Abstract
Aminoacylated (charged) transfer RNA isoacceptors read different messenger RNA codons for the same amino acid. The concentration of an isoacceptor and its charged fraction are principal determinants of the translation rate of its codons. A recent theoretical model predicts that amino-acid starvation results in ‘selective charging' where the charging levels of some tRNA isoacceptors will be low and those of others will remain high. Here, we developed a microarray for the analysis of charged fractions of tRNAs and measured charging for all Escherichia coli tRNAs before and during leucine, threonine or arginine starvation. Before starvation, most tRNAs were fully charged. During starvation, the isoacceptors in the leucine, threonine or arginine families showed selective charging when cells were starved for their cognate amino acid, directly confirming the theoretical prediction. Codons read by isoacceptors that retain high charging can be used for efficient translation of genes that are essential during amino-acid starvation. Selective charging can explain anomalous patterns of codon usage in the genes for different families of proteins.
Keywords: charging, codon usage, microarray, tRNA isoacceptor
Introduction
Aminoacylated transfer RNAs read codons in ribosome-bound messenger RNAs and transfer their 3′ attached amino acids to growing peptide chains. The rates of codon translation by aminoacyl-tRNAs are modulated by the total concentrations and the charged fractions of individual tRNAs. The total or relative concentrations of individual tRNAs in Escherichia coli and Bacillus subtilis, respectively, have been estimated for cells during exponential growth in various media (Dong et al, 1996; Dittmar et al, 2004). Experiments on several individual tRNAs (Yegian et al, 1966; Jakubowski & Goldman, 1984; Varshney et al, 1991; McClain et al, 1999; Sorensen, 2001) have suggested that the charged fractions of all tRNAs are about 80% during exponential growth, so that the rate of filling a ribosomal A site is nearly proportional to the total concentration of the tRNAs that read its codon. However, when the supply of an amino acid becomes limiting for protein synthesis (Elf et al, 2001, 2003), the charged levels of its cognate tRNAs (isoacceptors) may rapidly drop to near-zero values (Morris & DeMoss, 1965; Bock et al, 1966; Yegian & Stent, 1969; Sorensen, 2001). Amino-acid starvation in E. coli, during which the primary source for the missing amino acid is degradation of already existing proteins and peptides, leads to enhanced amino-acid substitution errors in nascent peptides (O'Farrell, 1978), stringent response (Cashel et al, 1996), elevated aminoacyl-tRNA synthetase levels (Grunberg-Manago, 1987) and transfer-messenger RNA activity (Pedersen et al, 2003). Interestingly, translation of selected mRNAs, such as those encoding amino-acid biosynthetic enzymes, proceeds fairly efficiently even during severe amino-acid limitation (Mandelstam, 1958; Brunschede & Bremer, 1971; O'Farrell, 1978).
In a previous work, Elf et al (2003) modelled how the charged fractions of tRNA isoacceptors that read different codons for the same amino acid respond when the supply of the amino acid becomes rate limiting for protein synthesis. A general prediction is that the charged levels of isoacceptors depend on the ratios between their total concentrations and the frequencies at which their cognate codons appear on the mRNAs in translating ribosomes. An isoacceptor with a low ratio will have a much lower charged fraction than an isoacceptor with a high ratio. Estimated tRNA concentrations and codon frequencies in E. coli (Ikemura, 1981; Dong et al, 1996) were used to predict the charged levels of members of isoacceptor families during limitation of their respective cognate amino acids. It was found that ‘starvation-sensitive codons', that is, those read by isoacceptors predicted to have very low charged levels during amino-acid starvation, are invariably used as control codons in transcriptional attenuation (Henkin & Yanofsky, 2002). Conversely, ‘starvation-insensitive codons', that is, those read by isoacceptors predicted to retain high charged levels during amino-acid limitation, are over-represented in genes for amino-acid synthetic enzymes. Until now, this theory had not been tested directly by measurements of the charged levels of tRNA isoacceptors during starvation for their cognate amino acids.
In the present work, we adapted the previously developed tRNA microarray system (Dittmar et al, 2004) for parallel assessment of the charged levels of all tRNAs in E. coli (Fig 1 and supplementary information online; see also http://www.ncbi.nlm.nih.gov/geo, accession number GSE2065). We tested, in particular, the predictions (Elf et al, 2003) of how the charged levels of isoacceptors in the leucine, threonine and arginine families respond to starvation for their cognate amino acids. The microarray data for tRNALeu isoacceptors were validated by northern blot analysis. We found that the theoretical model correctly identifies and ranks the starvation-insensitive isoacceptors, and that, in some cases, their measured charged fractions are lower than predicted.
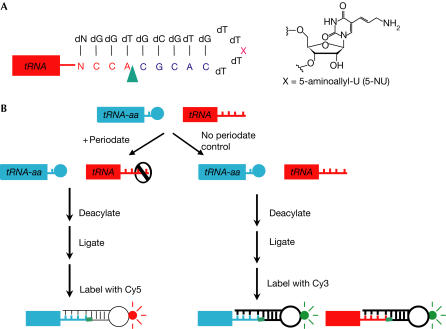
Figure 1.
Measuring tRNA charging levels by microarray. (A) The tagging oligonucleotide containing 5-aminoallyl-uridine (5-NU). tRNA is shown as a red box with four 3′ unpaired NCCA nucleotides. The ligation site is indicated by a green arrowhead. (B) The reaction scheme. Total RNA is split into two halves. One half is treated with periodate, which oxidizes the free 3′ ends of uncharged tRNAs (red), but leaves the charged tRNAs (blue) intact. Oxidized tRNAs cannot be ligated to the tagging oligonucleotide. The other half is not treated with periodate and serves as a control.
Results And Discussion
The microarray method for measuring charged fractions of tRNAs is based on the protection against periodate oxidation of the 3′ end of aminoacyl-tRNA by its covalently attached amino acid (Fig 1). Total RNA was isolated under mild acidic conditions, so that charged tRNA retains the amino acid. Each sample was split into two halves. One half was treated with periodate, which oxidizes the free 3′ ends of uncharged tRNAs, thus destroying their ability to be ligated with the oligonucleotide required for fluorescence labelling; this half was used to measure the amount of charged tRNA. The other half, without periodate treatment but otherwise handled identically, served as a control for the amount of total tRNA. Both samples were deacylated and then ligated with a tagging oligonucleotide, an RNA–DNA hybrid containing complementary nucleotides to the universal 3′ CCA of tRNA and 5-aminoallyl-uridine for fluorophore incorporation. Ligated tRNAs from the oxidized and control samples were reacted with succinimidyl esters of either the Cy5 or Cy3 fluorophore. The oxidized and the control samples were hybridized together on a microarray to determine the Cy5/Cy3 ratio for each tRNA. The charged fraction was obtained by normalizing the dye ratio to an added yeast tRNAPhe standard with a known charged level and by correcting for the background fluorescence caused by dye-reactive post-transcriptional modifications (supplementary information online).
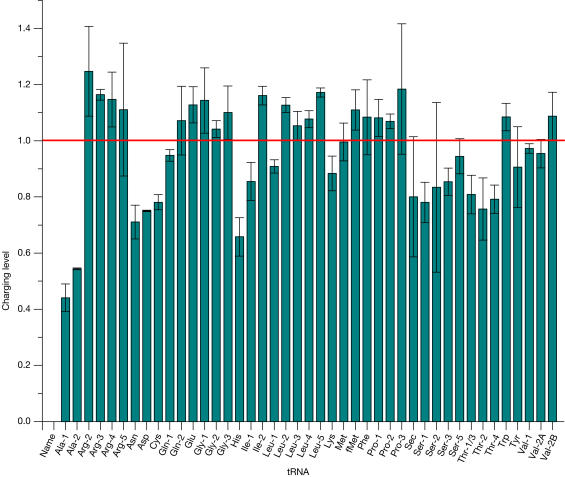
We expected that, before starvation, most tRNAs would be close to fully charged. This was indeed observed (Figs 2, 3), in line with literature reports on the charged levels of several tRNAs obtained from northern blots (Varshney et al, 1991; Kruger & Sorensen, 1998; McClain et al, 1999; Sorensen, 2001).
Figure 2.
Charged levels of tRNAs before starvation. The tRNA nomenclature is described in Table 1 and supplementary information online and is the same in the theoretical predictions (Elf et al, 2003). The error bars represent the standard deviation from dye-swap experiments where the periodate oxidized and the control samples are labelled with Cy5 and Cy3, or with Cy3 and Cy5, respectively. The large error bars for Arg-5, Ser-2 and selenocysteine-tRNA (Sec) are probably due to low fluorescence signals and low abundance for these tRNAs.
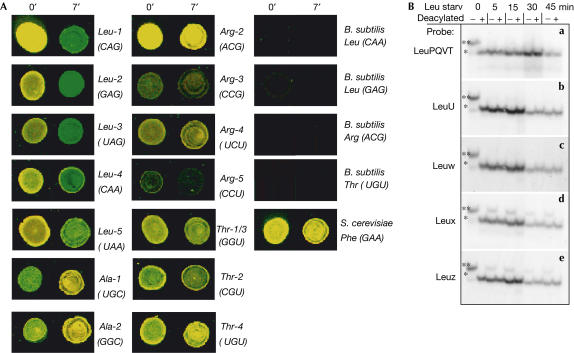
Figure 3.
RNA charging levels before and after starvation. (A) Representative microarray images from 0 and 7 min after leucine starvation. Several spots from Bacillus subtilis tRNA probes are also included to indicate the background of the array experiment. Charged yeast tRNAPhe has been added as a control. Total tRNA is labelled with Cy3 (green) and charged tRNA is labelled with Cy5 (red). In these images, overlap between Cy3 and Cy5 fluorescence is yellow representing full charging, whereas green represents decreased charging. (B) Northern blots showing the charged levels of tRNALeu isoacceptors during leucine starvation. Each panel represents a northern blot hybridized with specific 32P-labelled DNA probes. One and two asterisks indicate the position of the aminoacylated and deacylated tRNALeu, respectively.
E. coli has five tRNALeu isoacceptors that decode the six leucine codons, CUN and UUA/G. tRNALeu1 is the most abundant and decodes the most frequently used CUG codon. The model (Elf et al, 2003) predicts that, during leucine limitation, the charged levels for tRNALeu1 (CUG), tRNALeu2 (CUU/C) and tRNALeu3 (CUA/G) are less than 5% of their pre-starvation levels. In contrast, the charged levels for tRNALeu4 (UUG) and tRNALeu5 (UUA/G) are predicted to remain at between 40% and 80% of their pre-starvation levels (Table 1).
Table 1.
Summary of leucine, threonine and arginine starvation results
| tRNA isoacceptor | Anticodon | Codon | Predicted relative charged level following starvation | Observed relative charged levela |
|---|---|---|---|---|
| Leu-1(PQTV) |
CAG |
CUG |
0.05 |
0.086 (0.024)b |
| Leu-2(U) |
GAG |
CUU, CUC |
<0.02 |
0.042 (0.025) |
| Leu-3(W) |
UAG |
CUA, CUG |
0.04 |
0.039 (0.017) |
| Leu-4(X) |
CAA |
UUG |
0.8 |
0.24 (0.19) |
| Leu-5(Z) |
UAA |
UUA, UUG |
0.4 |
0.21 (0.11) |
| Thr-1/3(VT) |
GGU |
ACU, ACC |
<0.02 |
0.081 (0.08) |
| Thr-2(W) |
CGU |
ACG |
0.5 |
0.20 |
| Thr-4(U) |
UGU |
ACA, ACG, ACU |
0.1 |
0.09 |
| Arg-2(QVYZ) |
ACG |
CGU, CGC, CGA |
<0.02 |
0.22 (0.02)c |
| Arg-3(X) |
CCG |
CGG |
0.8 |
0.42 |
| Arg-4(U) |
UCU |
AGA, AGG |
0.9 |
0.43 |
| Arg-5(W) | CCU | AGG | 1.0 | NDd |
aAveraged 2–45 min after starvation.
bData in parentheses are those measured by northern blot (Figs 3,4 for tRNALeu1−5, and Sorensen (2001) for tRNAThr1 and tRNAArg2).
cThis high value from microarray measurement is probably derived from other effects as described in the text and supplementary information online. Our auxotrophic strain ceases to grow following arginine starvation, consistent with the low charged level measured by northern blot.
dNot determined due to very low signal intensity (Fig 3).
We measured the charged levels of all tRNAs following leucine starvation by microarray and northern blot (Figs 3, 4) for the relA+ strain CP78. This strain is auxotrophic for leucine, threonine and arginine, and has previously been used for starvation experiments (Sorensen, 2001). In amino-acid-containing media, its growth rate is comparable with that of wild type (Blumenthal et al, 1976). Removal of leucine, threonine or arginine leads to permanent starvation for either one of these amino acids, which makes it well suited for controlled experiments on charged tRNA levels under amino-acid limitation.
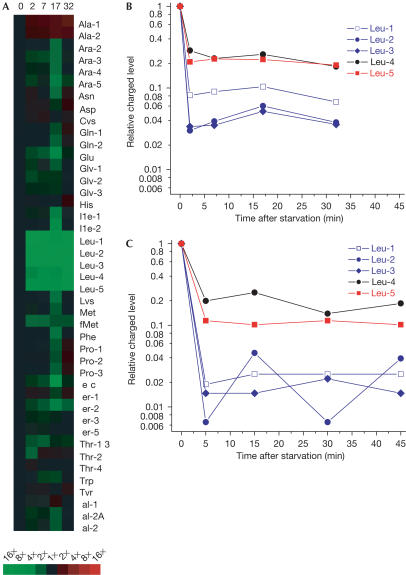
Figure 4.
Relative charged levels of tRNALeu isoacceptors during leucine starvation. (A) Relative Cy5/Cy3 ratios at 0, 2, 7, 17 and 32 min after starvation (from left to right) normalized to time zero, shown as TreeView image (Eisen et al, 1998). Red squares represent an increase and green squares represent a decrease according to the scale bar below. Relative charged levels of tRNALeu isoacceptors over time measured by microarrays (B) and by northern blot (C) are also shown. The charged levels for the tRNALeu isoacceptors before starvation determined by northern blot are as follows: LeuPQVT (Leu-1): 0.80; LeuU (Leu-2): 0.77; LeuW (Leu-3): 0.68; LeuX (Leu-4): 0.76; LeuZ (Leu-5): 0.84.
Both microarray and northern blot data show that tRNALeu4 (UUG) and tRNALeu5 (UUA/G) had much higher charged levels than the three other tRNALeu isoacceptors (CUN) following leucine starvation, in agreement with the theoretical prediction (Fig 4). There were, however, quantitative deviations: the measured charged fractions of tRNALeu4 and tRNALeu5 were between 10% and 30%, that is, significantly lower than predicted. In contrast, the low charged levels of tRNALeu2 and tRNALeu3 were very close to the theoretical predictions.
The microarray data also show that the charged levels of tRNAs for 17 of the remaining 19 amino acids changed by less than 1.5-fold over the whole time course of the experiment (Fig 4). The exceptions were the two tRNAAla isoacceptors and initiator tRNAfMet. During growth in rich media of a non-auxotrophic strain, tRNAAla2 was nearly fully charged (McClain et al, 1999). In our experiment, the charged levels of both tRNAAla isoacceptors were lower than the charged levels of other tRNAs before starvation (Fig 2), and increased about twofold after starvation (Figs 3, 4). Similar increases in the tRNAAla charged levels were observed after threonine or arginine starvation (data not shown), suggesting marginal starvation for alanine during exponential growth in MOPS minimal medium complemented with leucine, threonine and arginine before, but not after, removal of one of these auxotrophic amino acids. The charged fraction of tRNAfMet, in contrast, decreased about twofold after the onset of leucine starvation (Fig 4), but showed no change following starvation for threonine or arginine (data not shown).
We also measured the changes in the charged fractions of threonine or arginine isoacceptors following starvation of their respective amino acids (Table 1). Both tRNAThr1 and tRNAThr3 decode the most frequently used threonine codons, ACC and ACU, and were detected by a single probe on the tRNA microarray. The theory predicts tRNAThr1/3 to be sensitive to starvation, with charged fractions much lower than those of tRNAThr2(ACG) and tRNAThr4(ACA/G/U). The theory correctly identified the ranking order for charged levels: tRNAThr2>tRNAThr4>tRNAThr1/3. The charged level of tRNAThr4 was according to prediction, whereas the charged level of tRNAThr2 was lower but still more than twofold higher than the other isoacceptors. Besides tRNAAla, the charged levels of all other tRNA species changed less than 1.5-fold throughout the entire time course of the starvation experiment.
E. coli has four tRNAArg isoacceptors for reading the six arginine codons CGN and AGA/G. tRNAArg2 (CGA/U/C) is the most abundant and reads codons that are frequently used in highly expressed genes (Dong et al, 1996; Dittmar et al, 2004). The theory predicts that, following starvation for arginine, the charged fraction of tRNAArg2 will be very low, whereas the charged fractions of tRNAArg3 (CGG), tRNAArg4 (AGA/G) and tRNAArg5 (AGG) will remain high. Our experiments show that the ranking order for the charged levels of three of the arginine isoacceptors is correctly predicted: tRNAArg3∼tRNAArg4>tRNAArg2. The charged levels for the starvation-insensitive tRNAArg3 and tRNAArg4 were 40%–50% (Table 1). The signal for tRNAArg5 was low (Fig 3), which makes reliable estimation of its charged level during arginine starvation difficult. Surprisingly, the charged fraction of tRNAArg2 remained at about 20% after arginine starvation, much higher than predicted. However, previous northern blot data obtained under similar conditions show a much lower charged level of tRNAArg2 (Sorensen, 2001), in agreement with the theoretical prediction (Table 1). The highly charged fraction of tRNAArg2 measured by the microarray may either be due to crosshybridization between the probe for tRNAArg2 and an unidentified tRNA species or to other reasons (supplementary information online).
Why, then, are the predicted charged fractions of some isoacceptors significantly higher than the experimental estimates? One explanation for this could be errors in the estimates of total tRNA concentrations and of codon frequencies in actively translated mRNAs in E. coli on which the predictions have been made. This seems to be the case for the leucine-isoacceptor family, where recent northern blot experiments (M.A.S., unpublished results) indicate that the concentrations of tRNALeu4 and tRNALeu5 could have been previously overestimated (Dong et al, 1996). Alternatively, the assumption of uniform aminoacylation kinetics among all tRNA isoacceptors could be violated in some cases. A third explanation could be that ribosomes can move away from ‘hungry' codons in ways other than cognate codon reading, for example, by missense errors (O'Farrell, 1978), frame shifting (Gallant & Lindsley, 1993; Farabaugh & Bjork, 1999), by-passing (Gallant et al, 2003), cleavage of the mRNA in the A site by RelE and other endonucleases (Ivanova et al, 2004) and drop-off of peptidyl-tRNA (Menninger et al, 1994). Such non-canonical pathways were neglected in theory, but would reduce the differences between the charged fractions of the isoacceptor tRNAs during amino-acid limitation.
At the onset of amino-acid starvation, changes in the charged levels of tRNAs cognate to the rate-limiting amino acid occur rapidly, as the entire population of aminoacyl-tRNAs can turn over every second in rapidly growing E. coli (Jakubowski & Goldman, 1984). Differential charging of individual tRNA isoacceptors is therefore expected to become established in just a few seconds following amino-acid limitation, so that the steady-state assumption of the model (Elf et al, 2003) can also approximate variations in the charged levels of isoacceptors in non-auxotrophic cells. There, the charged levels are expected to first decrease, and then increase over a time period of about 10–50 min (Zaslaver et al, 2004) as the cells' capacity to synthesize amino acids increases towards its new steady-state value. Future experiments will tell whether the patterns of selective charging of tRNA isoacceptors, predicted by the model (Elf et al, 2003) and here verified by experiments, will also be apparent during starvation of non-auxotrophs.
The microarray technique for parallel detection of charged levels of tRNA isoacceptors is a powerful experimental tool that can be applied to tRNAs from any organism for which the genomic sequence is known. The selective responses of the charged levels of isoacceptors will show which tRNAs are sensitive to amino-acid limitation. This information will be useful in the search for stress-related genes, in particular for those encoding proteins with regulatory functions associated with amino-acid deficiency, in which starvation-insensitive codons are expected to be significantly over-represented.
Methods
Growth at 37°C was as described by Sorensen (2001). For microarray experiments, E. coli K12 CP78 cured for λ (N. Fiil): thr leu his argH thi mtl supE44 relA+spot+ λ− λs was first grown on agar plates containing MOPS minimal medium supplemented with 0.4% glycerol, 2.5 μg/ml thiamine, 5 μg/ml His and 50 μg/ml Arg, Leu and Thr. Overnight culture was diluted into 200 ml of the same medium for a starting A476 of ∼0.07. For northern blot experiments, cultures started from a single colony were grown exponentially for at least 10 generations before sample isolation. The growth rate was one doubling per 65–76 min and the zero time point refers to the sample before filtration. At an A476 of ∼0.7, a 200 ml portion of cells was collected on a nitrocellulose filter at 37°C, washed twice in one volume of the medium lacking the amino acid to be starved and resuspended in one volume of the same medium.
Microarray measurements. Microarray data are available on the GEO database (http://www.ncbi.nlm.nih.gov/geo, under accession number GSE2065). RNA was isolated using the trichloroacetic acid method (Kruger & Sorensen, 1998). Each sample was split into two halves. One half was subjected to periodate oxidation at 0.1 μg/μl total RNA, 100 mM KOAc/HOAc (pH 4.8) and 50 mM NaIO4; the control half was treated in the same way, except that 50 mM NaCl was used in place of NaIO4. Both samples were spiked with 0.066 μM of charged yeast tRNAPhe with its charged level (0.64–0.68) measured by thin-layer chromatography (Wolfson & Uhlenbeck, 2002). The mixtures were incubated at 22°C for 30 min and glucose was added to 100 mM. After another 5 min, the mixtures were run through a G25 spin column and precipitated twice with ethanol premixed with NaOAc/HOAc (pH 4.8).
Control and oxidized RNA samples were deacylated by incubation in 0.1 M Tris–HCl (pH 9.0) for 60 min at 37°C. Samples were ethanol precipitated and resuspended in H2O. Ligation to the tagging oligonucleotide was carried out at 0.15 μg/μl RNA in 66 mM Tris–HCl (pH 7.6), 6.6 mM MgCl2, 10 mM DTT, 66 μM ATP, 25% (v/v) DMSO, 7.5 μM tagging oligonucleotide and 1 μM T4 DNA ligase for at least 15 h at 16°C. This total RNA concentration falls in the range where the ligation product is linearly proportional to the total RNA concentration (data not shown). After ligation, samples were extracted with phenol:CHCl3 and precipitated with ethanol.
tRNA ligated with tagging oligonucleotide was reacted with Cy5 or Cy3 NHS-ester using the protocol supplied by the manufacturer (Amersham Biosciences, Piscataway, New Jersey, USA). To remove the intercalated excess dye, samples were resuspended in 6 M urea/10 mM EDTA/0.2 M NaCl, extracted with phenol:CHCl3, ethanol precipitated and resuspended in H2O to a final concentration of 0.2 μg/μl total RNA. The efficiency of labelling was analysed by denaturing PAGE followed by scanning with Molecular Dynamics Typhoon phosphorimager.
The design, printing, specificity tests of tRNA microarrays, the hybridization procedure and the data analysis were the same as described previously (Dittmar et al, 2004). Each array contained 18 replicates of 119 oligonucleotide probes complementary to all tRNAs from B. subtilis, E. coli and Saccharomyces cerevisiae. A single hybridization used 0.9 μg each of Cy3- and Cy5-labelled total RNAs.
Northern blot analysis. Northern blot analysis has been described by Sorensen (2001). The charged level in each sample was determined as the fraction of counts in the aminoacylated tRNALeu band divided by the sum of counts in the aminoacylated tRNALeu and deacylated tRNALeu bands. The following probes were used to detect tRNALeu isoacceptors: LeuPQVT (Leu-1): 5′-GTAAGGACACTAACACCTGAAGC; LeuU (Leu-2): 5′-TATTGGGCACTACCACCTCAAGG; LeuW (Leu-3): 5′-CTTGCGGCGCCAGAACCTAAATC; LeuX (Leu-4): 5′-TATTTCTACGGTTGATTTTGAA; LeuZ (Leu-5): 5′-AAAATCCCTCGGCGTTCGCGCT. The complementarities of these probes to the individual tRNAs, and their specificities, were verified by BLAST searches (www.ncbi.nlm.nih.gov/BLAST) of the E. coli K12 genome.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n2/extref/7400341s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M. Warrer for excellent technical assistance to M.A.S. We thank Dr C. Yanofsky, Dr G. Bjork, Dr J. Gallant and Dr P. Cluzel for insightful discussions. This work was supported in part by a National Institutes of Health grant (to T.P.) and by the Swedish Research Council (to M.E.). K.A.D. is a recipient of a National Science Foundation predoctoral fellowship.
References
- Blumenthal RM, Lemaux PG, Neidhardt FC, Dennis PP (1976) The effects of the relA gene on the synthesis of aminoacyl-tRNA synthetases and other transcription and translation proteins in Escherichia coli A. Mol Gen Genet 149: 291–296 [DOI] [PubMed] [Google Scholar]
- Bock A, Faiman LE, Neidhardt FC (1966) Biochemical and genetic characterization of a mutant of Escherichia coli with a temperature-sensitive valyl ribonucleic acid synthetase. J Bacteriol 92: 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunschede H, Bremer H (1971) Synthesis and breakdown of proteins in Escherichia coli during amino-acid starvation. J Mol Biol 57: 35–57 [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry D, Hernandez V, Vinella D (1996) The stringent response. In Escherichia coli and Salmonella typhimurium, Neidhardt FC (ed) pp 1458–1496. Washington, DC: ASM Press [Google Scholar]
- Dittmar KA, Mobley EM, Radek AJ, Pan T (2004) Exploring the regulation of tRNA distribution on the genomic scale. J Mol Biol 337: 31–47 [DOI] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Berg OG, Ehrenberg M (2001) Comparison of repressor and transcriptional attenuator systems for control of amino acid biosynthetic operons. J Mol Biol 313: 941–954 [DOI] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M (2003) Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300: 1718–1722 [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ, Bjork GR (1999) How translational accuracy influences reading frame maintenance. EMBO J 18: 1427–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J, Lindsley D (1993) Ribosome frameshifting at hungry codons: sequence rules, directional specificity and possible relationship to mobile element behaviour. Biochem Soc Trans 21: 817–821 [DOI] [PubMed] [Google Scholar]
- Gallant J, Bonthuis P, Lindsley D (2003) Evidence that the bypassing ribosome travels through the coding gap. Proc Natl Acad Sci USA 100: 13430–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M (1987) Regulation of the expression of aminoacyl-tRNA synthetases and translation factors. In Escherichia coli and Salmonella typhimurium, Neidhardt FC (ed) pp 1386–1409. Washington, DC: ASM Press [Google Scholar]
- Henkin TM, Yanofsky C (2002) Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. BioEssays 24: 700–707 [DOI] [PubMed] [Google Scholar]
- Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151: 389–409 [DOI] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M (2004) Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol 338: 33–41 [DOI] [PubMed] [Google Scholar]
- Jakubowski H, Goldman E (1984) Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J Bacteriol 158: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger MK, Sorensen MA (1998) Aminoacylation of hypomodified tRNAGlu in vivo. J Mol Biol 284: 609–620 [DOI] [PubMed] [Google Scholar]
- Mandelstam J (1958) Turnover of protein in growing and non-growing populations of Escherichia coli. Biochem J 69: 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain WH, Jou YY, Bhattacharya S, Gabriel K, Schneider J (1999) The reliability of in vivo structure–function analysis of tRNA aminoacylation. J Mol Biol 290: 391–409 [DOI] [PubMed] [Google Scholar]
- Menninger JR, Coleman RA, Tsai LN (1994) Erythromycin, lincosamides, peptidyl-tRNA dissociation, and ribosome editing. Mol Gen Genet 243: 225–233 [DOI] [PubMed] [Google Scholar]
- Morris DW, DeMoss JA (1965) Role of aminoacyl-transfer ribonucleic acid in the regulation of ribonucleic acid synthesis in Escherichia coli. J Bacteriol 90: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH (1978) The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14: 545–557 [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M (2003) The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112: 131–140 [DOI] [PubMed] [Google Scholar]
- Sorensen MA (2001) Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(−) strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J Mol Biol 307: 785–798 [DOI] [PubMed] [Google Scholar]
- Varshney U, Lee CP, RajBhandary UL (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem 266: 24712–24718 [PubMed] [Google Scholar]
- Wolfson AD, Uhlenbeck OC (2002) Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc Natl Acad Sci USA 99: 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegian CD, Stent GS (1969) An unusual condition of leucine transfer RNA appearing during leucine starvation of Escherichia coli. J Mol Biol 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Yegian CD, Stent GS, Martin EM (1966) Intracellular condition of Escherichia coli transfer RNA. Proc Natl Acad Sci USA 55: 839–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U (2004) Just-in-time transcription program in metabolic pathways. Nat Genet 36: 486–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information