 500 ng FEN-1-depleted F17 and 500 ng FEN-1-depleted F17 supplemented with 100 ng of purified recombinant human (h)FEN-1. NE, nuclear extract.
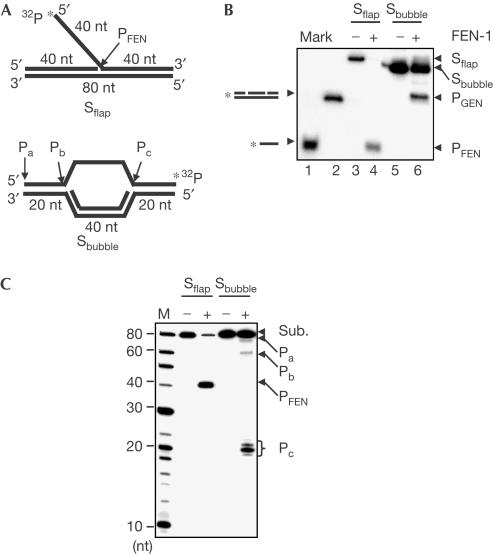
We then prepared a DNA bubble structure (Fig 2A), which resembles the stalled replication fork structure previously observed (Sogo et al, 2002). Incubating FEN-1 with DNA flap substrates generated characteristic flap cleavage products (Fig 2B, lane 4). Likewise, FEN-1 cleavage of DNA bubble structures generated a product (Fig 2B, lane 6), which co-migrates with the duplex DNA control mimicking the predicted GEN product (Fig 2B, lane 2). Notably, the cleavage of DNA flap substrate by FEN-1 alone was more efficient than that observed with the bubble substrate. We further mapped the cleavage site of DNA bubble structures by FEN-1 with a 15% sequencing gel. The result showed that the GEN activity of FEN-1 cleaved DNA bubble structures at ssDNA–dsDNA junctions producing products Pb and Pc (Fig 2B). Pb, being further processed by the FEN activity, was less than Pc. The exonuclease activity of FEN-1 also cleaved the 5′ end of bubble structures generating Pa (Fig 2B). Taken together, our data suggested that FEN-1 had intrinsic GEN activity, which cleaves DNA replication fork model structures in vitro. This was consistent with our previous finding showing that CRN-1, the FEN-1 homologue in Caenorhabditis elegans, cleaves DNA duplex structures with gaps mimicking the one in the apoptosis pathway (Parrish et al, 2003).
Figure 2.
Cleavage of DNA bubble and flap substrates by FEN-1. (A) DNA flap (oligos 7, 9, 15) and bubble substrates (oligos 7, 8, 13). (B) Analysis of FEN-1 cleavage of DNA substrates with native PAGE. A 2 pmol portion of human FEN-1 was incubated with 1 pmol of indicated DNA substrates. DNA substrates and products were analysed with native PAGE. Lanes 1 and 2 are ssDNA (oligo 16) and dsDNA (oligos 7, 8, 11, 12) markers, respectively. (C) Mapping of cleavage site of DNA substrates by FEN-1. Similar reactions to those in (B) were analysed with sequencing gel. Lane 1 is the molecular marker.
WRN significantly stimulates the GEN activity
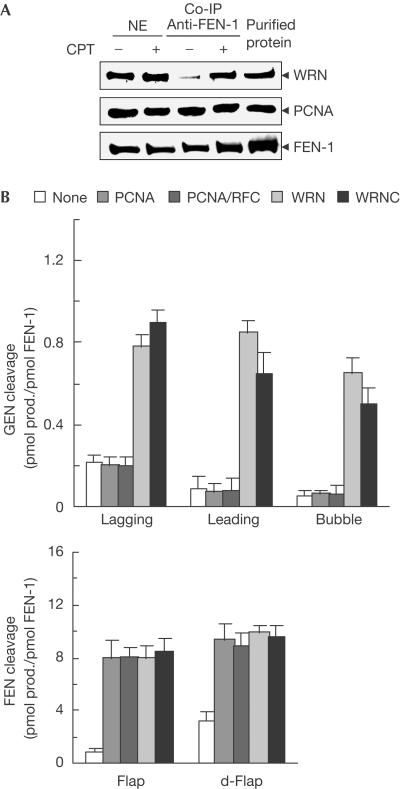
Next, we examined whether FEN-1 forms a complex with proteins that are involved in resolving stalled replication forks, such as Werner syndrome protein (WRN; Wu & Hickson, 2002), in response to DNA replication arrest. We assessed the amount of WRN and proliferative cell nuclear antigen (PCNA) that complex with FEN-1 in HeLa cells in response to camptothecin (CPT) treatment by using the co-immunoprecipitation assay. CPT specifically blocks replication forks by stabilizing the DNA–topoisomerase-1 complex (Hsiang et al, 1985). Our data showed that in response to CPT treatment, the FEN-1–WRN complex increased markedly, whereas the PCNA–FEN-1 complex showed little change (Fig 3A). There was little change in the levels of nuclear FEN-1 and WRN in response to CPT treatment, indicating that the increase of the FEN-1–WRN complex did not result from an increase of WRN or FEN-1 expression. Our observation was consistent with the study showing that exposure of cells to mitomycin C, a DNA crosslinker, induces FEN-1 and WRN colocalization (Sharma et al, 2004). In our in vitro endonuclease assays, the GEN activity of FEN-1 weakly cleaved lagging, leading or bubble substrate at rates of 0.22, 0.08 or 0.06 pmol product/pmol FEN-1, respectively, in the absence of WRN (Fig 3B; supplementary Fig S1 online). The cleavage of 5′ flap or double flap substrate by FEN-1 was at a significantly higher rate of 0.8 or 3.2 pmol product/pmol FEN-1, respectively. However, WRN significantly increased GEN cleavage of lagging, leading and bubble substrate to 0.8, 0.9 and 0.7 pmol product/pmol FEN-1, respectively (Fig 3B; supplementary Fig S1 online). WRN stimulated FEN-1 cleavage of 5′ flap substrates as previously reported (Brosh et al, 2001). In addition, the GEN and FEN stimulation fold increased as the concentration of FEN-1 decreased (data not shown). The stimulation did not require the helicase or 3′ exonuclease activities of WRN, as WRNC—the WRN carboxy-terminal region that interacts with FEN-1 (Brosh et al, 2001)—showed similar stimulation as the full-length WRN (Fig 3B; supplementary Fig S1 online). Interestingly, PCNA failed to stimulate FEN-1's GEN cleavage of either DNA substrates with gaps in the presence of the clamp loader replication factor C (RFC), which ensures the appropriate load of PCNA onto bubble substrates, but it strongly stimulated the FEN cleavage of the flap substrate (Fig 3B; supplementary Fig S1 online).
Figure 3.
WRN interacts with FEN-1 and stimulates its GEN activity. (A) Immunoprecipitation of FEN-1. Nuclear extracts from CPT-treated or untreated HeLa S3 cells were incubated with purified polyclonal anti-FEN-1-bound protein A–agarose. The precipitated FEN-1, WRN and PCNA were detected with monoclonal anti-FEN-1, anti-WRN and anti-PCNA, respectively. The input levels of FEN-1, WRN and PCNA were also examined. The purified recombinant FEN-1, WRN and PCNA were included as a control. (B) WRN and PCNA effects on GEN and FEN activities of FEN-1. The GEN activity was assayed by incubation of 1 pmol of FEN-1 with 1 pmol of 5′ and 3′ double-stranded flap (lagging (oligos 1, 2, 4, 6) and leading (oligos 1, 3, 4, 5) substrates, respectively) or bubble substrates in the absence or presence of 5 pmol of PCNA, WRN, WRNC or 50 U RFC. FEN activity was assayed by incubation of 0.1 pmol of FEN-1 with 1 pmol of normal flap and double flap (d-flap, oligos 7, 10, 15) substrates in the absence or presence of 5 pmol of PCNA, WRN, WRNC or 50 U RFC. The DNA substrates and products were resolved with sequencing gel and quantified with the Imagine Quante program. Values are an average of three independent experiments.
RPA recruits FEN-1 to DNA bubble structures
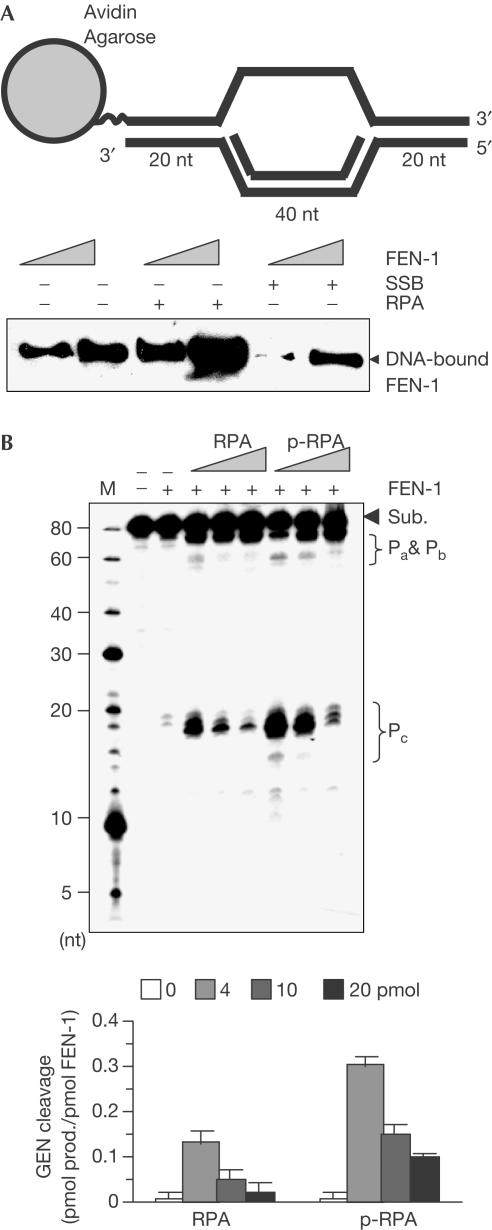
Next, we investigated whether RPA, a eukaryotic ssDNA-binding and FEN-1 interaction protein (Khlimankov et al, 2002), affected the binding and cleavage of DNA bubble structures by FEN-1 in vitro. Our data showed that RPA strongly stimulated FEN-1 binding to the DNA bubble structure, whereas Escherichia coli ssDNA-binding protein (SSB) inhibited the interaction (Fig 4A). Nuclease assays showed that in the presence of 4 pmol RPA, FEN-1 cleavage of DNA bubble structures was stimulated; however, the stimulation decreased at 10 and 20 pmol RPA (Fig 4B). We further investigated the effects of phosphorylated RPA (p-RPA) on the GEN activity of FEN-1, because it was reported that replication fork arrest led to phosphorylation of RPA (Shao et al, 1999). We found that p-RPA strongly stimulates the GEN activity (Fig 4B). The stimulation by p-RPA was more effective than p-RPA. We also noted that RPA/p-RPA enhanced the cleavage at all Pa, Pb and Pc sites, which represents a unique cleavage pattern of bubble structures by the GEN activity of FEN-1 (Fig 4B). These results suggested that the concentration and the phosphorylation of RPA were important in modulation of the GEN activity of FEN-1.
Figure 4.
RPA effects on FEN-1 bubble DNA substrate binding and cleavage. (A) RPA stimulates FEN-1 binding to DNA bubble structures. DNA bubble substrate-bound agarose beads were left uncoated or coated with RPA or E. coli SSB. The beads were then incubated with 5 and 10 pmol of human (h)FEN-1. Binding of FEN-1 to the agarose beads was evaluated with western blotting using antibody against FEN-1. (B) RPA and p-RPA concentration effect on the GEN activity of FEN-1. A 2 pmol portion of FEN-1 was incubated with 1 pmol of DNA substrates in the presence of 4, 10 and 20 pmol of RPA or p-RPA for 30 min at 30°C. GEN cleavage was quantified (lower panel).
The GEN resolves stalled replication forks
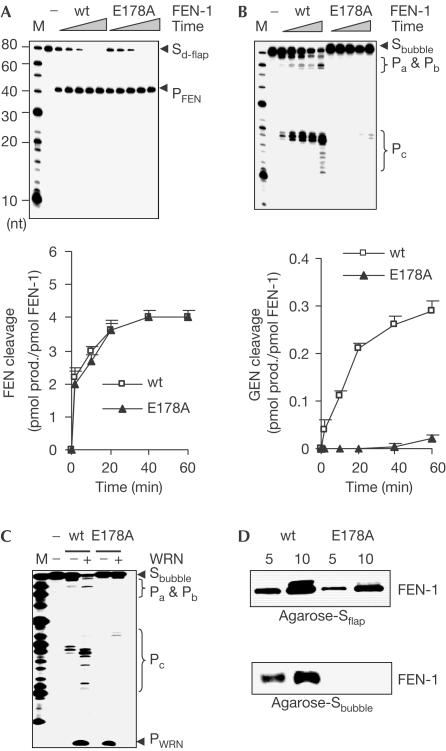
To address whether the GEN activity of FEN-1 is involved in processing stalled replication forks in vivo, we sought to identify a mutation that specifically eliminates the GEN activity of FEN-1. By screening a pool of more than 50 point mutations of FEN-1, we found that the E178A mutation in FEN-1 abolished more than 95% of the GEN activity, but retained the FEN activity cleaving 5′ double flap substrates (Fig 5A,B). This result indicated that the two endonuclease activities could be segregated. Even in the presence of WRN, the GEN cleavage of bubble structures by E178A was less than 3% (Fig 5C). Substrate-binding assays indicated that E178A retained the ability to bind flap substrates, but lost binding capacity to DNA bubble structures (Fig 5D). It suggested that FEN-1 might have distinct substrate-binding modes, depending on the substrate structure. With flap substrates, FEN-1 recognized and tracked down the 5′ ssDNA flap strand to the ssDNA–dsDNA junction for cleavage (Murante et al, 1995), whereas in DNA substrates with gaps lacking 5′ free DNA end, FEN-1 directly bound to the ssDNA region.
Figure 5.
E178A is a GEN-deficient FEN-1 mutant. (A,B) Cleavage of DNA double flap and bubble substrates by wt FEN-1 and E178A. A 0.5 or 5 pmol portion of FEN-1 proteins was incubated with 2 pmol of DNA double flap (A) or bubble substrates (B), respectively, for 2, 10, 20, 40 and 60 min at 37°C. GEN and FEN cleavages were quantified (lower panel). (C) WRN effect on wt FEN-1 and E178A cleavage of DNA bubble structures. A 2 pmol portion of FEN-1 was incubated with 1 pmol of bubble structure DNA in the absence or presence of 10 pmol WRN. (D) DNA substrate binding by wt FEN-1 and E178A. Agarose-bound biotinylated DNA flap or bubble substrate was incubated with 5 or 10 pmol of FEN-1 proteins. FEN-1 bound to agarose was evaluated with western blotting.
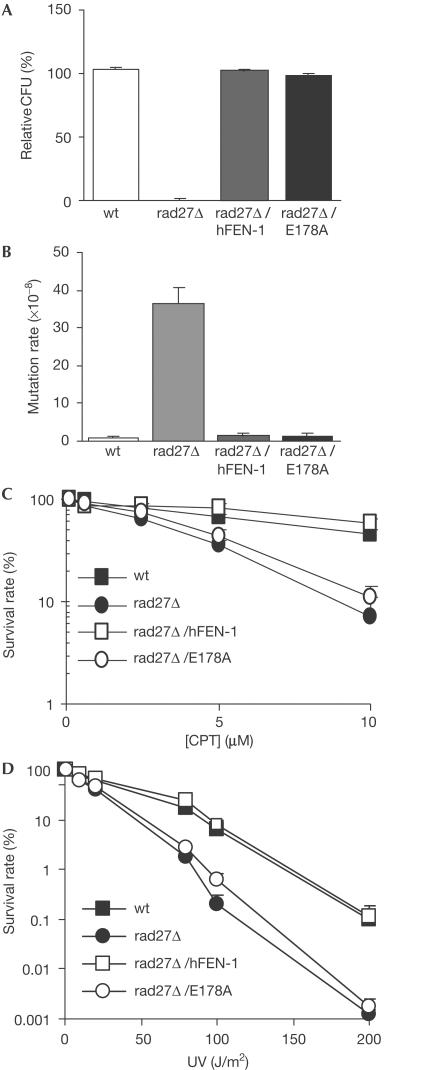
To test the specific function of the GEN activity of FEN-1 in resolving stalled replication forks, we complemented rad27 null mutant (rad27Δ) yeast with wild-type (wt) and GEN-deficient (E178A) human FEN-1. The rad27Δ strain expressing E178A (rad27Δ/E178A) grew as well as the wt strain at 30°C and at the restrictive 37°C (Fig 6A). Furthermore, the spontaneous Lys+ reverse mutation rate of the rad27Δ/E178A strain was similar to that of the wt strain, but was about 30-fold less than that of the rad27Δ strain (Fig 5B). Of these Lys+ reverse mutations in the rad27Δ/E178A strain, similar to the wt strain, only 7% of the mutations were duplication mutations, compared with 67% in the rad27Δ strain. This suggested that E178A, with a similar FEN activity as the wt enzyme, could fully substitute for RAD27's function in RNA–DNA primer removal. The rad27Δ strain had similar CPT sensitivity to that seen in the sgs1 null strain lacking the yeast WRN homologue (data not shown). Furthermore, the rad27Δ/E178A strain showed similar CPT sensitivity to that of the rad27Δ strain, whereas the rad27Δ/human FEN-1 and the wt strains were both resistant to CPT treatment (Fig 6C). Ultraviolet (UV) irradiation also causes DNA crosslinks and stalls the DNA replication fork (Limoli et al, 2002). Deletion of rad27 caused yeast to be sensitive to UV irradiation (Reagan et al, 1995). Here, we found that the GEN activity is critical in FEN-1-mediated UV resistance, because the rad27Δ/E178A strain showed similar UV sensitivity to that of the rad27Δ strain, whereas the rad27Δ/human FEN-1 and the wt yeast strains showed similar resistance to UV irradiation (Fig 6D).
Figure 6.
Complementation study of wt FEN-1 and the GEN-deficient mutant E178A. (A) Temperature sensitivity of different yeast strains. The temperature sensitivity was expressed as relative colony-forming units (CFU) grown at 37°C over those grown at 30°C. (B) Lys+ reverse mutation rate of different yeast strains. Values represent an average of the mutation rate for 28 independent colonies of each strain. (C) CPT sensitivity of different yeast strains. The CPT sensitivity was measured as survival of cells treated with indicated concentration of CPT over survival of untreated cells. (D) UV irradiation sensitivity of different yeast strains. UV sensitivity was measured as survival of cells exposed to the indicated intensity of UV irradiation over nonexposure survival of cells.
Our current study suggests that FEN-1 is involved in processing stalled replication forks through a cellular mechanism that is alternative to the recently identified Rad51C-mediated Holliday junction cleavage (Liu et al, 2004). FEN-1, in complex with replication fork repair protein WRN, directly cleaves stalled replication forks by the GEN activity to initiate the recombination repair pathway. It is consistent with previous studies showing that in yeast, FEN-1 is synthetically lethal with nuclease complex Mus81–Mms4 (Tong et al, 2001), which cleaves the leading template strand of a synthetic replication fork to generate DSBs in vitro (Kaliraman et al, 2001). Conversely, the suggestion that the FEN-1–WRN complex initiates recombination seems to be contradictory to the observations that WRN and FEN-1 are both anti-recombination proteins (Tishkoff et al, 1997; Gangloff et al, 2000). However, WRN may both promote and suppress DNA recombination as its prokaryotic homologue, E. coli Rec Q helicase, has been shown to both initiate and inhibit DNA recombination depending on the growth condition (Harmon & Kowalczykowski, 1998). Similarly, under normal growth condition, FEN-1, with the assistance of PCNA, efficiently cleaves RNA–DNA flaps to avoid DSB generation during DNA replication, thus suppressing DNA recombination (Tishkoff et al, 1997). In response to DNA damage during DNA replication, FEN-1 interacts with WRN to cleave the replication forks efficiently, switching its function to initiation of BIR.
Methods
Yeast strains, genetic manipulation and genotoxic assays. The rad27Δ, rad27Δ/human FEN-1 and rad27Δ/E178A Saccharomyces cerevisiae strains were constructed as previously reported (Frank et al, 1998). The growth rate and temperature sensitivity were measured as previously reported (Frank et al, 1998). The Lys2-Bgl mutation rate and spectrum analysis were carried out as previously reported (Tishkoff et al, 1997). In the mutation spectrum analysis, DNA sequences of the Lys2 gene from 28 independent Lys2+ revertants from each genetic background were analysed. The UV sensitivity was measured as the survival rate of yeast cells on exposure to UV irradiation as described previously (Reagan et al, 1995). The CPT sensitivity was determined using a modified procedure as described previously (Pouliot et al, 1999). Experiments were repeated at least three times independently.
Fractionation of native GEN from HeLa S3 cells. Nuclear extracts of HeLa S3 cells (National Cell Culture Center) were prepared and fractionated by ammonium sulphate precipitation as previously described (Ramilo et al, 2002). The GEN fraction was resuspended in buffer A (25 mM Hepes, pH 7.5, 0.1 mM EDTA, 2 mM dithiothreitol, protein inhibitor cocktails (Roche Applied Science, Indianapolis, IN, USA) and 25 mM KCl) and applied onto an ssDNA cellulose column (Sigma, St Louis, MO, USA), followed by washing and eluting with buffer A containing 0–1 M NaCl. The GEN fraction was further fractionated by a fast-performance liquid chromatography system harbouring a HiTrap-Q-HP or HiTrap-SP-HP column (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer's instructions. Each fraction was analysed with SDS–PAGE and nuclease assay.
Recombinant protein purification, nuclease activity and DNA binding assay. Expression and purification and preparation of recombinant human FEN-1, RPA, WRN and WRNC were performed on the basis of previously published protocols (Brosh et al, 2001; Yannone et al, 2001; Zheng et al, 2002). RPA was phosphorylated by DNA–protein kinase and purified as previously described (Oakley et al, 2003). RFC was a gift from the Hurwitz laboratory (Memorial Sloan-Kettering Cancer Center, New York, NY, USA). Synthetic DNA substrates were prepared and nuclease activity assays conducted as previously described (Zheng et al, 2002) and oligo sequences of DNA substrates are listed in supplementary Table S1 online and specified in the figure legends. Briefly, FEN-1 was incubated with indicated DNA substrates in 50 mM Tris–HCl, pH 8.0, 50 mM NaCl and 5 mM Mg2+ at 37°C for 30 min if not specified. Nuclease assays with RPA/p-RPA were carried out at 30°C to avoid potential transient breathing of DNA duplex region caused by RPA. For native PAGE analysis, DNA substrates and products were extracted with an equal volume of phenol and chloroform (1:1), followed by analysis with 5% native PAGE. For sequencing PAGE analysis, DNA substrate and products were denatured with 95% formamide, 20 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol and analysed with 15% denaturing PAGE. The interaction of FEN-1 with DNA substrates was analysed as previously described (Zheng et al, 2002).
Co-immunoprecipitation of FEN-1–WRN and FEN-1–PCNA complexes. HeLa cells were synchronized at the G1/S phase boundary by serum starvation for 72 h and then released into S phase by replacement of fresh DMEM containing 10% serum. Cells were treated or untreated with 10 μM CPT (Sigma) for 6 h. The nuclear proteins were extracted and immunoprecipitated according to a previously published procedure with purified polyclonal anti-FEN-1 antibody (Partridge et al, 2003).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n1/extref/7400313s1.pdf).
Supplementary Material
Supplementary Fig S1
Acknowledgments
We thank Dr J. Hurwitz for providing purified recombinant RFC. We thank Dr M. Lieber for his expert advices for using the appropriate DNA bubble structures mimicking DNA replication forks. We also thank Dr S. Alas and Dr P. Singh in the Shen laboratory for their discussions and technical assistance. This work was supported by NIH grants R01CA085344 and R01CA073763 to B.H.S.
References
- Brosh RM Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA (2001) Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J 20: 5791–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM (2001) Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu Rev Genet 35: 53–82 [DOI] [PubMed] [Google Scholar]
- Frank G, Qiu J, Somsouk M, Weng Y, Somsouk L, Nolan JP, Shen B (1998) Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J Biol Chem 273: 33064–33072 [DOI] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25: 192–194 [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev 12: 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang YH, Hertzberg R, Hecht S, Liu LF (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem 260: 14873–14878 [PubMed] [Google Scholar]
- Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ (2001) Functional overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes Dev 15: 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlimankov D, Rechkunova NI, Khodyreva SN, Petruseva IO, Nazarkina Zh K, Belousova EA, Lavrik OI (2002) Interaction of replication protein A and flap endonuclease 1 with DNA duplexes containing a nick or flap. Mol Biol (Mosk) 36: 1044–1053 [PubMed] [Google Scholar]
- Limoli CL, Laposa R, Cleaver JE (2002) DNA replication arrest in XP variant cells after UV exposure is diverted into an Mre11-dependent recombination pathway by the kinase inhibitor wortmannin. Mutat Res 510: 121–129 [DOI] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O'Regan P, West SC (2004) RAD51C is required for Holliday junction processing in mammalian cells. Science 303: 243–246 [DOI] [PubMed] [Google Scholar]
- Michel B (2000) Replication fork arrest and DNA recombination. Trends Biochem Sci 25: 173–178 [DOI] [PubMed] [Google Scholar]
- Murante RS, Rust L, Bambara RA (1995) Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem 270: 30377–30383 [DOI] [PubMed] [Google Scholar]
- Oakley GG, Patrick SM, Yao J, Carty MP, Turchi JJ, Dixon K (2003) RPA phosphorylation in mitosis alters DNA binding and protein–protein interactions. Biochemistry 42: 3255–3264 [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Yang C, Shen B, Xue D (2003) CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J 22: 3451–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JJ, Lopreiato JO Jr, Latterich M, Indig FE (2003) DNA damage modulates nucleolar interaction of the Werner protein with the AAA ATPase p97/VCP. Mol Biol Cell 14: 4221–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, Nash HA (1999) Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286: 552–555 [DOI] [PubMed] [Google Scholar]
- Ramilo C, Gu L, Guo S, Zhang X, Patrick SM, Turchi JJ, Li G-M (2002) Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Mol Cell Biol 22: 2037–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan MS, Pittenger C, Siede W, Friedberg EC (1995) Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol 177: 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y (1999) Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J 18: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM Jr (2004) WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol Biol Cell 15: 734–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Tishkoff DX, Filosi N, Gaida GM, Kolodner RD (1997) A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88: 253–263 [DOI] [PubMed] [Google Scholar]
- Tong AH et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2002) RecQ helicases and cellular responses to DNA damage. Mutat Res 509: 35–47 [DOI] [PubMed] [Google Scholar]
- Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ (2001) Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem 276: 38242–38248 [DOI] [PubMed] [Google Scholar]
- Zheng L, Li M, Shan J, Krishnamoorthi R, Shen B (2002) Distinct roles of two Mg2+ binding sites in regulation of murine flap endonuclease-1 activities. Biochemistry 41: 10323–10331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig S1