Abstract
The F-actin crosslinker filamin from Dictyostelium discoideum (ddFLN) has a rod domain consisting of six structurally similar immunoglobulin domains. When subjected to a stretching force, domain 4 unfolds at a lower force than all the other domains in the chain. Moreover, this domain shows a stable intermediate along its mechanical unfolding pathway. We have developed a mechanical single-molecule analogue to a double-jump stopped-flow experiment to investigate the folding kinetics and pathway of this domain. We show that an obligatory and productive intermediate also occurs on the folding pathway of the domain. Identical mechanical properties suggest that the unfolding and refolding intermediates are closely related. The folding process can be divided into two consecutive steps: in the first step 60 C-terminal amino acids form an intermediate at the rate of 55 s−1; and in the second step the remaining 40 amino acids are packed on this core at the rate of 179 s−1. This division increases the overall folding rate of this domain by a factor of ten compared with all other homologous domains of ddFLN that lack the folding intermediate.
Keywords: atomic force microscope, filamin, folding kinetics
Introduction
An actin-crosslinking protein is constantly subject to mechanical forces as the cytoskeleton rearranges itself during cell division or movement (Pollard & Borisy, 2003). In the F-actin crosslinker filamin from Dictyostelium discoideum (ddFLN), among the six immunoglobulin rod domains, domain 4 shows significantly lower unfolding forces than all the other domains. In addition, this domain also unfolds via an intermediate where the 60 carboxy-terminal residues form a folded core (Schwaiger et al, 2004). In an earlier study, low unfolding forces suggested a role for domain 4 (ddFLN4) as an extensible link that is able to buffer high-tension forces before they can do damage to the structural integrity of the cytoskeletal meshwork (Schwaiger et al, 2004). A key question that has remained unresolved is whether the occurrence of a stable unfolding intermediate has any influence on the folding properties of this domain. So far, many mechanical single-molecule studies have given detailed insight into unfolding pathways along the N–C-terminal reaction coordinate in the high-dimensional protein energy landscape biased by an external force (Brockwell et al, 2003; Carrion-Vazquez et al, 2003; Williams et al, 2003; Zhuang & Rief, 2003; Schlierf et al, 2004). What can we learn about folding pathways of proteins from single-molecule mechanical experiments?
To detect intermediates in classical double-jump stopped-flow studies, experimenters switch solvent conditions rapidly from denaturing to renaturing for a varying time span, and then back to denaturing to follow the accumulation of intermediate states (Eaton et al, 2000). Inspired by this technique, we have developed a single-molecule mechanical double-jump experiment, where the applied force allows switching rapidly between denaturing and renaturing conditions. Here, we use this technique to follow, in detail, the folding pathway and kinetics of the fast-folding immunoglobulin domain ddFLN4.
Results And Discussion
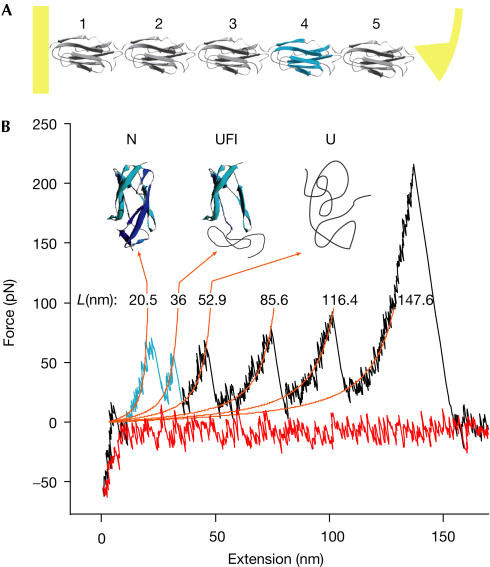
The experimental geometry of mechanical unfolding experiments is shown in Fig 1A. All mechanical experiments described here were performed with a rod construct from ddFLN comprising immunoglobulin rod domains 1–5 (ddFLN1–5, ddFLN4 is coloured blue in Fig 1A). A mechanical unfolding sample trace in which four of the five immunoglobulin domains unfold is given in Fig 1B. Most domains unfold in a clear two-state manner and only domain 4 shows a characteristic double peak (marked in blue) due to the presence of an intermediate along its unfolding pathway (Schwaiger et al, 2004). The first unfolding peak reflects detachment and unfolding of strands A and B, whereas strands C–G remain structured (Schwaiger et al, 2004). The second peak marks complete unfolding (Fig 1B).
Figure 1.
Single-molecule mechanics of ddFLN1–5. (A) Structure of the ddFLN1–5 construct comprising immunoglobulin domains 1–5 including domain 4 (blue). (B) Force–extension curve of ddFLN1–5 unfolding. A total of four immunoglobulin domains are unfolded. Domain 4 unfolding (highlighted in blue) proceeds by means of a stable unfolding intermediate, whereas other immunoglobulin domains are two-state unfolders. (Nuclear magnetic resonance structures are adapted from Fucini et al (1997) and Schwaiger et al (2004).) Orange lines are worm-like chain fits using P=0.5 nm and contour lengths as indicated above the peaks (Bustamante et al, 1994).
Refolding of ddFLN4 in single-molecule mechanical experiments occurs at a faster rate compared with all other immunoglobulin domains in ddFLN1–5 if the polypeptide chain is relaxed to almost zero tension (Schwaiger et al, 2004). Stretch and relax experiments can provide direct insight into the folding pathway of ddFLN4. In the experimental series shown in Fig 2A, a single ddFLN4 domain is stretched and relaxed repeatedly at high pulling velocities (2 μm/s) so that the time allowed for the polypeptide strand to refold is short (<50 ms).
Figure 2.
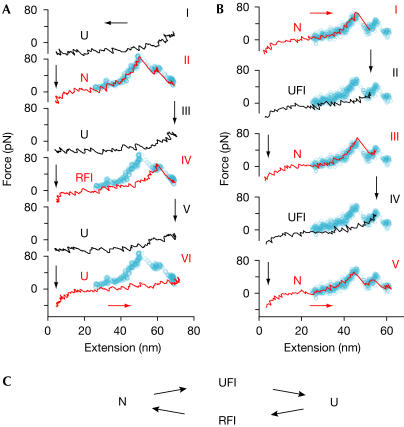
Domain 4 of ddFLN has a stable refolding intermediate. (A) Mechanical refolding experiments with ddFLN4. Three typical outcomes from a series of 50 stretch and relax cycles with a single ddFLN4 domain. Black traces represent relaxation curves and red traces stretching curves. Starting from the unfolded state U (traces I, III and V), ddFLN4 can either fold back to the native state N (II), to a folding intermediate RFI (IV) or not fold at all (VI). The blue pattern superimposed in the background is a complete unfolding pattern of ddFLN4. (B) Refolding experiments from the unfolding intermediate. (I) Unfolding trace, starting from the native state N and ending in the unfolding intermediate state UFI. (II) Domain 4 is relaxed and allowed to refold. (III) Subsequent stretching shows that ddFLN4 has folded back to the native state N from UFI. This experiment is repeated in traces IV and V. (C) Kinetic scheme of the mechanical unfolding and refolding pathways of ddFLN4.
Three different outcomes of ddFLN4 refolding experiments can be observed: complete refolding (Fig 2A, trace II), refolding only to an intermediate structure (Fig 2A, trace IV) or no refolding within the short time span (Fig 2A, trace VI). Obviously, an intermediate state is populated not only during mechanical unfolding experiments but also during refolding. A priori, there is no reason to assume that this refolding intermediate is identical to the mechanical unfolding intermediate we observe in Fig 1. We will therefore refer to the intermediate state populated during mechanical unfolding from the native state as UFI and to the intermediate populated during refolding experiments as RFI (Fig 2C).
We cannot yet decide whether RFI is on or off the folding pathway. However, we can directly show that, from UFI as a starting conformation, the domain refolds quickly to the native state N. Such a series is shown in Fig 2B. In the first trace (Fig 2B, trace I), we unfold a completely folded ddFLN4 domain to UFI. Instead of further rupturing the domain, we now relax the polypeptide chain to a near zero force (Fig 2B, trace II) and immediately stretch it again (Fig 2B, trace III). In such experiments, we observe that the domain always folds into its native conformation at a timescale of <50 ms.
Investigation of protein refolding kinetics of multidomain constructs in mechanical experiments has produced considerably (up to 30 times) lower refolding rates than solution studies for domain I27 of titin. Carrion-Vazquez et al (1999) have explained this discrepancy by entropic costs of tethering. Small mechanical forces can drastically slow refolding. Also, in multidomain constructs, gradual shortening of the polypeptide chain due to the sequential refolding of the domains leads to increasing force constraints for those domains that fold later in the process. As the force–distance relation of a polypeptide chain in the relevant force range below 5 pN is not known and may be sequence specific, such effects cannot be accurately accounted for.
Quantitative investigation of the refolding of ddFLN4 therefore relies on three important prerequisites: (i) refolding experiments have to be performed with only a single domain, (ii) switching between unfolding and refolding conditions has to occur fast, and (iii) residual strain within the polypeptide chain during refolding must be minimized. We thus developed a mechanical pulse protocol that allows rapid switching between renaturing and denaturing conditions by subjecting the protein to a mechanical force.
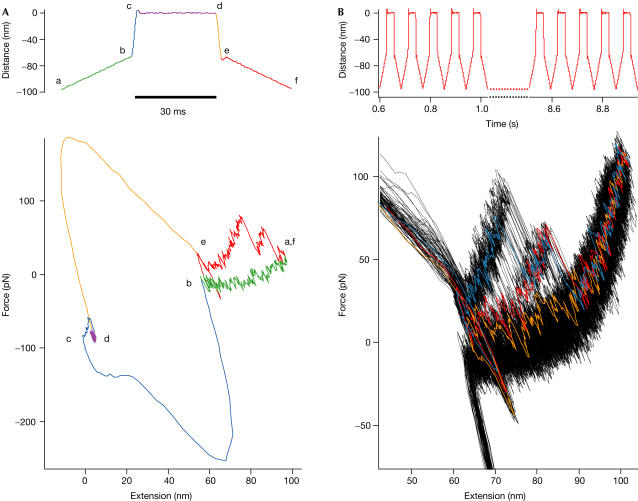
In the initial cycle, the tip indents into the surface to contact a single molecule. After a waiting time of 1 s, it is then retracted to about 100 nm above the surface. At this distance, three domains of the protein will be unfolded. In many cases, domain 4 will be among those three domains. As two of the domains fold slowly compared with ddFLN4, they serve as sacrificial domains that will stay unfolded throughout the experiment and provide a long-enough polypeptide polymer spacer such that the entropic costs of tethering for refolding of ddFLN4 will be minimal. Now, periodic folding–refolding cycles can be started. The time course of this pulse protocol is shown in Fig 3A (upper panel). From the fully extended position (position a), the polypeptide chain is relaxed to a distance of about 70 nm above the surface (position b) at a speed of 2 μm/s. At this position, the polypeptide chain is still sufficiently strained so that refolding does not occur. From here, the tip is then rapidly approached to the surface (position c) within 2 ms to start refolding. To minimize tensile strain on the polypeptide chain, the tip indents slightly into the surface with a force of <150 pN. After a variable waiting time of 5–40 ms, during which the protein is allowed to attempt refolding under minimal strain (positions c and d), the position is rapidly (2 ms) switched back to position e, again 70 nm above the surface. At this position, refolding will be mechanically quenched and the amount of structure that has refolded can now be probed in a force versus distance curve at 2 μm/s up to the initial position (f).
Figure 3.
Double-jump mechanical single-molecule experiment. (A) Time course of the mechanical extension (upper left) and corresponding force-extension curve of a single double jump. (B) Time course of the mechanical extension for a typical experiment running through 50 unfolding–refolding cycles (upper right). Lower right: superposition of the corresponding force-extension traces. Sample traces for the three possible outcomes of the refolding experiments are coloured in blue (complete refolding), red (refolding to the intermediate RFI) and orange (no refolding).
The rapid switching phases are essential: if approach and retraction phases took an essential fraction of the total refolding time allowed, the protein would refold under ill-defined force conditions and quantitative analysis would be impossible. During the switching phases (b,c and d,e), the cantilever probe is subject to large hydrodynamic forces visible in the sample traces (Fig 3A, lower panel). These phases will therefore impose a ‘blind window' on our experiment.
The protocol described above is a close single-molecule analogue of bulk double-jump stopped-flow techniques. The superposition of 50 cycles from a single ddFLN4 domain shown in Fig 3B (lower panel) demonstrates that we can obtain statistically significant amounts of data and force distributions from one individual protein domain. So far, in single-molecule force measurements, unbinding or unfolding force data from different individual domains are compiled into histograms. Such measurements assume that all molecules behave the same and any different local environments are generally averaged over.
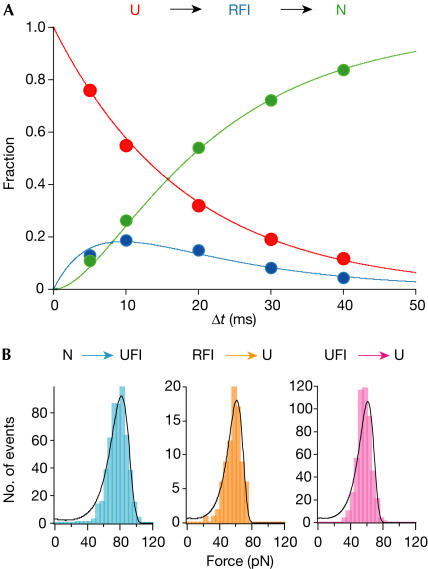
Varying the time allowed for ddFLN4 to refold from 5 to 40 ms and counting the events that lead to either complete refolding (green circles), refolding to the intermediate (blue circles) or no refolding (red circles) now allows us to reconstruct the complete refolding pathway of ddFLN4 (Fig 4A, graph). The data closely follow a kinetic scheme in which the refolding intermediate RFI is obligatory (Fig 4A, top) and is populated at a rate of 55±4 s−1 from the unfolded state. The transition from the intermediate to the completely folded conformations occurs at 179±20 s−1 (see Methods). Models in which the intermediate lies off-pathway fail to describe our data. The total folding rate we find for ddFLN4 of 42 s−1 is among the fastest observed for an immunoglobulin domain so far. This is even more surprising because in our experimental geometry the molecule is tethered between tip and substrate, and refolding should rather be slower than in solution studies.
Figure 4.
Single-molecule folding and unfolding kinetics of ddFLN4. (A) Fraction of observed events for mechanical refolding as a function of the allowed refolding time (red: no refolding; green: complete refolding; blue: refolding to RFI). For each time point at 5, 10, 20, 30 and 40 ms, 1,377, 404, 417, 536 and 270 events were analysed, respectively. The lines are fits according to the kinetic folding scheme shown at the top with k1=55±4 s−1 and k2=179±20 s−1 (see Methods). (B) Histograms of unfolding forces for unfolding from native state N to UFI (left), from RFI to unfolded state U (middle) and from UFI to U (right). The solid lines in the left and middle histograms are fits according to a two-state model (see text and Methods).
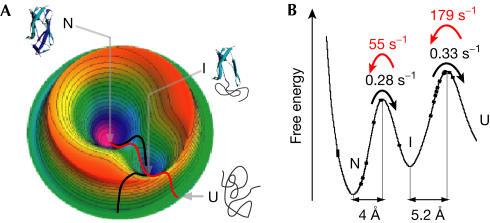
An obligatory intermediate occurs both on the mechanical unfolding pathway (UFI) and on the refolding pathway (RFI; Fig 2C). However, we have experimental evidence that RFI and UFI are closely related if not identical. Although single-molecule mechanical experiments do not provide direct structural information, they provide many parameters that sensitively depend on protein structure (Li et al, 2000). The core of folded amino acids constituting those amino acids that form the structured portion of the unfolding intermediate UFI has been mapped previously (Schwaiger et al, 2004). By analysing the length gain upon unfolding, we find that the number of amino-acid residues constituting RFI is identical to the number we obtain for UFI (60 residues). Other parameters that sensitively depend on protein structure are the average unfolding force and the width of the measured force distribution. We find that the unfolding force distribution is identical for UFI (Fig 4B, right histogram) and RFI (Fig 4B, middle histogram). The shape of the unfolding force distribution is determined by the unfolding rate at zero force koff and the position of the transition state along the direction of the force applied Δx (Evans & Ritchie, 1999). From the fit to the measured unfolding force distribution of RFI (solid line in Fig 4B, middle histogram), we obtain values of koff=0.33 s−1 and Δx=5.25±0.4 Å. The same parameters exactly describe the unfolding force distribution of UFI (solid line in Fig 4B, right histogram). As unfolding force distributions generally change strongly on changes in structure or on mutations (Li et al, 2000), we conclude that, at the resolution of our experiments, RFI and UFI are indistinguishable. Our finding that refolding from UFI to the native state is fast (Fig 2B) supports this conclusion. The left histogram in Fig 4B shows the force distribution for the unfolding transition from the native state to the intermediate state of ddFLN4. In this transition, strands A and B detach from the barrel (Schwaiger et al, 2004; Fig 1B). From the fit, we obtain values of koff=0.28 s−1 and Δx=4±0.35 Å.
Our results of the folding and unfolding pathway of ddFLN4 are summarized in Fig 5. The same intermediate state is populated during refolding and unfolding (Fig 5A). However, the pathways over the transition barriers and the associated transition states are probably different during mechanical unfolding as compared with refolding at zero force. Fig 5B shows a cross-section through the energy landscape along the pathway of mechanical unfolding. The unfolding rates over the barrier at zero force and the position of the transition state along the axis of pulling were obtained from the histograms in Fig 4B.
Figure 5.
Mechanical unfolding and refolding pathways in the energy landscape. (A) Schematic view of the ddFLN4 folding and unfolding energy landscape. The native state N and the intermediate state I correspond to minima in the energy landscape. The mechanical unfolding pathway (black line) and the folding pathway (red) are probably different. However, they both proceed through the same intermediate state. (B) Projection of the mechanical unfolding pathway on the mechanical reaction coordinate (N–C-terminal distance).
The mechanical refolding experiments provide a direct comparison of the folding kinetics between ddFLN4 and the other immunoglobulin domains of the rod. In our double-jump experiments, together with ddFLN4, we always stretch two sacrificial immunoglobulin domains that serve as polymer spacers. We, therefore, have a direct comparison between the ddFLN4 folding rate and those of the sacrificial domains. Refolding of one of the sacrificial domains would lead to only one unfolding peak in the subsequent unfolding pattern, lacking the intermediate state UFI. This happens in less than 20% of the events in which we observe complete refolding of a domain. If we attribute all these events to refolding of one of the two sacrificial domains, we can conclude that refolding of ddFLN4 is at least ten times faster than refolding of all other domains in ddFLN1–5. Folding of ddFLN4 is therefore a clear example that the presence of a stable folding intermediate can accelerate folding drastically as compared with homologous domains lacking such a stable intermediate.
Interestingly, Plaxco et al (1997) have found that the fibronectin type III domain FN10 from fibronectin folds much faster than the structurally homologous domain FN9. Later, Cota & Clarke (2000) reported the presence of a folding intermediate in FN10. Although structural information for the folding intermediate of FN10 is not available, we suggest that, similar to ddFLN4, also in FN10 the presence of the folding intermediate is the main determinant of fast folding for this domain.
Fernandez & Li (2004) have recently reported a continuum of intermediates during mechanical refolding of ubiquitin. In contrast, we observe a discrete, structurally well-defined intermediate consistent with the view that protein-folding landscapes are dominated by discrete, native-like, partially folded intermediates (Baldwin & Rose, 1999).
Is there a potential physiological implication for the fast-folding kinetics and the obligatory folding intermediate of ddFLN4? The combination of low unfolding forces of ddFLN4 and the fast-refolding kinetics we observe in this study suggests a role for ddFLN4 as a reversible, extensible mechanical link in the ddFLN rod (Schwaiger et al, 2004). Another intriguing possibility is a potential role as a molecular strain sensor that can sense highly strained conformations of the cytoskeleton. Partial unfolding of ddFLN4 on external strain may expose a cryptic binding site that allows a specific binding partner (Ingham et al, 1997; Sechler et al, 2001; Oberhauser et al, 2002). As soon as the tension drops, fast refolding would ensure closing of the binding site. Identification of specific binding partners to the ddFLN4 intermediate will be an important task for the future.
Methods
Cloning and protein expression. The ddFLN1–5 construct, containing five immunoglobulin rod domains, was purified as described earlier (Schwaiger et al, 2004).
Force spectroscopy of single proteins. All single-molecule force measurements were performed with a custom-built atomic force microscope apparatus under conditions identical to those described by Schwaiger et al (2004; also see supplementary information online).
Single-domain mechanical double-jump experiments. For a detailed description, see main text and Fig 3.
Modelling of unfolding force distributions. In Fig 4B, the corrected unfolding force values for the transitions N → UFI, U → RFI and UFI → U are compiled into histograms (see supplementary information online). To determine unfolding rates koff and positions of transition state Δx for the transitions N → UFI and U → RFI (left and middle histograms), the respective unfolding force distributions were fitted with a two-state model including the worm-like chain elasticity of the polypeptide spacers as introduced by Evans & Ritchie (1999) using a fixed contour length.
Kinetic schemes to model refolding. To model refolding kinetics, we assumed an irreversible consecutive three-state transition  Solving the rate equations leads to [U](t)=exp(−k1t) [F](t)=(k2/(k1–k2))exp(−k1t)−((k2/(k1–k2))−1)exp(−k2t), and [RFI](t)=1−[U](t)–[F](t), where [U], [F] and [RFI] are the probabilities of observing the unfolded, the completely folded or the intermediate conformation, respectively.
Solving the rate equations leads to [U](t)=exp(−k1t) [F](t)=(k2/(k1–k2))exp(−k1t)−((k2/(k1–k2))−1)exp(−k2t), and [RFI](t)=1−[U](t)–[F](t), where [U], [F] and [RFI] are the probabilities of observing the unfolded, the completely folded or the intermediate conformation, respectively.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n1/extref/6-7400317-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by an SFB 413 grant of the Deutsche Forschungsgemeinschaft.
References
- Baldwin RL, Rose GD (1999) Is protein folding hierarchic? II. Folding intermediates and transition states. Trends Biochem Sci 24: 77–83 [DOI] [PubMed] [Google Scholar]
- Brockwell DJ, Paci E, Zinober RC, Beddard GS, Olmsted PD, Smith DA, Perham RN, Radford SE (2003) Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat Struct Biol 10: 731–737 [DOI] [PubMed] [Google Scholar]
- Bustamante C, Marko JF, Siggia ED, Smith S (1994) Entropic elasticity of λ-phage DNA. Science 265: 1599–1600 [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM (1999) Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci USA 96: 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion-Vazquez M, Li H, Lu H, Marszalek PE, Oberhauser AF, Fernandez JM (2003) The mechanical stability of ubiquitin is linkage dependent. Nat Struct Biol 10: 738–743 [DOI] [PubMed] [Google Scholar]
- Cota E, Clarke J (2000) Folding of β-sandwich proteins: three-state transition of a fibronectin type III module. Protein Sci 9: 112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton WA, Munoz V, Hagen SJ, Jas GS, Lapidus LJ, Henry ER, Hofrichter J (2000) Fast kinetics and mechanisms in protein folding. Annu Rev Biophys Biomol Struct 29: 327–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K (1999) Strength of a weak bond connecting flexible polymer chains. Biophys J 76: 2439–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JM, Li H (2004) Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science 303: 1674–1678 [DOI] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA (1997) The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol 4: 223–230 [DOI] [PubMed] [Google Scholar]
- Ingham KC, Brew SA, Huff S, Litvinovich SV (1997) Cryptic self-association sites in type III modules of fibronectin. J Biol Chem 272: 1718–1724 [DOI] [PubMed] [Google Scholar]
- Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM (2000) Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol 7: 1117–1120 [DOI] [PubMed] [Google Scholar]
- Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM (2002) The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol 319: 433–447 [DOI] [PubMed] [Google Scholar]
- Plaxco KW, Spitzfaden C, Campbell ID, Dobson CM (1997) A comparison of the folding kinetics and thermodynamics of two homologous fibronectin type III modules. J Mol Biol 270: 763–770 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Schlierf M, Li H, Fernandez JM (2004) The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci USA 101: 7299–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M (2004) A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol 11: 81–85 [DOI] [PubMed] [Google Scholar]
- Sechler JL, Rao H, Cumiskey AM, Vega-Colon I, Smith MS, Murata T, Schwarzbauer JE (2001) A novel fibronectin binding site required for fibronectin fibril growth during matrix assembly. J Cell Biol 154: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PM, Fowler SB, Best RB, Toca-Herrera JL, Scott KA, Steward A, Clarke J (2003) Hidden complexity in the mechanical properties of titin. Nature 422: 446–449 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Rief M (2003) Single-molecule folding. Curr Opin Struct Biol 13: 88–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information