Abstract
Some bacterial genomes contain an incomplete set of genes encoding phosphoribosyl isomerases, raising the question of whether there exists broadened substrate specificity for the missing gene products. To investigate the underlying molecular principles of this hypothesis, we have determined the crystal structure of the bifunctional enzyme PriA from Streptomyces coelicolor at 1.8 Å resolution. It consists of a (βα)8-barrel fold that is assembled by two symmetric (βα)4 half-barrels. The structure shows how its active site may catalyse the isomerization reactions of two different substrates, and we provide a plausible model of how the smaller of the two substrates could be bound in two different orientations. Our findings expand the half-barrel ancestor concept by demonstrating that symmetry-related half-barrels could provide a smart solution to cope with dual substrate specificity. The data may help to unravel molecular rationales regarding how organisms with miniature genomes can keep central biological pathways functional.
Keywords: phosphoribosyl isomerase, evolution of (βα)8-barrels, tryptophan and histidine biosynthesis
Introduction
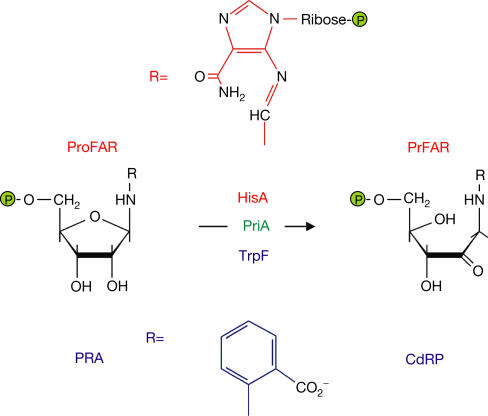
The (βα)8-barrel scaffold represents one of the most versatile protein folds that occur in nature (Wierenga, 2001). In the histidine biosynthesis pathway, at least two reactions are catalysed by (βα)8-barrel enzymes (Lang et al, 2000). Comparison of the sequences and structures of N′-((5′-phosphoribosyl)-formimino)-5-aminoimidazole-4-carboxamide ribonucleotide (ProFAR) isomerase (HisA) with the cyclase subunit of the imidazole glycerol phosphate synthase bienzyme complex (HisF) has shown a two-fold repeat structure in both enzymes, suggesting that a common ancestor might have evolved from a half-barrel protein (Lang et al, 2000). These findings were further supported by the detection of residual ‘partner' activities in the enzymes (Lang et al, 2000) and by the reconstitution of separate half-barrels into a complete catalytically active (βα)8-barrel enzyme (Hoecker et al, 2001). Furthermore, close structural and functional relationships between two (βα)8-barrel isomerases from histidine and tryptophan biosynthesis, HisA and N-(5′-phosphoribosyl)anthranilate (PRA) isomerase (TrpF), respectively, have been demonstrated (Juergens et al, 2000; Lang et al, 2000). Both enzymes catalyse isomerization reactions by the Amadori rearrangement mechanism (Juergens et al, 2000; Henn-Sax et al, 2002; Fig 1). Studies using a directed in vitro evolution approach have shown that the exchange of a single residue position results in the conversion of HisA into an enzyme having TrpF activity (Juergens et al, 2000; Leopoldseder et al, 2004).
Figure 1.
HisA and TrpF catalyse similar reactions in histidine and tryptophan biosynthesis. HisA and TrpF catalyse the isomerizations of the aminoaldoses ProFAR and PRA to the aminoketoses N′-((5′-phosphoribulosyl)formimino)-5-aminoimidazole-4-carboxamide ribonucleotide (PRFAR) and 1-(o-carboxyphenylamino)-1-deoxyribulose 5-phosphate (CdRP). The PriA protein catalyses both reactions in Streptomyces coelicolor and Mycobacterium tuberculosis. These organisms lack a trpf gene.
Comparative genomics approaches have allowed the identification of missing genes and, together with results from metabolic reconstruction, have suggested mechanisms whereby the viability of the respective organisms may have been maintained (Osterman & Overbeek, 2003). Recently, it has been reported that in the genomes of Mycobacterium tuberculosis and Streptomyces coelicolor, a single gene found in the his operon (priA) is solely responsible for HisA and TrpF activities (Fig 1; Barona-Gomez & Hodgson, 2003). It was proposed that a priA-like gene product could be a common ancestor of HisA and TrpF (Barona-Gomez & Hodgson, 2003), supporting an earlier hypothesis (Juergens et al, 2000; Lang et al, 2000) and making priA an ideal target for in vitro evolution approaches (J. Kuper, R. Sterner and M. Wilmanns, personal communication). In this study, we provide structural and functional evidence that phosphoribosyl isomerase (PriA) from S. coelicolor could indeed be an ancestor-like enzyme still present in extant single-substrate isomerases. We present a model for how the two-fold repeated structure of ancient (βα)4 half-barrels may have evolved a two-fold superimposed active site, thereby avoiding nonproductive binding of the smaller of the two possible substrates.
Results And Discussion
Overall structure of PriA
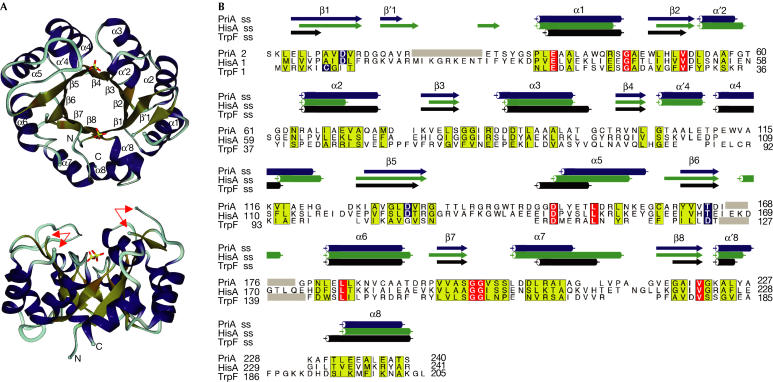
PriA is a monomeric enzyme the structure of which shows a classical (βα)8-barrel fold (Fig 2). A further three short α-helices (α′2, α′4 and α′8) are found within loops at the carboxy-terminal face of the (βα)8-barrel. Two long loops, connecting β1/α1 and β6/α6 at the C-terminal PriA barrel face, are disordered partially in the present crystal form (Fig 2A,B). Two sulphate ions, originating from the crystallization buffer, are bound to loops 3/4 and 7/8, mimicking the phosphate groups of the respective enzyme substrate (Wilmanns et al, 1991; Lang et al, 2000; Vega et al, 2003), thus defining the boundaries of the active site.
Figure 2.
Structure of PriA. (A) Ribbon representation of the PriA structure (top view and side view). Secondary structural elements (α-helices and β-strands) are coloured in blue and bronze, respectively, and are numbered. The sulphate ions, bound to the PriA active site, are shown in stick mode, using atom type colours (sulphur, yellow; oxygen, red). The two loop segments missing in the structure of PriA are indicated by arrows. The N- and C-termini are labelled. (B) Structure-based sequence alignment of TrpF (1LBM; Henn-Sax et al, 2002) and HisA (1QO2; Lang et al, 2000) from Thermotoga maritima, and PriA from S. coelicolor. Yellow boxes show residues with similar properties (Livingstone & Barton, 1993). Invariant residues are in red boxes. Residues that are involved in the catalytic reactions are shown in blue boxes. The positions of the secondary structure elements of the three proteins are shown above the sequences, in the same order. Note that loops missing in the structures were excluded from the alignment and are marked with grey bars. (A) Prepared with DINO (http://www.dino3d.org) and rendered with POVRAY 3.6 (http://www.povray.org). The alignment was prepared with the software SSM (Krissinel & Henrick, 2003), and subsequently coloured with the program ALSCRIPT (Barton, 1993). The secondary structural elements were determined with the software STRIDE (Frishman & Argos, 1995).
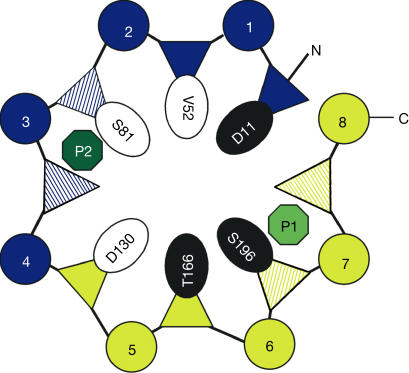
As it has been suggested that PriA functions as an isomerase with dual substrate specificity (Barona-Gomez & Hodgson, 2003), we investigated its structural similarity with the known ProFAR (HisA) and PRA (TrpF) isomerase structures (Fig 2B) using secondary structure matching (SSM; Krissinel & Henrick, 2003). We found that the structures of these three representative enzymes show a high degree of structural similarity (r.m.s.d. Cα: 1.7 Å, PriA-HisA; 2.5 Å, PriA-TrpF). Structural comparison and phylogenetic tree analysis revealed a closer relationship between HisA and PriA than between TrpF and PriA (data not shown). The structure of PriA can be dissected into two half-barrels that can be superimposed with an r.m.s.d. of 1.6 Å, similar to that of HisA (Lang et al, 2000). The similarity of the two half-barrel structures, creating +4 equivalence of secondary structural elements, is reflected by equivalent positions of D11/D130 in loops 1/5, V52/T166 in loops 2/6, S81/S196 in loops 3/7 and the two bound sulphate ions (Fig 3).
Figure 3.
Schematic representation of the two (βα)4-barrels of PriA in blue and yellow, respectively. β-Strands are depicted as triangles and α-helices as circles. The two phosphate binding sites are indicated by P1 and P2. The striped triangles represent positions involved in phosphate binding. The black and white residues represent the symmetry-related equivalent half-barrel residue groups considered to be important for bisubstrate catalytic activities (see text).
The bisubstrate active site of PriA
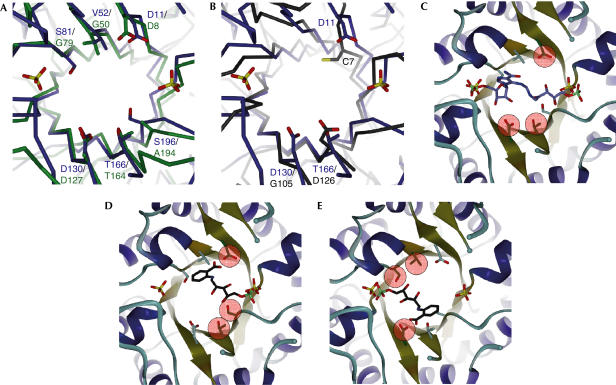
An interesting consideration regards the evolution of substrate specificity from ancestral enzymes with broad specificity. One plausible assumption is that the catalytic mechanism of these enzymes has been preserved, whereas the substrate specificity has changed (Jensen, 1976; Gerlt & Babbitt, 2001). PriA is a paradigm of an enzyme that can be used to test this hypothesis, taking into consideration that the enzymatically catalysed isomerizations of ProFAR (histidine biosynthesis) and PRA (tryptophan biosynthesis) have been proposed to follow basically the same general acid–base reaction mechanism (Henn-Sax et al, 2002; Leopoldseder et al, 2004; Fig 1). A superposition of the active sites of PriA and HisA shows that the three residues important for HisA catalytic activity from loops 1 (D8), 5 (D127) and 6 (T164; Henn-Sax et al, 2002) are identical to those in PriA (Fig 4A). The structural similarity between the PriA and TrpF active sites is, however, less obvious. The general base residue C7 from the C-terminus of β-strand 1 from TrpF is in a similar, but not identical, position with respect to D8/D11 in HisA/PriA (Fig 4B). The second residue essential for TrpF catalysis is D126 from loop 6, which superimposes with T166 in PriA. Furthermore, the aspartate residue from loop 5 (D130 in PriA), which, according to previous experiments, abolishes PRA isomerase activity of HisA but seems to be essential for ProFAR isomerase activity (Juergens et al, 2000; Leopoldseder et al, 2004), is present in the PriA structure but absent in TrpF.
Figure 4.
Active site of PriA. (A) Superimposition of the active sites of PriA (blue) and HisA (green). Active-site residues and the two bound sulphate ions are shown in stick mode. The carbon atoms are in the colour of the corresponding Cα trace. Oxygen molecules are coloured red and sulphur in yellow. (B) Superimposition of the active sites of PriA (blue) and TrpF (black). Active-site residues and the two bound sulphate ions are shown in stick mode using atom-type colours. (C) PriA active sites with the HisA product analogue rPRFAR. The rPRFAR coordinates have been taken from the superimposed HisF structure (not shown) in the presence of rPRFAR (Chaudhuri et al, 2001). Residues most probably involved in catalysis are highlighted green, surrounded by red circles, in this panel and in all following panels with modelled complexes. (D) PriA active site with the TrpF product analogue rCdRP. The rCdRP coordinates were obtained from the superimposed TrpF structure in complex with rCdRP (Henn-Sax et al, 2002). (E) PriA active site with the TrpF product analogue rCdRP modelled binding to the second phosphate binding site. The rCdRP coordinates were obtained from the TrpF structure in complex with rCdRP (Henn-Sax et al, 2002) superimposed with the C-terminal half-barrel of PriA. Complete PriA was then superimposed using the coordinates obtained from the C-terminal half-barrel. (A–E) Prepared with DINO (http://www.dino3d.org) and rendered with POVRAY 3.6 (http://www.povray.org).
To catalyse the isomerization of the two substrates, ProFAR and PRA, the PriA active site must be able to accommodate both structures. As the structure of HisA with a reaction analogue is not available, we used the structure of the subsequent histidine biosynthesis enzyme, HisF, in complex with its substrate analogue rPRFAR (Chaudhuri et al, 2003), which can be regarded as a product analogue for modelling of the HisA-like catalysed reaction. The two phosphate groups of rPRFAR are in the same positions as the two sulphate ions that are bound to PriA (Fig 4C). The catalytic HisA residues D8 and D127 are in identical positions in PriA (Fig 4A), suggesting that they serve the same function as general base and general acid, respectively, in PriA.
If the PriA active site is superimposed with that of the TrpF structure that contains the product analogue rCdRP (Henn-Sax et al, 2002), its phosphate group is in the same position as the sulphate ion that is bound to the C-termini of PriA strands β7 and β8 (Fig 4D). TrpF does not have a second phosphate-binding site. This may be due to the absence of the G/A/S-G-G-V/I phosphate-binding motif (Wilmanns et al, 1991) in loop 3, which is found in HisA/PriA, as well as in loop 7 of the structures of all three enzymes under investigation (Fig 2B). In TrpF, C7 and D126 have been suggested to function as the general base and the general acid, respectively (Henn-Sax et al, 2002; Leopoldseder et al, 2004). In PriA, D11 is the closest residue that may act as the general base. This observation is supported by the TrpF-based PriA–rCdRP model (Fig 4D), wherein the D11 carboxylate group and the C2′ atom of rCdRP are about 4 Å apart. However, the identity of the residue that may function as the general acid is less clear. D126 in loop 6 of the TrpF structure, which has been suggested as the general acid in this enzyme (Henn-Sax et al, 2002), aligns with T166 of PriA, making it one possible candidate (Fig 4D).
Although it has been shown that PriA has both TrpF and HisA activities in vivo, by assessing functional complementation, these data have not been confirmed in vitro by measuring catalytic activities of the purified enzymes (Barona-Gomez & Hodgson, 2003). Table 1 shows the results of steady-state kinetic studies showing that recombinantly expressed and purified PriA is able to catalyse the isomerization of both PRA and ProFAR in vitro, directly demonstrating its bisubstrate specificity. The catalytic efficiency kcat/KMPRA (3 μM−1 s−1) is comparable with that of TrpF from Thermotoga maritima and Escherichia coli (Sterner et al, 1996), indicating efficient processing of PRA. The value for kcat/KMProFAR (0.03 μM−1 s−1) is lower by about two orders of magnitude as compared with the kcat/KMPRA or kcat/KMProFAR values of the T. maritima or E. coli HisA homologues (Henn-Sax et al, 2002) due to a significantly higher Michaelis constant. This indicates that the bisubstrate specificity is achieved at least partly at the expense of efficient binding of ProFAR to PriA.
Table 1.
Steady-state kinetic constants of PriA from S. coelicolor
| Assayed activity (substrate) | KM (μM) | kcat (s−1) | kcat/KM (μM−1 s−1) |
|---|---|---|---|
| ProFAR |
28 |
0.9 |
0.03 |
| PRA | 4 | 12 | 3 |
The standard errors on all constants are less than ±25%.
Molecular insights into bisubstrate specificity of PriA
What is the molecular basis underlying the ability of PriA to retain its ability to catalyse the isomerization reactions of both substrates, ProFAR and PRA, in contrast with the single-substrate specificities of HisA and TrpF? Although both enzymatic reactions have the same catalytic mechanism, the different structures of the two substrates lead to different active-site architecture requirements. Previous directed evolution experiments showed that the removal of a negative charge at the C-terminus of strand β5 generates TrpF activity in HisA as well as in HisF. In the wild-type HisA and HisF enzymes, this negative charge seems to impede the binding of the negatively charged TrpF substrate PRA due to electrostatic repulsion (Leopoldseder et al, 2004). Although the overall structural similarity is comparable in all three enzymes, the PriA active site shows closer similarity to that of HisA than to that of TrpF, generally allowing it to accommodate both substrates.
One outstanding question regards the ability of PriA to avoid the postulated inhibition by nonproductive binding of the TrpF substrate PRA by interactions with both PriA phosphate binding sites. By analogy to previous in vitro evolutionary studies on HisA, the presence of D130 in PriA (D127 in HisA) at the C-terminus of strand β5 should abolish TrpF-like PRA isomerase activity (Leopoldseder et al, 2004). An attractive, but hypothetical, model would be to consider a dual PRA isomerase active site in PriA, exploiting the recently detected half-barrel symmetry (Lang et al, 2000). In this type of model, the second phosphate group binding site in PriA, formed by loops 3 and 4, could become a productive binding site for PRA as well (Fig 4E). In turn, D130 from the C-terminus of strand β5 would replace the function of D11 from the C-terminus of strand β1, which is symmetrically related to the two half-barrels (Lang et al, 2000). The two symmetry-related and superimposed active sites for PRA isomerization catalysis would be completed by a set of two further catalytic competent residues in identical half-barrel positions, T166 (loop 6)/S196 (loop 7) and V52 (loop 2)/S81 (loop 3), respectively (Figs 3, 4D,E). Interestingly, serine residues are found neither in loop 3 nor in loop 7 of HisA, providing a possible rationale as to why the proposed second substrate orientation in PriA, which uses V52, S81 and D130 for catalysis according to previous directed evolution experiments, may be ‘forbidden' in HisA.
Speculations
In summary, the structure of the PriA enzyme from S. coelicolor provides an excellent model to study the evolution of enzyme specificity. It may be that organisms with small genomes have less-specific enzymes to accommodate the function of a smaller genetic portfolio. Our new structural findings for PriA show that the previously identified prerequisites for the isomerase substrate specificity towards PRA by removal of the conserved aspartate in loop 5 (Leopoldseder et al, 2004) may have been bypassed during natural evolution. The established model for the evolution of some of these (β/α)8-barrel enzymes from an ancestral half-barrel (Lang et al, 2000) may serve as a basis for the unique bisubstrate specificity observed in PriA.
Methods
Expression, purification and crystallization of PriA. The priA gene (SCO2050) was obtained from a cosmid described by Redenbach et al (1996) through PCR, introducing the restriction sites for subcloning using the primers 5′-CCG GTA GTC ATG AGC AAG CTC GAA CTC C-3′ and 5′-GGT GTC CGC TCG AGC GAC GTA GCC TCC AAG-3′. The amplified gene was subcloned into pTYB4 (NEB, Impact™ CN system, Frankfurt, Germany) using BspHI and XhoI restriction sites. ER2566 cells were grown aerobically in Luria–Broth medium containing the appropriate antibiotics at 37°C until they reached an OD600 of 0.5. At this point, the culturing temperature was lowered to 15°C and priA gene expression was induced with 0.3 mM isopropyl β-thiogalactoside. After 14 h of induction, the cells were harvested and resuspended in lysis buffer (50 mM Tris–HCl (pH 8.5), 10% (v/v) glycerol, 1 M NaCl, 0.02% (v/v) α-mono-thio-glycerol). All subsequent steps were carried out at 0–4°C. Cells were sonicated (Bandelin Sonopuls, Berlin, Germany), and the supernatant was obtained by centrifugation at 18,000g for 30 min. Intein-tagged PriA was affinity purified and Intein cleavage was induced following the manufacturer's protocols. PriA protein was subjected to size-exclusion chromatography using a Superdex200 HR16/60 column (APB Biotech, Freiburg, Germany) equilibrated with crystallization buffer (50 mM Tris–HCl (pH 8), 200 mM NaCl, 2 mM dithiothreitol). Monomeric fractions were concentrated to 10 mg/ml. Crystallization trials were carried out using vapour diffusion in hanging-drop plates. Needle-shaped crystals were obtained in 1.6 M ammonium sulphate buffered to pH 5 with 100 mM citrate buffer at 21°C.
Data collection and refinement. X-ray data were collected on flash-cooled crystals up to 1.8 Å at beamline X11 at EMBL/DESY, Hamburg, Germany. Mother liquor, to which 25% (v/v) glycerol was added, was used as the cryoprotectant. Data were indexed with MOSFLM (Leslie, 1992) and processed with SCALA (Evans, 1997) from the CCP4 suite (CCP4, 1994). Data and refinement statistics are shown in supplementary information online. The initial phases were obtained from molecular replacement using MOLREP (Vagin & Teplyakov, 1997), and using Protein Data Bank (PDB) entry 1H5Y (Banfield et al, 2001) as a search model. The structure was refined by alternating rounds of manual model building in COOT (http://www.ysbl.york.ac.uk/~emsley/coot/) and refinement with REFMAC (Murshudov et al, 1997). Atomic coordinates and structure factors have been deposited in the PDB as entry 1VZW.
Steady-state enzyme kinetics. The isomerization of PRA into CdRP was followed at 25°C by a fluorimetric assay, using 50 mM Tris–HCl (pH 7.5), 0.5 mM MgCl2 and a molar excess of the helper enzymes anthranilate phosphoribosyl transferase and indole glycerol phosphate synthase (Hommel et al, 1995; Juergens et al, 2000). The progress curves were recorded and analysed with the integrated form of the Michaelis–Menten equation using the program COSY to obtain kcat and KMPRA (Eberhard, 1990). ProFAR isomerase activity was measured as described elsewhere (Juergens et al, 2000), using 50 mM Tris–acetate (pH 8.5) in the presence of 200 mM ammonium acetate and an excess of the HisF subunit of the imidazole glycerol phosphate synthase to rapidly convert PRFAR into imidazole glycerol phosphate (ImGP) and aminoimidazole carboxamide ribonucleotide (AICAR). This avoided product inhibition and allowed us to use the decrease of absorbance at 300 nm to follow the reaction (Δɛ 300(ProFAR−AICAR)=5.637 mM−1 cm−1). The dependence of initial velocities on the substrate concentration was recorded and analysed with the direct linear plot method, yielding values for KMProFAR and kcat.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n2/extref/7400330s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr M. Groves and Professor R. Sterner for helpful discussions and critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (grants WI 1058/5-3, WI 1058/5-4 and WI 1058/6-1 to M.W.).
References
- Banfield MJ, Lott JS, Arcus VL, McCarthy AA, Baker EN (2001) Structure of HisF, a histidine biosynthetic protein from Pyrobaculum aerophilum. Acta Crystallogr D 57: 1518–1525 [DOI] [PubMed] [Google Scholar]
- Barona-Gomez F, Hodgson DA (2003) Occurrence of a putative ancient-like isomerase involved in histidine and tryptophan biosynthesis. EMBO Rep 4: 296–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chaudhuri BN, Lange SC, Myers RS, Chittur SV, Davisson VJ, Smith JL (2001) Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (β/α)8 barrel joins two active sites. Structure (Camb) 9: 987–997 [PubMed] [Google Scholar]
- Chaudhuri BN, Lange SC, Myers RS, Davisson VJ, Smith JL (2003) Toward understanding the mechanism of the complex cyclization reaction catalyzed by imidazole glycerolphosphate synthase: crystal structures of a ternary complex and the free enzyme. Biochemistry 42: 7003–7012 [DOI] [PubMed] [Google Scholar]
- Eberhard M (1990) A set of programs for analysis of kinetic and equilibrium data. Comput Appl Biosci 6: 213–221 [DOI] [PubMed] [Google Scholar]
- Evans PR (1997) Scala. Joint CCP4 & ESF-EACBM Newslett 33: 22–24 [Google Scholar]
- Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23: 566–579 [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC (2001) Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem 70: 209–246 [DOI] [PubMed] [Google Scholar]
- Henn-Sax M, Thoma R, Schmidt S, Hennig M, Kirschner K, Sterner R (2002) Two (βα)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms and common strategies for protecting their labile substrates. Biochemistry 41: 12032–12042 [DOI] [PubMed] [Google Scholar]
- Hoecker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R (2001) Dissection of a (βα)8-barrel enzyme into two folded halves. Nat Struct Biol 8: 32–36 [DOI] [PubMed] [Google Scholar]
- Hommel U, Eberhard M, Kirschner K (1995) Phosphoribosyl anthranilate isomerase catalyzes a reversible amadori reaction. Biochemistry 34: 5429–5439 [DOI] [PubMed] [Google Scholar]
- Jensen RA (1976) Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30: 409–425 [DOI] [PubMed] [Google Scholar]
- Juergens C, Strom A, Wegener D, Hettwer S, Wilmanns M, Sterner R (2000) Directed evolution of a (βα)8-barrel enzyme to catalyze related reactions in two different metabolic pathways. Proc Natl Acad Sci USA 97: 9925–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2003) Protein structure comparison in 3D based on secondary structure matching (SSM) followed by Ca alignment, scored by a new structural similarity function. In Proceedings of the 5th International Conference on Molecular Structural Biology, Vienna, Kungl AJ, Kungl PJ (eds), 88 [Google Scholar]
- Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M (2000) Structural evidence for evolution of the β/α barrel scaffold by gene duplication and fusion. Science 289: 1546–1550 [DOI] [PubMed] [Google Scholar]
- Leopoldseder S, Claren J, Jurgens C, Sterner R (2004) Interconverting the catalytic activities of (βα)(8)-barrel enzymes from different metabolic pathways: sequence requirements and molecular analysis. J Mol Biol 337: 871–879 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 & ESF-EAMCB Newslett Protein Crystallogr 26 [Google Scholar]
- Livingstone CD, Barton GJ (1993) Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci 9: 745–756 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Osterman A, Overbeek R (2003) Missing genes in metabolic pathways: a competitive genomics approach. Curr Opin Chem Biol 7: 238–251 [DOI] [PubMed] [Google Scholar]
- Redenbach M, Kieser HM, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood DA (1996) A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol 21: 77–96 [DOI] [PubMed] [Google Scholar]
- Sterner R, Kleemann GR, Szadkowski H, Lustig A, Hennig M, Kirschner K (1996) Phosphoribosyl anthranilate isomerase from Thermotoga maritima is an extremely stable and active homodimer. Protein Sci 5: 2000–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Vega MC, Lorentzen E, Linden A, Wilmanns M (2003) Evolutionary markers in the (β/α)8-barrel fold. Curr Opin Chem Biol 7: 694–701 [DOI] [PubMed] [Google Scholar]
- Wierenga RK (2001) The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett 492: 193–198 [DOI] [PubMed] [Google Scholar]
- Wilmanns M, Hyde CC, Davies DR, Kirschner K, Jansonius JN (1991) Structural conservation in parallel β/α-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry 30: 9161–9169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information