Abstract
Expression of p16INK4a is elevated during ageing and replicative senescence. Here, we report the presence of an instability determinant within the 3′-untranslated region (UTR) of the p16 messenger RNA in WI-38 human diploid fibroblasts. The p16 3′UTR was found to be a specific target of AUF1, an RNA-binding protein implicated in promoting mRNA decay. Both AUF1 levels and AUF1–p16 mRNA associations were strikingly more abundant in early-passage than late-passage fibroblast cultures. Moreover, short interfering RNA-based reductions in AUF1 levels increased the stability of p16 3′UTR-containing transcripts, elevated the expression of p16 and accentuated the senescence phenotype. Together, our findings show that p16 mRNA turnover decreases during replicative senescence and that the instability-conferring region is located within the 3′UTR of p16, as well as identifying AUF1 as a critical mediator of these regulatory events.
Keywords: INK4a, mRNA turnover, AUF1, post-transcriptional regulation
Introduction
Human diploid fibroblasts (HDFs) proliferate in culture for a finite number of population doublings (pdl), eventually arresting in the G1 phase of the cell division cycle, but remaining viable in a metabolically active state known as replicative senescence (Hayflick, 1965; Campisi, 1997). Contributing to the growth inhibition of senescent cells are decreased levels of cell cycle regulatory proteins such as cyclin A, cyclin B1, cyclin H, CAK, Cdc2, E2F-1, E2F-2, DP-1, DHFR and PCNA, as well as increased abundance of cyclin-dependent kinase (CDK) inhibitors, particularly p16INK4a and p21WAF1 (hereafter p16 and p21, respectively), and a correspondingly reduced CDK activity (Wong & Riabowol, 1996; Cristofalo et al, 1998; Stein & Dulic, 1998; Lee et al, 1999). In turn, the activities of tumour suppressors pRB (regulated by CDKs) and p53 are constitutively elevated in senescent cells (Atadja et al, 1995; Thomas et al, 2003). Together, these findings provide support for the widely held hypothesis that cellular senescence constitutes a state of protection against tumorigenesis, particularly in young organisms (Krtolica et al, 2001).
The contribution of post-transcriptional gene regulatory events (such as messenger RNA processing, mRNA turnover, translation and protein stability) during replicative senescence is becoming increasingly apparent (Rattan, 1996; Brewer, 2002; Zhu et al, 2002). The involvement of mRNA turnover was reported for transcripts encoding cyclins A and B1, which were shown to be targets of binding and stabilization by the RNA-binding protein (RBP) HuR during the proliferative phases of the cell division cycle and in early-passage HDFs (Wang et al, 2000b, 2001). The 3′-untranslated regions (UTRs) of mRNAs encoding cyclins A and B1 contain A+U-rich elements (AREs), the best-understood cis-acting determinants of transcript stability (Xu et al, 1997). Several other RBPs have been reported to bind to AREs and cause transcript destabilization (Wilson & Brewer, 1999); among them, AUF1 (hnRNP D) has been the most extensively studied. AUF1 is expressed as a family of four protein isoforms (p37, p40, p42 and p45) arising through alternative splicing. Some differences in the activity of the various AUF1 isoforms have been documented, including ARE-mediated mRNA stabilization (Xu et al, 2001), but they generally seem to enhance the degradation of target mRNAs (Loflin et al, 1999). AUF1-dependent mRNA decay is closely linked to the ubiquitination and proteasome targeting of AUF1 (Laroia et al, 1999), and to the association of AUF1 with the exosome, an important RNA degradation machinery in the cell (Chen et al, 2001). AREs that influence transcript stability have also been described in several mitogenic (c-fos, c-jun, c-myc, egr-1), immune response (interleukins, interferons), stress response (p21, hsp70, MnSOD, catalase) and cell cycle regulatory genes (p21, cdc25).
The presence of an ARE in the p16 3′UTR led us to postulate that its expression might be influenced by alterations in mRNA stability. In particular, we sought to investigate whether p16 expression during cellular senescence was influenced by altered mRNA turnover, and used the WI-38 HDF model of replicative senescence to test this hypothesis. Here, we report the identification of a region within the 3′UTR of the p16 mRNA that confers transcript instability in early-passage HDFs, and we provide evidence that AUF1 binds to this instability region and promotes transcript decay.
Results And Discussion
Elevated p16 mRNA levels in senescent WI-38 HDFs
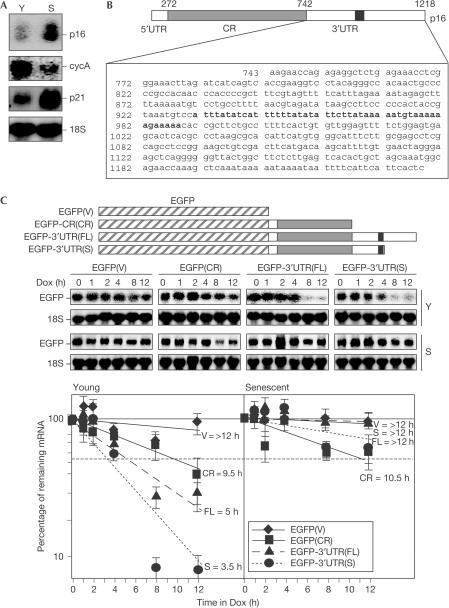
In keeping with earlier reports (Hara et al, 1995), steady-state p16 mRNA levels were markedly higher in WI-38 HDFs that had undergone senescence (S) in culture (Fig 1A). Previously, comparisons of young (Y) and senescent WI-38 cells (20–28 and 55–60 pdl, respectively), showed that the senescent populations had considerably lower CDK activity, diminished rates of [3H]thymidine incorporation, increased G1-phase cells and greatly elevated activity of a neutral, senescence-associated β-galactosidase (SA-β-gal) that is used as a biomarker of replicative senescence (Dimri et al, 1995; Wang et al, 2001). In addition, senescent cells exhibited higher levels of cyclin D1 and p21, and reduced levels of cyclin A, cyclin B1 and c-fos (Fig 1A; Wang et al, 2001).
Figure 1.
The p16 3′UTR confers instability to chimeric transcripts in young WI-38 cells. (A) Northern blot analysis of p16, cyclin A and p21 expression in young (Y, pdl 25) and senescent (S, pdl 60) WI-38 HDFs. (B) Schematic representation of the p16 mRNA, depicting the 3′UTR ARE (black), and the constructs prepared for chimeric RNA analysis. (C) Young and senescent WI-38 cells were transiently transfected with the constructs shown in (B) (1 μg each) along with the tet repressor plasmid (20 μg), and then cultured for an additional 20 h. Following addition of Dox (1 μg/ml), northern blotting was performed to assess the half-life of each transcript (supplementary information online).
Given the presence of a distinct ARE in the p16 mRNA (Fig 1B), we postulated that its senescence-dependent expression might be influenced by changes in mRNA stability. As previously suggested (Hara et al, 1995), examination of the p16 mRNA half-life in HDFs after addition of actinomycin D showed that this mRNA was highly stable in senescent populations, but its very low abundance in young HDFs made it difficult both to measure and to compare with the senescent populations (data not shown), and therefore alternative approaches were used, as described below.
p16 3′UTR confers mRNA instability in young HDFs
To test whether the half-life of p16 mRNA was altered with cellular senescence, we used a transcriptional pulse strategy based on the Tet-regulatory system (Xu et al, 1998; Lin et al, 2000). The effect of sequences within the p16 mRNA on transcript stability was studied by engineering the tTA-regulated constructs pTRE-d2EGFP(CR), pTRE-d2EGFP-3′UTR(FL) and pTRE-d2EGFP-3′UTRs(S) (Fig 1C; supplementary information online) for transfection into either early- or late-passage (young and senescent, respectively) WI-38 cells. Typically, in this transcriptional-pulse method, plasmid pTet-Off is introduced first and clones that both express high levels of tTA and potently suppress reporter gene transcription are selected by addition of tetracycline or doxycycline (Dox). Establishing and characterizing such clonal populations was not possible with WI-38 HDFs, as the lengthy processes of selection and clonal expansion rendered HDFs senescent, thereby precluding the critical testing of early-passage populations. Therefore, we adopted an alternative transfection strategy whereby each transfection group received 20 μg of plasmid pTet-Off along with 1 μg of the pTRE-d2EGFP-derived expression vectors. Following a 20-h transcriptional pulse, Dox was added to shut off the tTA-driven transcription, and the rates of clearance of the four transcripts (control transcript EGFP(V) and chimeric transcripts EGFP(CR), EGFP-3′UTR(FL) and EGFP-3′UTR(S)) in HDFs were monitored by northern blotting (Fig 1C). In young cells, chimeric transcripts EGFP(V) and EGFP(CR) were very stable, with half-lives of >12 and ∼9.5 h, respectively, whereas chimeric transcripts containing the 3′UTR were much less stable, with half-lives of 5 and 3.5 h measured for EGFP-3′UTR(FL) and EGFP-3′UTR(S), respectively. By contrast, all of the transcripts were more stable in senescent cells, including the chimeric transcripts comprising the 3′UTR, the half-lives (>12 h) of which were strikingly higher than in the young group. These observations suggested that the 3′UTR conferred instability to the p16 mRNA, mapped an instability region to the 247-base 3′UTR segment proximal to the coding region (CR) and further suggested that p16 mRNA was preferentially unstable in young HDFs.
AUF1 binding to the p16 3′UTR is senescence dependent
Given the concerns associated with the simultaneous transfection method used with WI-38 HDFs (where neither the tTA levels nor the transcriptional suppression by Dox could be directly tested in transfected cells), we sought additional evidence that the p16 3′UTR influenced mRNA stability.
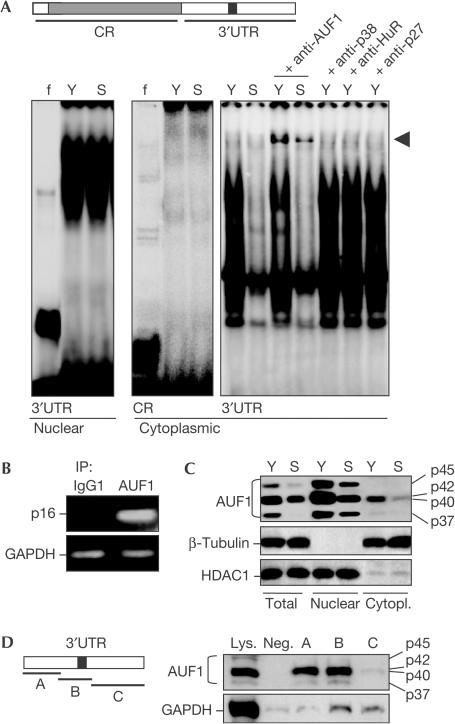
First, to ascertain the ability of cellular proteins to bind the p16 mRNA, radiolabelled RNAs corresponding to either its CR or 3′UTR were prepared for analysis by RNA electrophoretic mobility shift assay (REMSA). As shown using the 3′UTR transcript, REMSA signal patterns and intensities using nuclear lysates prepared from young (Y) and senescent (S) WI-38 cells were comparable (Fig 2A); a strong binding activity, irrespective of pdl, was also observed when using the p16 CR (not shown). Interestingly, however, proteins present in cytoplasmic lysates prepared from young WI-38 cells showed a much more extensive binding to radiolabelled transcripts than did proteins present in cytoplasmic lysates from senescent cells. The RBP AUF1 was part of these senescence-dependent (p16 3′UTR–protein) complexes, as evidenced by the ability of an anti-AUF1 antibody to supershift some of the protein–RNA associations (arrowhead), whereas antibodies recognizing other proteins, including HuR, did not produce supershifts (Fig 2A).
Figure 2.
Binding of proteins to p16 transcripts. (A) REMSA analyses were performed using nuclear or cytoplasmic protein lysates (10 μg each) obtained from young and senescent WI-38 cells. RNA probes encompassed the CR or the 3′UTR. Supershift REMSA was performed with antibodies recognizing AUF1, HuR, p38 or p27. (B) Whole-cell lysates (400 μg) were prepared from young HDFs, and endogenous target transcripts were detected by RT–PCR assay of the corresponding IP material; PCR products corresponding to GAPDH mRNA (which bound IP materials at background levels, thus serving as loading control) and p16 mRNA were visualized on agarose gels. (C) Western blot of AUF1 levels in total (10 μg), cytoplasmic (40 μg) and nuclear (10 μg) lysates prepared from either young or senescent WI-38 cells. AUF1 isoforms (p37, p40, p42 and p45) are indicated. Cytoplasm-specific β-tubulin and nucleus-specific HDAC1 (histone deacetylase 1) signals showed the quality and loading of the cytoplasmic and nuclear preparations, respectively. (D) Pull-down assay (supplementary information online) using biotinylated fragments A–C (left) to detect bound cytoplasmic AUF1 by western blotting (right). A 5 μg portion of whole-cell lysate (Lys.) and biotinylated GAPDH (negative control, Neg.) was included.
Further demonstration of the existence of (AUF1–p16 mRNA) complexes was obtained through immunoprecipitation (IP) assays under conditions that preserved endogenous protein–RNA interactions, followed by reverse transcriptase–PCR (RT–PCR) amplification of the IP material using sequence-specific primers. As shown, the p16 mRNA was prominently bound to AUF1, as a significant amount of p16 PCR product was amplified from anti-AUF1 IP reactions (Fig 2B). By contrast, IPs using control IgG1 showed undetectable signals; a control PCR product corresponding to the highly abundant GAPDH mRNA (not an AUF1 target) served to monitor background mRNA binding to IP materials (Fig 2B).
AUF1 was markedly higher in young compared with senescent WI-38 HDFs (Fig 2C). This difference was evident for all AUF1 isoforms in both the cytoplasmic and nuclear compartments. To explain the p16 3′UTR regions recognized by AUF1, biotinylated transcripts spanning segments of the 3′UTR (A, B, C) were prepared and tested in pull-down assays (Fig 2D; supplementary information online). As shown by western blotting of AUF1 in the pull-down materials, fragments A and B (but not fragment C or a negative control (a biotinylated GAPDH transcript, Neg.)) were strong targets of AUF1. Together, these results showed that AUF1, a protein reported to enhance mRNA degradation, was more abundant in the cytoplasm of early-passage HDFs and was capable of binding the p16 3′UTR.
p16 3′UTR confers instability to chimeric transcripts
The second set of experimental data supporting the unstable nature of the p16 mRNA was obtained using an immortal cell system in which stable expression of tTA was attainable. Briefly, stably transfected H2 cells (supplementary information online) expressed high levels of tTA (Lin et al, 2000). Transient transfection of pTRE-d2EGFP-derived constructs (Fig 1C) into H2 cells was followed by the addition of Dox and measurement of the stability of the chimeric transcripts (Fig 3A). Although their half-lives were found to be generally lower than those of transcripts tested after transfection of HDFs (Fig 1C), their relative stabilities were similar to those observed in young HDFs: transcripts EGFP and EGFP(CR) had the longest half-lives (∼3.5 h), whereas chimeric transcripts bearing either the full-length (FL) or partial (S) p16 3′UTR had significantly shorter half-lives (2 and 1.7 h, respectively).
Figure 3.
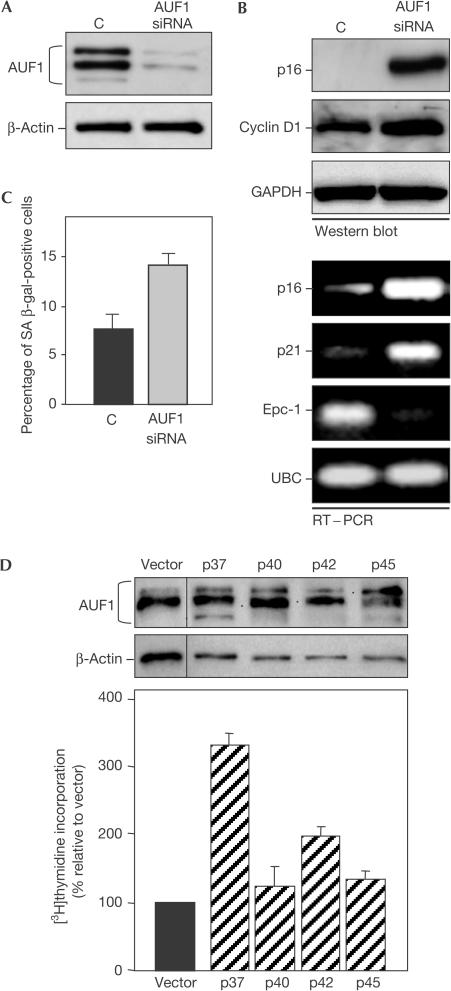
p16 3′UTR confers instability to chimeric transcripts in lung carcinoma cells. (A) Following transient transfection of H2 cells (constitutively expressing the tet repressor) with the constructs depicted in Fig 1B (1 μg each), Dox was added, RNA was collected at the times shown and the rates of clearance of each transcript (mRNA half-life) were monitored by northern blotting, as described (supplementary information online). (B) Western blot of AUF1 expression in H2 cells 3 days after transfection with plasmid pSILENCER-AUF15 (AUF1 siRNA), expressing an siRNA that targeted all four AUF1 isoforms, or control plasmid pSILENCER (lane C); β-actin, loading control. (C) At 3 days after transfection of H2 cells as explained in (B), plasmids expressing either EGFP(V) or EGFP-3′UTR(S) were transfected and the stability of the chimeric RNAs was tested (explained in (A)) in both the C and the AUF1 siRNA treatment groups.
The H2 cell system also allowed direct investigation of whether AUF1 influenced the stability of the p16 mRNA. To study this possibility, we developed a construct expressing short interfering RNA (AUF1 siRNA) targeting all AUF1 isoforms that reduced AUF1 expression to ∼15% of the levels seen in control vector (C) transfections (Fig 3B). As shown, the stability of control EGFP transcript was comparable in both the C and AUF1 siRNA populations, whereas the stability of chimeric EGFP-3′UTR(S) transcript was markedly lower in the C populations (Fig 3C). Together, these findings further supported the notions that the proximal p16 3′UTR conferred transcript instability and that AUF1 contributed to mRNA destabilization.
Modulating AUF1 levels alters cellular senescence
Given that AUF1 was capable of regulating the stability and steady-state levels of p16 mRNA (Figs 1, 2, 3), we set out to test directly whether AUF1 might be capable of controlling p16 expression in WI-38 HDFs. The efficiency of transfection into WI-38 cells was only ∼15–20% (not shown), and therefore a co-transfection strategy was devised whereby pEGFP-C1 expression vector was introduced along with either vector plasmid or expression plasmids to modulate AUF1 expression. Two sequential co-transfections were performed (1 and 4 days after plating); on day 7, cells were sorted and the EGFP-positive populations (C and AUF1 siRNA) were collected for analysis. AUF1 levels were significantly lower in the AUF1 siRNA group by 3 days after re-plating the EGFP-positive population (Fig 4A). As shown, p16 expression was strikingly higher in the AUF1 siRNA group (Fig 4B). The levels of senescence markers cyclin D1 protein and p21 mRNA were also elevated, whereas those of Epc-1 mRNA were reduced, as anticipated (Fig 4B). Importantly, SA-β-gal activity was significantly higher in EGFP-positive cells from the AUF1 siRNA group than the control (C) population (Fig 4C). Conversely, co-transfection of pEGFP-C1 with isoform-specific AUF1 expression constructs, particularly p37 and p42 (supplementary information online), moderately increased [3H]thymidine uptake (Fig 4D), supporting the notion that AUF1 expression promoted cell proliferation, as seen in young HDFs.
Figure 4.
AUF1 modulates WI-38 senescent phenotype and p16 expression. Following a sequential co-transfection of WI-38 cells with an EGFP-expressing plasmid along with either a control plasmid (lane C) or an AUF1 siRNA-expressing vector, EGFP-positive cells were sorted by flow cytometry and used to assess the expression of AUF1 (A) and additional genes (B) by either western blotting or RT–PCR, as well as β-galactosidase-positive cells (C). (D) WI-38 HDFs were co-transfected with an EGFP-expressing plasmid and each of four isoform-specific AUF1 expression vectors and AUF1 levels (top, dots indicate individual overexpressed AUF1 isoforms) and [3H]thymidine incorporation (graph) were assessed. Vector, insertless plasmid. Graphs represent the means+s.e.m. of 3–5 independent assessments.
It is interesting to note that p37 and p42 had been found to show the highest affinity for AREs and the greatest destabilizing effects on ARE-bearing mRNAs (Loflin et al, 1999). It is also worth remarking that p45 was not detectable in cytoplasmic WI-38 lysates or in complexes with biotinylated p16 3′UTR (Fig 2 C,D), whereas p37 and p40/p42 were readily visible in each instance, suggesting a preferential role for the latter isoforms in mediating p16 mRNA decay in young HDFs. However, given the complex biology of AUF1, with isoform-specific differences in their binding affinities (Wagner et al, 1998), subcellular localization (Sarkar et al, 2003) and cell type-dependent mRNA degradation (Raineri et al, 2004) and stabilization (as reported for NIH3T3 cells; Xu et al, 2001), further work is warranted to understand fully the relevance of the findings reported here and the specific effects of AUF1 on p16 mRNA turnover.
Concluding remarks
Taken together, these findings indicate that the reduction in AUF1 expression occurring with replicative senescence contributes to the stabilization and increased expression of p16 in senescent cells and the implementation of the senescence phenotype. p16 and p14ARF (another potent tumour suppressor) are encoded by the INK4a/ARF locus and share most of the 3′UTR, raising the possibility of a joint post-transcriptional regulation for both genes during senescence and tumorigenesis (additional discussion on this point is covered in supplementary information online). The differential expression of AUF1 in early- versus late-passage HDFs probably influences the expression of additional senescence-associated genes (e.g. the ARE-containing p21, cyclin D1 and p53 mRNAs, all targets of AUF1; Lal et al, 2004) and may also be linked to tumorigenesis (Gouble et al, 2002; supplementary information online). Together with earlier studies showing that HuR expression was reduced during replicative senescence (Wang et al, 2001) and work implicating hnRNP A1 and A2 (Zhu et al, 2002) in the stabilization of senescence-associated mRNAs, our findings underscore the contribution of mRNA turnover in regulating gene expression patterns during replicative senescence. We anticipate that a more complete understanding of AUF1 function will explain crucial links underlying the regulation of genes that control cellular senescence, immortalization, tumour suppression and organismal senescence.
Methods
Cell culture. Culture and transfection of early-passage (∼20–28 pdl) and late-passage (∼55–60 pdl) human WI-38 diploid fibroblasts (Coriell Cell Repositories, Camden, NJ, USA) as well as H2 cells has been described (Wang et al, 2001; supplementary information online). SA-β-gal activity was assessed using a Senescence β-Galactosidase Staining Kit (Cell Signaling Technology, Beverly, MA, USA); blue cell percentages were calculated five independent times after blind scoring by two individuals.
Northern and western blotting, and mRNA half-life measurements. Northern and western blotting reagents as well as transcription shut-off assays are described (supplementary information online; Xu et al, 1998). The p16 sequences of interest (Fig 1) were excised from pCMV-p16 (Serrano et al, 1995) and subcloned into pTRE-d2EGFP (BD Biosciences Clontech, Palo Alto, CA, USA; supplementary information online). In WI-38 HDFs, 20 μg of pTet-Off plasmid (Clontech) along with 1 μg of each of the four EGFP-expressing constructs was simultaneously transfected. H1299 cells stably expressing tTA (H2 cells; Lin et al, 2000) were transiently transfected with each of the EGFP-expressing constructs; 24 h after transfection, expression of transcripts EGFP(V), EGFP(CR), EGFP-3′UTR(FL) and EGFP-3′UTR(S) was shut off by adding Dox (1 μg/ml) and transcript half-lives were measured (supplementary information online).
Binding assays: REMSA, REMSA supershift, IP of RNA–protein complexes. Whole-cell, cytoplasmic and nuclear lysates were prepared as described (Wang et al, 2000a). For in vitro synthesis of all transcripts, plasmid pCMV-p16 was used as template in PCR reactions to amplify the CR or 3′UTR of the p16 mRNA, as described (Wang et al, 2000a). Oligomers and methodologies used for REMSA and REMSA supershift are described (supplementary information online). For IP of endogenous RNA–protein complexes, 3 mg of whole-cell lysates was subjected to IP, washes and RT–PCR as described (Lal et al, 2004; supplementary information online).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v6/n2/extref/7400346s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M. Serrano for plasmid pCMV-p16, G. Brewer for the isoform-specific AUF1 expression vectors and R.P. Wersto and the Flow Cytometry Unit (RRB, NIA) for their assistance with cell sorting.
References
- Atadja P, Wong H, Garkavtsev I, Veillette C, Riabowol K (1995) Increased activity of p53 in senescing fibroblasts. Proc Natl Acad Sci USA 92: 8348–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G (2002) Messenger RNA decay during aging and development. Ageing Res Rev 1: 607–625 [DOI] [PubMed] [Google Scholar]
- Campisi J (1997) The biology of replicative senescence. Eur J Cancer 33: 703–709 [DOI] [PubMed] [Google Scholar]
- Chen CY et al. (2001) AU-binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Cristofalo VJ, Volker C, Francis MK, Tresini M (1998) Age-dependent modifications of gene expression in human fibroblasts. Crit Rev Eukaryot Gene Expression 8: 43–80 [DOI] [PubMed] [Google Scholar]
- Dimri GP et al. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D (2002) A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res 62: 1489–1495 [PubMed] [Google Scholar]
- Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G (1995) Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 16: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37: 614–636 [DOI] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J (2001) Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA 98: 12072–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23: 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ (1999) Control of mRNA decay by heat shock-ubiquitin–proteasome pathway. Science 284: 499–502 [DOI] [PubMed] [Google Scholar]
- Lee C-K, Kloop RG, Weindruch R, Prolla TA (1999) Gene expression profile of aging and its retardation by caloric restriction. Science 285: 1390–1393 [DOI] [PubMed] [Google Scholar]
- Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M (2000) Down-regulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol 20: 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loflin P, Chen CY, Shyu A-B (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev 13: 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri I, Wegmueller D, Gross B, Certa U, Moroni C (2004) Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Research 32: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan SI (1996) Synthesis, modifications, and turnover of proteins during aging. Exp Gerontol 31: 33–47 [DOI] [PubMed] [Google Scholar]
- Sarkar B, Lu JY, Schneider RJ (2003) Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem 278: 20700–20707 [DOI] [PubMed] [Google Scholar]
- Serrano M, Gomez-Lahoz E, DePinho RA, Beach D, Bar-Sagi D (1995) Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science 267: 249–252 [DOI] [PubMed] [Google Scholar]
- Stein GH, Dulic V (1998) Molecular mechanisms for the senescent cell cycle arrest. J Invest Dermatol Symp Proc 3: 14–18 [PubMed] [Google Scholar]
- Thomas DM, Yang HS, Alexander K, Hinds PW (2003) Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol Ther 2: 124–130 [PubMed] [Google Scholar]
- Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G (1998) Structure and genomic organization of the human AUF1 gene. Genomics 48: 195–202 [DOI] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook NJ, Gorospe M (2000a) HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin S, Caldwell MC, Furneaux H, Gorospe M (2000b) HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 19: 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M (2001) Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 21: 5889–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Brewer G (1999) The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol 62: 257–291 [DOI] [PubMed] [Google Scholar]
- Wong H, Riabowol K (1996) Differential CDK-inhibitor gene expression in aging human diploid fibroblasts. Exp Gerontol 31: 311–325 [DOI] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB (1997) Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol 17: 4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB (2001) Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol 21: 6960–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Loflin P, Chen C-Y, Shyu A-B (1998) A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res 26: 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Xu G, Ghandhi S, Hubbard K (2002) Modulation of the expression of p16INK4a and p14ARF by hnRNP A1 and A2 RNA-binding proteins: implications for cellular senescence. J Cell Physiol 193: 19–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information