Abstract
Eukaryotic transcriptional regulation often involves regulatory elements separated from the cognate genes by long distances, whereas appropriately positioned insulator or enhancer-blocking elements shield promoters from illegitimate enhancer action. Four proteins have been identified in Drosophila mediating enhancer blocking—Su(Hw), Zw5, BEAF32 and GAGA factor. In vertebrates, the single protein CTCF, with 11 highly conserved zinc fingers, confers enhancer blocking in all known chromatin insulators. Here, we characterize an orthologous CTCF factor in Drosophila with a similar domain structure, binding site specificity and transcriptional repression activity as in vertebrates. In addition, we demonstrate that one of the insulators (Fab-8) in the Drosophila Abdominal-B locus mediates enhancer blocking by dCTCF. Therefore, the enhancer-blocking protein CTCF and, most probably, the mechanism of enhancer blocking mediated by this remarkably versatile factor are conserved from Drosophila to humans.
Keywords: Drosophila, CTCF, enhancer blocking, Fab-8, Abdominal-B
INTRODUCTION
Expression of the eukaryotic genome is controlled by enhancer and silencer elements, both of which can mediate their function from a distance. Insulator elements with enhancer-blocking activity curb enhancer activity, such that only appropriate promoters are activated (Ishii & Laemmli, 2003). The proteins that mediate insulator function have been identified for only a few Drosophila insulator sequences. These are Zw5, BEAF-32 (Zhao et al, 1995; Gaszner et al, 1999), GAGA factor (Ohtsuki & Levine, 1998; Belozerov et al, 2003) and Su(Hw) (Gerasimova et al, 1995).
Another perspective on the requirement of insulators comes from the fact that many genes are controlled by several regulatory elements needed for tissue- and cell-specific expression. For example, the Drosophila gene Abdominal-B (Abd-B) contains an extended 3′ regulatory region that is functionally subdivided into distinct enhancer domains. Functional separation of the enhancer sequences is achieved by intervening insulators such as Frontabdominal (Fab)-7 and Fab-8 (Hagstrom et al, 1996; Barges et al, 2000). Although both elements have been shown to mediate enhancer-blocking function, the protein involved in this activity has not been described.
In sharp contrast to Drosophila, the genome of vertebrates is much more expanded, due primarily to larger distances between genes. Therefore, the need for insulators to separate genes may not seem as pronounced as it is in Drosophila. Indeed, until now, only a single protein, CTCF, has been identified to mediate enhancer-blocking activity (Ohlsson et al, 2001). Binding sites for CTCF have been shown to be involved in gene activation (Vostrov & Quitschke, 1997), gene repression (Baniahmad et al, 1990; Lobanenkov et al, 1990) and enhancer blocking (Bell et al, 1999; Hark et al, 2000; Kanduri et al, 2000; Szabo et al, 2000; Filippova et al, 2001; Lutz et al, 2003; Tanimoto et al, 2003). Furthermore, vertebrate- and mammalian-specific functions, such as X-chromosome inactivation and control of the epigenetic DNA methylation state, seem to involve CTCF (Lee, 2003).
Obviously, the function of enhancer blocking has developed during evolution such that Drosophila uses several proteins and mechanisms for enhancer blocking and insulation (Kuhn et al, 2003). However, none of the known Drosophila insulator proteins has a counterpart found to be conserved in vertebrates. Rather, vertebrates use CTCF, which has not yet been found in Drosophila. Here, we characterize a Drosophila orthologue of CTCF with similarities to many of the features identified for vertebrate CTCF. Furthermore, we show that a previously characterized Drosophila insulator, Fab-8, mediates enhancer blocking by CTCF in Drosophila as well as in vertebrate cells. Thus, the enhancer-blocking protein CTCF and, probably, the mechanisms of CTCF-driven enhancer blocking are both conserved from Drosophila to humans.
RESULTS AND DISCUSSION
Vertebrate and Drosophila CTCF are similar
FlyBase data entries and cDNA sequence analysis (see supplementary information online) revealed an open reading frame (ORF) coding for a protein similar to vertebrate CTCF with respect to the overall structure. dCTCF contains all of the expected 11 zinc fingers (Zn-fingers), separated by both standard and noncanonical inter-finger linkers (Fig 1A). Furthermore, most of the crucial DNA base recognition residues at positions −1, 2, 3 and 6 are identical. Variation in position 6 for fingers #6 and #9 generate a change from alanine or serine to methionine, which is of no consequence for the DNA-binding specificity, as the recognition code is not changed (Suzuki et al, 1994).
Figure 1.
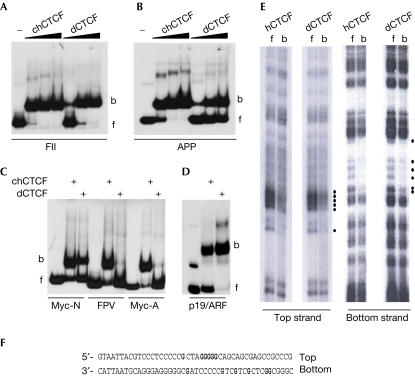
Drosophila CTCF encodes a protein similar to human CTCF. (A) Sequence alignment of the Zn-finger domain of dCTCF and hCTCF. Similar and identical residues (shading), zinc-coordinating amino acids (red) and amino acids with identical binding site recognition (yellow) are indicated. (B) In situ hybridization of Drosophila egg chamber with DIG-labelled antisense dCTCF RNA. Polyploid nurse cell nuclei are surrounded by dark staining of cytoplasmic dCTCF RNA (top), nuclei are identified by Hoechst staining (middle) and in the overlay (bottom). The egg chamber to the right is surrounded by follicle cells (Hoechst-stained nuclei). (C,D) dCTCF detection in syncytial blastoderm stage 3 with the rabbit anti-dCTCF-C antibody and visualization by avidin-peroxidase at two different magnifications (scale bars (B–D), 40 μm). (E) CV-1 and COS-1 cells were transfected with DNA constructs expressing the GAL DNA-binding domain (GAL), a fusion of the Zn-finger domain plus the C-terminus of chicken CTCF, a similar fusion of dCTCF or of v-erbA. Relative CAT activity is calculated with the GAL transfection defined as 100%.
Similarities in Zn-fingers do not necessarily imply similarities in function. Therefore, we examined whether dCTCF can act as a transcriptional repressor, as has been demonstrated previously for vertebrate CTCF (Burcin et al, 1997). The strongest repressive function has been shown to reside within the combined carboxy-terminal plus Zn-finger domains (Lutz et al, 2000). Equivalent regions of Drosophila and chicken CTCF (chCTCF) were fused to the yeast GAL4 transcription factor DNA-binding domain. Both Drosophila and chicken GAL4–CTCF fusions repressed reporter gene activity to a similar extent in two different cell lines (Fig 1E) and in a way comparable with the previously characterized strong repressor GAL4-v-erbA362 (Baniahmad et al, 1992). These results clearly indicate that dCTCF, like its vertebrate counterpart, has transcriptional repressor activity.
In vertebrates, CTCF is ubiquitously expressed (Burke et al, 2002), apparently functioning as a global transcriptional regulator in all cell types (Ohlsson et al, 2001). In comparison, we monitored dCTCF RNA expression levels at various stages of fly development. Using in situ hybridization, we found that dCTCF RNA is present in the cytoplasm of the nurse cells within the fly egg chamber (Fig 1B), transported into and distributed uniformly in the developing oocyte and in 0–24 h embryos as a maternal RNA (not shown). Later stages show expression in all tissues and stages (see supplementary information online), revealing that dCTCF is a ubiquitous factor as in vertebrates. Location of dCTCF protein is clearly nuclear, exemplified by the nuclear staining of syncytial blastoderm embryos with dCTCF-specific antibodies (Fig 1C,D).
To extend the comparison of vertebrate and Drosophila CTCF, we tested in vitro-translated Drosophila and vertebrate CTCF for binding to several previously characterized vertebrate CTCF targets (CTS). The sequences tested included the CTS of the FII insulator element of the β-globin gene, the APP gene, the myc genes and the mouse ARF promoter (Fig 2A–D). With the exception of the two myc FPV and A sites, all the other sequences bound chicken and Drosophila CTCF similarly. Binding and enhancer-blocking results with additional CTS are summarized in supplementary Table I online.
Figure 2.
Similar binding site specificity on vertebrate target sites. Zn-finger domains of chCTCF and dCTCF were in vitro translated and incubated with DNA. (A–D) Indicated DNA fragments were tested in electrophoretic mobility shift assay (EMSA), the β-globin FII insulator sequence (Bell et al, 1999), the CTCF-binding site in the APP gene (Vostrov & Quitschke, 1997), the sites N, FPV and A of the myc genes (Filippova et al, 1996; Lutz et al, 2003) and in the mouse ARF promoter (Filippova et al, 2002). (E) DMS methylation interference analysis was carried out with the β-globin FII insulator fragment labelled on the top or the bottom strand. After separation of bound (b) and unbound fragments (f), the DNA was analysed on a sequencing gel. The G nucleotides interfering with binding after methylation are indicated by dots, and in the sequence (F) by bold lettering.
We used a methylation-interference assay to determine whether both proteins contact the same guanidine nucleotides on a given target DNA site (Fig 2E). Both Drosophila and human CTCF were found to contact the same nucleotides on the β-globin FII insulator fragment. These results indicate that, despite considerable overall sequence divergence, fly and human CTCF show a striking degree of functional conservation with respect to DNA binding.
Enhancer blocking of the Fab-8 insulator by CTCF
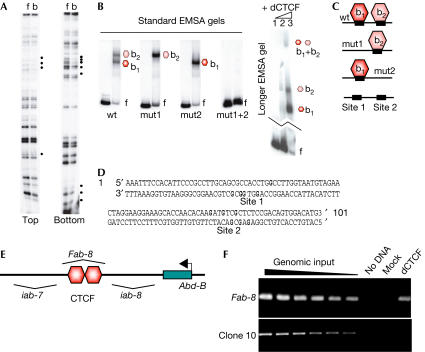
To identify potential Drosophila CTCF regulatory targets, we performed an in vitro screen for CTCF-binding sites (supplementary information online) and found the Fab-8 element for which enhancer-blocking and boundary function have been shown (Barges et al, 2000). This sequence is situated in the Abd-B locus, separating and insulating enhancer domains infraabdominal-7 (iab-7) from iab-8. As the protein involved in this mechanism was unknown, and as vertebrate CTCF mediates enhancer-blocking activity (Ohlsson et al, 2001), we tested whether dCTCF might have a similar role in the context of the Fab-8 element. In vitro binding of dCTCF to Fab-8, as determined by methylation interference (Fig 3A), suggested two binding sites for CTCF. We generated binding site mutations resulting in single-site mutations (mut1 or mut2) and in a double-site mutation (mut1+2) used for electrophoretic mobility shift assay (EMSA; Fig 3B). The wild-type Fab-8 element generates two retarded bands corresponding to a different mobility of the same DNA molecule occupied by CTCF at one of the two closely spaced CTS sequences. These different mobilities are probably caused by a site-specific DNA bending (Arnold et al, 1996), which has also been observed on other dual binding sites, such as the H19 locus (Kanduri et al, 2000). Excess protein generated a slow mobility complex only resolved after a long run of the gel (Fig 3B), reflecting binding of CTCF to both sites.
Figure 3.
In vitro and in vivo binding of dCTCF to the Fab-8 element. (A) DMS methylation interference was determined with the in vitro-translated dCTCF Zn-finger domain. Bound (b) and unbound fragments (f) labelled on the top or the bottom strand were analysed on a sequencing gel. Two groups of interfering nucleotides (marked with dots) are indicated (sites 1 and 2). (B) EMSA analysis of dCTCF binding. In vitro-translated, full-length dCTCF was incubated with the Fab-8 wild-type fragment and with the mutants mut1, mut2 and mut1+2. Interpretation of the single and double occupancy of dCTCF on the respective fragments is shown in (C), double occupancy can only be visualized with excess of protein (long gel, lane 3, (B)). The migration difference of b1 and b2 complexes is probably caused by site-specific DNA bending (see text). (D) Contact G nucleotides in the Fab-8 sequence (bold) are converted to AT in mutants mut1 and mut2 (see also supplementary information online). (E) Diagram (not to scale) of the Fab-8 location downstream of the Abdominal-B gene, flanked by the iab-7 and iab-8 regulator sequences. (F) In vivo binding of dCTCF to Fab-8. Chromatin from Drosophila embryos was precipitated with the rabbit anti-dCTCF-C antibody (dCTCF) and tested by PCR for the presence of Fab-8 sequences. Increasing amounts of genomic input are included for standardization. A sequence (clone 10) without dCTCF binding, mock precipitation and no DNA are included as negative controls with no signal.
To test in vivo dCTCF binding to this important element, we prepared crosslinked chromatin from Drosophila embryos and precipitated CTCF-occupied sites with the anti-dCTCF-C antibody. PCR primers for the Fab-8 sequence identified specifically precipitated chromatin, whereas primers against a different non-dCTCF-binding site (clone 10) and mock-precipitated chromatin resulted in no signal (Fig 3F).
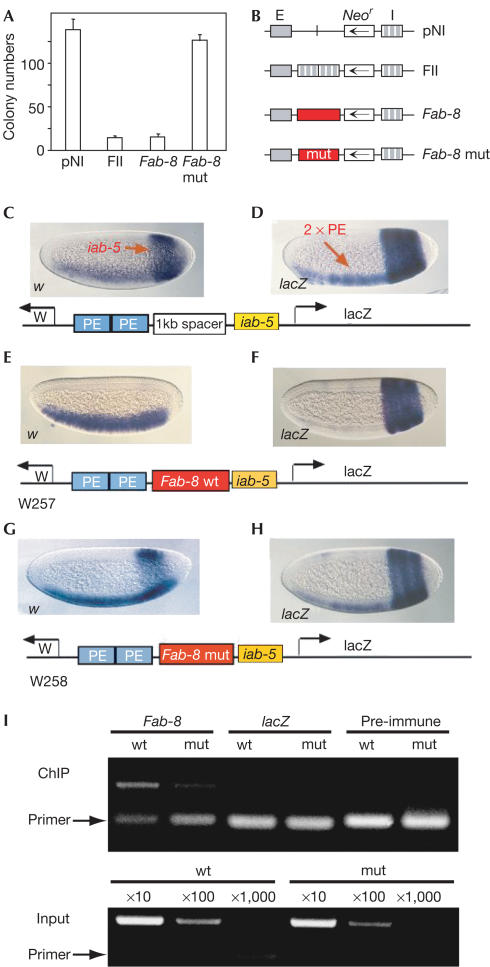
To test the functional similarity between dCTCF and vertebrate CTCF, we analysed enhancer blocking of Fab-8 in vertebrate K562 cells. In comparison to the known enhancer-blocking effect mediated by the FII sequence (Bell et al, 1999), a similar reduction in colony numbers mediated by Fab-8 was seen (Fig 4A). More importantly, specific abrogation of CTCF binding by the double mutation, mut1+2, resulted in loss of enhancer blocking.
Figure 4.
CTCF-dependent enhancer blocking of Fab-8 in vertebrate cells and in Drosophila. (A) K562 cells were transfected with the indicated DNA constructs (B) and colony number was determined after neomycin selection. The pNI and FII constructs (Chung et al, 1993) were used as controls. Transcription of the neomycin gene (Neo) is driven by the β-globin enhancer (E). In the FII construct, two copies of the β-globin insulator (I) separate the enhancer from Neo. The Fab-8 fragment mediates enhancer blocking, whereas mutation of both CTCF-binding sites (Fab-8 mut) does not. (C–H) Fab-8 enhancer blocking in Drosophila depends on dCTCF-binding sites. Transgenic embryos with the indicated vectors were hybridized with either digoxygenin-labelled white (w) in (C,E,G) or lacZ antisense RNA probes (D,F,H). (C,D) white and lacZ expression is visualized when a 1.6 kb λ DNA fragment was inserted between the 2 × PE and iab-5 enhancers (see text). (E,F) The Fab-8 DNA insert between 2 × PE and iab-5 enhancer was analysed in 13 independent lines. Expression of the white reporter gene is restricted to the ventral mesoderm and of the lacZ gene to the abdomen. With the CTCF sites mutagenized, 22 independent lines were generated: white gene activity in the abdomen (G) and lacZ expression in the ventral mesoderm (H) are seen. (I) Chromatin was prepared from embryos carrying transgenes with either Fab-8 wt or the mut1+2 CTCF-binding sites (mut). Primers that specifically recognize transgenic (not genomic) sequences were used. ChIP using anti-CTCF-C antibody shows an increased occupancy at wild-type versus mutant sequences, in contrast to pre-immune serum and to lacZ sequences. For standardization, input dilutions were subjected to semiquantitative PCR as well.
The crucial test for enhancer-blocking activity of dCTCF had to be carried out in flies. Therefore, we used a vector with two regulatory regions containing the iab-5 enhancer from the Abd-B locus and two copies of the minimal twist enhancer, PE, directing an additive pattern of expression when placed between divergently transcribed white and lacZ reporter genes (Lin et al, 2003). As shown in Fig 4C,D, the iab-5 enhancer directs expression in the posterior one-third of the blastoderm stage embryo, whereas the 2 × PE enhancer activates transcription in the ventral-most region where twist is normally expressed. Enhancer elements are enhancing both the white gene as well as the lacZ gene. Altered patterns of transcription were observed when the 1 kb spacer sequence was replaced by the 680 bp Fab-8 element. On the white promoter, the iab-5 activity was completely abolished (shown as the lack of staining in the iab-5 activity region), while the 2 × PE enhancer was still activating the white gene (Fig 4E). The lacZ promoter, conversely, could be activated only by the proximal iab-5 but not by the distal 2 × PE (Fig 4F). This result suggests that the Fab-8 fragment blocks the respective distal enhancer for both the white and the lacZ promoters. When the CTCF sites were mutagenized (mut1+2 as in Fig 3), the iab-5 activity on white was partly restored (Fig 4G). Similarly, the 2 × PE element again directed the transcription of the lacZ gene (Fig 4H). Chromatin/CTCF immunoprecipitation revealed specific CTCF binding to the Fab-8 element of the enhancer-blocking vector, whereas binding to the Fab-8 mut element was clearly reduced (Fig 4I). This correlation between strong CTCF binding and full enhancer-blocking function (Fig 4E,F) indicates that the activity of Fab-8 is at least partly mediated by CTCF and that dCTCF, similar to vertebrate CTCF, confers enhancer blocking.
Thus, at least one enhancer-blocking protein (CTCF) in Drosophila and vertebrates is conserved with a similar enhancer-blocking function. In addition to enhancer blocking, mammalian CTCF has gained functions involving the control of epigenetic states in the context of imprinted genes and X-chromosome inactivation (Lee, 2003; Lewis & Murrell, 2004).
METHODS
EMSA. Full-length CTCF and 11 Zn-finger fragments were subcloned into pET 16B vector by PCR-designed primers. Drosophila CTCF proteins were produced using the TnT in vitro coupled transcription–translation kit (Promega Co., Madison, WI, USA). Probes for EMSA were obtained by PCR, gel purified, end-labelled by T4 kinase and used in DNA binding as described (Filippova et al, 1996).
Repression and enhancer blocking. Repression analysis in COS-1 and CV-1 cells was performed as described earlier (Lutz et al, 2000). The colony formation assay in K562 cells has been carried out essentially as in Chung et al (1993) and Lutz et al (2003). The Drosophila enhancer-blocking assay was carried out as described (Lin et al, 2003).
For information regarding primers for methylation interference, site-directed mutagenesis, plasmid construction, peptides for antibody production, immunocytology, western and chromatin immunoprecipitation, as well as Drosophila target-site library construction, see supplementary information online.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n2/extref/7400334s1.pdf, http://www.nature.com/embor/journal/v6/n2/extref/7400334s2.pdf).
Supplementary Material
Supplementary Figure 1 and Table 1
Supplementary Information: Materials and Methods
Acknowledgments
We thank G. Gilfillan for embryo chromatin, and Ru Zhang for western analysis, H.C. Morse III, A. Paululat and M. Mohan for reading and discussions. This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (to V.L.) and the Deutsche Forschungsgemeinschaft (to R.R.).
References
- Arnold R, Burcin M, Kaiser B, Muller M, Renkawitz R (1996) DNA bending by the silencer protein NeP1 is modulated by TR and RXR. Nucleic Acids Res 24: 2640–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad A, Steiner C, Kohne AC, Renkawitz R (1990) Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514 [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Kohne AC, Renkawitz R (1992) A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J 11: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F (2000) The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P, Cai HN (2003) A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22: 3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R (1997) Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol 17: 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke LJ, Hollemann T, Pieler T, Renkawitz R (2002) Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech Dev 113: 95–98 [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ (2001) CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet 28: 335–343 [DOI] [PubMed] [Google Scholar]
- Filippova GN et al. (2002) Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res 62: 48–52 [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P (1999) The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer–promoter interaction. Genes Dev 13: 2098–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position–effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 10: 3202–3215 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Ishii K, Laemmli UK (2003) Structural and dynamic functions establish chromatin domains. Mol Cell 11: 237–248 [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Kuhn EJ, Viering MM, Rhodes KM, Geyer PK (2003) A test of insulator interactions in Drosophila. EMBO J 22: 2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2003) Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr Biol 13: R242–R254 [DOI] [PubMed] [Google Scholar]
- Lewis A, Murrell A (2004) Genomic imprinting: CTCF protects the boundaries. Curr Biol 14: R284–R286 [DOI] [PubMed] [Google Scholar]
- Lin Q, Wu D, Zhou J (2003) The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development 130: 519–526 [DOI] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5: 1743–1753 [PubMed] [Google Scholar]
- Lutz M et al. (2000) Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res 28: 1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R (2003) Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J 22: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M (1998) GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev 12: 3325–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Gerstein M, Yagi N (1994) Stereochemical basis of DNA recognition by Zn fingers. Nucleic Acids Res 22: 3397–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR (2000) Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10: 607–610 [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Sugiura A, Omori A, Felsenfeld G, Engel JD, Fukamizu A (2003) Human β-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol Cell Biol 23: 8946–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW (1997) The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem 272: 33353–33359 [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 and Table 1
Supplementary Information: Materials and Methods