Abstract
Kaede is a natural photoconvertible fluorescent protein found in the coral Trachyphyllia geoffroyi. It contains a tripeptide, His 62-Tyr 63-Gly 64, which acts as a green chromophore that is photoconvertible to red following (ultra-) violet irradiation. Here, we report the molecular cloning and crystal structure determination of a new fluorescent protein, KikG, from the coral Favia favus, and its in vitro evolution conferring green-to-red photoconvertibility. Substitution of the His 62-Tyr 63-Gly 64 sequence into the native protein provided only negligible photoconversion. On the basis of the crystal structure, semi-rational mutagenesis of the amino acids surrounding the chromophore was performed, leading to the generation of an efficient highlighter, KikGR. Within mammalian cells, KikGR is more efficiently photoconverted and is several-fold brighter in both the green and red states than Kaede. In addition, KikGR was successfully photoconverted using two-photon excitation microscopy at 760 nm, ensuring optical cell labelling with better spatial discrimination in thick and highly scattering tissues.
Keywords: fluorescent protein, mutagenesis, optical labelling, photoconversion, two-photon excitation
Introduction
Fluorescent proteins, such as green fluorescence protein (GFP) from the jellyfish Aequoria victoria and GFP-like fluorescent proteins from Anthozoa species, allow the direct genetic encoding of fluorescence, facilitating their wide usage in molecular and cell biology (Tsien, 1998; Zhang et al, 2002; Miyawaki et al, 2003). An emerging property of some fluorescent proteins is that their maturation and subsequent fluorescence can be ‘photoactivated' by illumination at specific wavelengths, which is advantageous because it allows fluorescence to be turned on with high spatio-temporal resolution. During the past few years, three new fluorescent proteins that undergo photochemical modification in or near the chromophore have been developed (Ando et al, 2002; Patterson & Lippincottschwartz, 2002; Chudakov et al, 2003). They enable selective activation of fluorescence signals after specific illumination, and can be used to fluorescently mark individual cells, organelles or proteins.
A green-emitting fluorescent protein cloned from the stony coral Trachyphyllia geoffroyi was serendipitously discovered by Ando et al (2002) to be useful as an optical cell marker. The fluorescent protein, named Kaede, emits bright green fluorescence after synthesis, but changes efficiently to a bright and stable red fluorescence on irradiation with UV or violet light. This photoconversion results in a >2,000-fold change in the red/green fluorescence ratio. Selective exposure of some cells expressing the protein to (ultra-) violet light allows for cellular marking, with the added benefit that the excitation wavelength required for fluorescence does not induce photoconversion.
Kaede contains a unique tripeptide, His 62-Tyr 63-Gly 64, which acts as a green chromophore that can be photoconverted to red. A recent study by Mizuno et al (2003) provided insights into the molecular mechanisms of Kaede photoconversion. UV irradiation causes peptide cleavage between Nα and Cα of His 62 as a result of a β-elimination reaction. Following cleavage, the imidazole ring of His 62 becomes a functional part of the red chromophore 2-[(1E)-2-(5-imidazolyl)ethenyl]-4-(p-hydroxybenzylidene)-5-imidazolinone. Active involvement of His 62 in this photochemical reaction was suggested. Substitution of tyrosine, tryptophan, aspartate or arginine for His 62 led to dimmed or abolished fluorescence, whereas substitution of Tyr 63 or Gly 64 yielded green-emitting mutants that did not show photo-induced cleavage or photoconversion. Thus, His 62 is required for initiating the β-elimination reaction. Conversely, mutations outside the chromophore-forming region, such as A69S, generated mutants that fluoresced green but were not capable of photoconversion. It is therefore concluded that His 62 is not sufficient for the reaction; photo-induced peptide cleavage and the resulting expansion of π-conjugation seem to require a strict three-dimensional structure for catalysis.
These findings about Kaede led us to attempt to engineer an ordinary fluorescent protein into a photoconvertible one. Our strategy was to replace the original chromophore-forming tripeptide of a GFP-like protein with His 62-Tyr 63-Gly 64, and then introduce numerous additional mutations to generate a photoconvertible mutant. As a starting template, we used a novel fluorescence protein from stony coral, from which highly diffractive crystals could be formed, allowing us to direct the mutagenesis of specific residues for chromophore environment optimization.
Results and Discussion
A new green-emitting fluorescent protein from coral
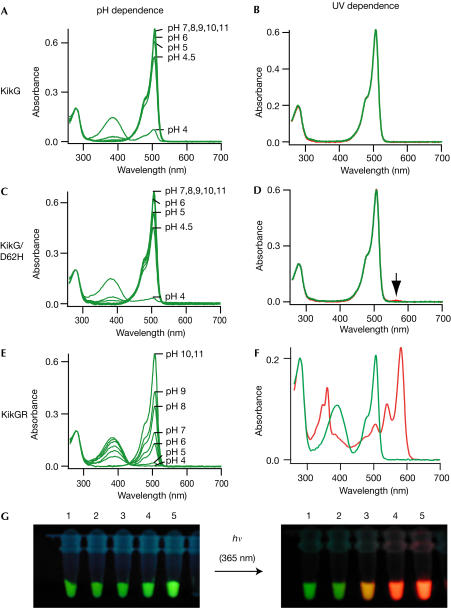
We obtained a full-length complementary DNA carrying a 684 base pair (bp) coding region from the stony coral Favia favus. Escherichia coli expressing the protein showed strong green fluorescence. We dubbed the protein KikG, because it was derived from Favia, the Japanese name of which is ‘Kikume-ishi', and because it emitted green fluorescence. The amino-acid sequence of KikG is shown in Fig 1. KikG showed a principal absorption maximum at 507 nm with a high molar extinction coefficient (ɛ=81 × 103 M−1 cm−1) at pH 7.4 (Fig 2A). At acidic pH, the 507 nm peak decreased, whereas another broad peak at around 390 nm increased, probably corresponding to the ionized and neutral chromophore forms, respectively. The apparent pKa was ∼4.2. The excitation and emission spectra showed maxima at 507 and 517 nm, respectively. Whereas the ionized form is highly fluorescent (fluorescence quantum yield (ΦFL)=0.68), the neutral form did not emit significant steadystate fluorescence when excited at pH 4.0.
Figure 1.
Amino-acid sequence (single-letter code) alignment of DsRed and Kaede with KikG and a derived mutant, KikGR. The numbering is on the basis of KikG. Regions forming βsheets are indicated by black bars. The residues forming the interior of the β-can fold are shaded. Of these, the residues that were subjected to site-directed random mutagenesis are indicated in dark grey shading. Residues responsible for chromophore synthesis are indicated by asterisks.
Figure 2.
Spectral properties of KikG variants. (A,C,E) pH dependence of absorption spectra of KikG (A), KikG/D62H (C) and KikGR (E). (B,D,F) Absorption spectrum at pH 7.0 acquired before (green line) and after (red line) irradiation with ∼365 nm light (1.8 mW cm−2) for KikG (B), KikG/D62H (D) and KikGR (F). (G) Fluorescence of protein solutions of KikG mutants acquired immediately after (left) and several minutes after (right) excitation with ∼365 nm light. 1, KikG; 2, KikG/D62H; 3, KikG/M40V/D62H/I198M; 4, KikG/M10I/L12V/M40V/V60A/D62H/Y119N/P144S/R197L/I198M; 5, KikGR.
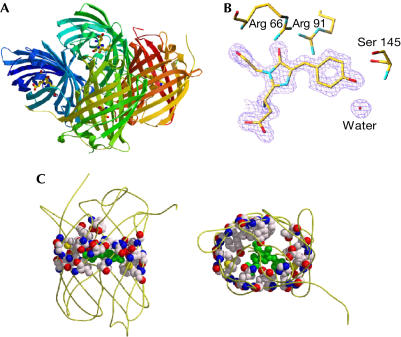
The crystal structure of KikG has been determined at 1.6 Å resolution. A well-packed tetramer structure generated by means of crystallographic symmetry (Fig 3A) shows common features to that of DsRed, including the ‘squat rectangular prism' structure (Yarbrough et al, 2001) with two interfaces for oligomerization mediated by hydrophobic interactions and hydrogen bonding. The overall structure of the KikG protomer was similar to that of Aequoria GFP, DsRed and other previously solved GFP-like proteins (Örmo et al, 1996; Yang et al, 1996; Wall et al, 2000; Yarbrough et al, 2001), characterized by an 11stranded β-can with a central α-helix holding a chromophore. The electron density map (Fig 3B) indicates the presence of the 5-[(4-hydroxypheny)methylene]-imidazolinione chromophore system, which adopts a planar conformation. The chromophore is tightly packed by the surrounding residues, including Arg 66, Arg 91 and Ser 145 (Fig 3B), and water molecules.
Figure 3.
KikG structure (Protein Data Bank accession code 1XSS). (A) The quaternary, tetrameric structure of KikG, with each protomer represented in a different colour. The chromophore is shown in a ball-andstick representation. (B) The refined model of the KikG chromophore is superimposed on a portion of the electron density map. Some surrounding side chains and a water molecule are shown. (C) The residues selected for random mutagenesis are represented in balls. Left, side view; right, top view. Graphics are outputs of Molscript and Raster3D softwares.
Structure-assisted evolution of photoconvertibility
KikG did not demonstrate any spectral alternation on irradiation with (ultra-) violet light (Fig 2B,G). Aiming to obtain a photoconvertible version of KikG, we replaced Asp 62 with His, which is required in that position to initiate the β-elimination reaction observed in the photoconversion of Kaede (Ando et al, 2002). Although the resulting mutant, KikG/D62H (Fig 1), emitted stable green fluorescence, only a slight increase in the absorption spectrum at 570 nm (Fig 2D, indicated by an arrow) was observed after irradiation with UV light (1.8 mW cm−2) for over 5 h.
We then mutated amino-acid residues surrounding the chromophore, selecting those predicted to favour photo-induced cleavage and cause colour conversion. On the basis of the KikG crystal structure, we selected pairs of amino-acid residues with side chains orientated towards the chromophore on each of the 11 β-strands of KikG/D62H (Fig 3C; supplementary information online). A degenerative primer was designed for each strand so that the two residues would be randomly replaced with other amino acids. We also used a combination of several primer sets to introduce random mutations at several selected sites simultaneously. Plates of E. coli cells transformed with mutagenized plasmid DNAs were screened for the development of red fluorescence under a dissection microscope equipped with a 75 W xenon lamp and rhodamine filters following illumination at 365 nm (1.8 mW cm−2) for 1 h. Out of approximately 6,000 colonies, one colony emitting red fluorescence was identified. Sequence analysis showed that the protein carried two mutations, M40V and I198M. Although our site-directed random mutagenesis approach was performed using KikG/M40V/D62H/I198M, the products were also subjected to error-prone PCR to introduce additional variation at other positions that might affect the photoconversion reaction or protein folding. Each mutagenesis cycle introduced amino-acid changes by both site-directed random mutagenesis and error-prone PCR. The directed evolution of KikG towards a photoconvertible protein was achieved after 30 cycles. The introduction of eight mutations, M10I, M40V, V60A, D62H, K70E, Y119N, R197Q and I198M, transformed KikG into an efficient highlighter, which we refer to as ‘KikGR' (Figs 1 and 2F,G).
The spectral properties of the green and red states of KikGR (referred to as green KikGR and red KikGR, respectively) were analysed. The absorption spectrum of green KikGR measured at pH 7.4 (Fig 2E) demonstrates two principal absorption peaks at 390 nm (ɛ=14.5 × 103 M−1 cm−1) and 507 nm (ɛ=28.2 × 103 M−1 cm−1), corresponding to the neutral and ionized chromophore forms. Although the absorption spectrum was similar to that of KikG, the absorption of green KikGR proved to be more pHsensitive: pKa=7.8 (green KikGR; Fig 2E) versus pKa=4.2 (KikG; Fig 2A). The fluorescence emission spectrum (ΦFL=0.7) of green KikGR was also indistinguishable from that of KikG. At pH 7.4, red KikGR showed two principal absorption peaks at 583 nm (ɛ=32.6 × 103 M−1 cm−1) and 360 nm (ɛ=21.7 × 103 M−1 cm−1; Fig 2F). At acidic pH, these peaks both decreased at an apparent pKa of ∼5.5 (data not shown), indicating that they derive from the ionized form of the red chromophore. Red KikGR showed an emission peak at 593 nm and two excitation peaks at 583 nm (ΦFL=0.65) and 360 nm (ΦFL=0.92). As photoconversion of KikGR was highly dependent on irradiation wavelength (350–420 nm) and pH (efficient at lower pH), as reported for Kaede (Ando et al, 2002), photoconversion probably initiates from excitation of the neutral chromophore form. Therefore, the acquisition of a high pKa may be necessary to evolve efficient photoconvertibility.
To determine whether there was an increase in the quantum yield for photoconversion (ΦPC), we compared ΦPC between KikGR and two intermediate mutants. ΦPC values, determined using the extinction coefficient and photoconversion rate on irradiation with violet light of these mutants, were 2.7 × 10−3 (KikG/M40V/D62H/I198M), 3.5 × 10−3 (KikG/M10I/L12V/M40V/V60A/D62H/Y119N/P144S/R197L/I198 M) and 4.7 × 10−3 (KikGR), respectively.
We thus concluded that both the increase in the quantum yield of photoconversion and the pKa of chromophore protonation were responsible for the evolution of photoconvertibility.
Performance as a highlighter in cellular applications
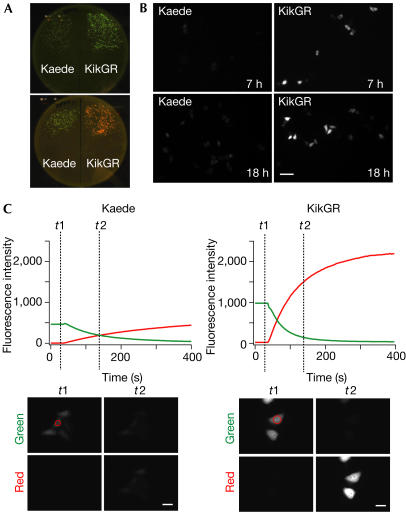
To determine the performance of KikGR in biological applications, we performed a comparative analysis with Kaede. Bacterial colonies expressing KikGR emitted brighter green fluorescence than those expressing Kaede (Fig 4A, top). Furthermore, these colonies turned to red more rapidly when placed near a laboratory window (Fig 4A, bottom). These trends were also observed in mammalian cells. HeLa cells expressing KikGR showed significant green fluorescence several hours after transfection, which increased several-fold with time (Fig 4B, right). Under the same conditions, the green fluorescence of Kaede became detectable 12 h after transfection (Fig 4B, left). At 1 day after transfection, the cells were subjected to continuous illumination at 400 nm, with intermittent monitoring of the green and red fluorescence signals. KikGR showed an approximately threefold faster photoconversion than Kaede (Fig 4C). Photoconversion is not detrimental to the cells, as highlighted cells go on to divide at a similar rate as non-highlighted cells (supplementary Fig S1 online). The brighter fluorescence of KikGR compared with Kaede was observed not only in HeLa cells, but also in human embryonic kidney (HEK) 293 cells and hippocampal primary neurons (data not shown).
Figure 4.
Performance of KikGR in cells. (A) Escherichia coli colonies expressing Kaede (left) and KikGR (right) on a bacterial plate. Transformed cells were incubated in the dark for 12 h (top), and then left for an hour near a window to allow for photoconversion (bottom). Fluorescence images were taken using a home-made plate image analyser (Sawano & Miyawaki, 2000). (B) HeLa cells expressing Kaede (left) or KikGR (right) at 7 h (top) and 18 h (bottom) after transfection. Scale bar, 100 μm. (C) HeLa cells expressing Kaede or KikGR were photoconverted with violet light (400 nm). Green and red fluorescence intensities were measured with excitation at 475 and 550 nm, respectively, and are plotted against time (top). The green and red fluorescence images at t1 and t2 are shown (bottom). Scale bar, 30 μm. The images in (A–C) were acquired and are displayed under the same conditions for Kaede- and KikGR-expressing cells.
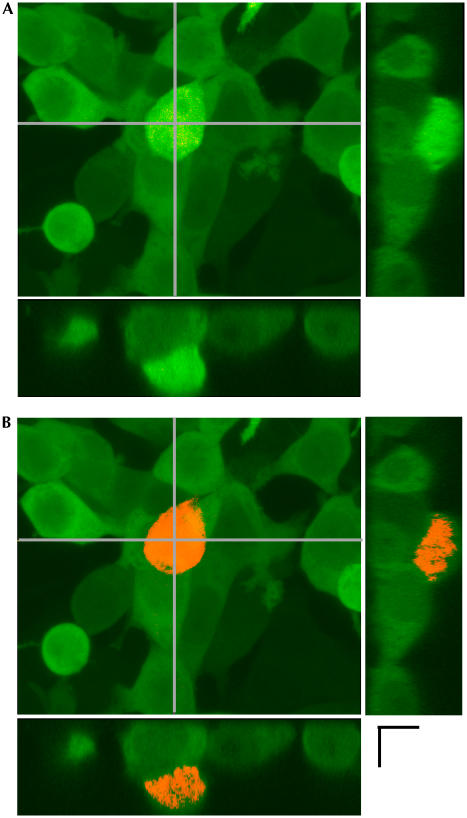
These results validated the use of KikGR for both bright cell labelling and efficient optical marking. Moreover, photoconversion by two-photon excitation using an infrared ultrashort-pulsed laser was possible with KikGR. The nonlinear properties of two-photon excitation allow spatial resolution of photoconversion, permitting the labelling of a specific single cell within densely or three-dimensionally packed cells (Fig 5). Such spatial resolution is impossible with photoconversion by means of single-photon excitation.
Figure 5.
Optical labelling by two-photon excitation. (A) HEK-293 cultures expressing KikGR were imaged using an FV500 Olympus confocal microscope equipped with Ar (488 nm) and He/Ne (543 nm) lasers. Transverse images were reconstituted using the Fluoview software. (B) KikGR photoconversion was restricted to only one cell after a Tisapphire ultra-short pulse laser (760 nm, 80 MHz). Scale bar, 10 μm.
Although the molar extinction coefficient of KikGR at pH 7.4 (ɛ=28.2 × 103 M−1 cm−1) is about threefold lower than that of Kaede (ɛ=98.0 × 103 M−1 cm−1), KikGR fluorescence was several-fold brighter than Kaede fluorescence in cells. Two possibilities might explain this result: either KikGR protein is expressed at higher levels or chromophore formation of KikGR is more efficient. Following expression of KikGR or Kaede with an epitope tag in cells, we estimated the total amount of protein, including unfolded proteins, by immunocytochemistry with antibodies against the tag. KikGR showed higher total protein amounts and a higher ratio of fluorescence to the amount of tag, suggesting that both effects contribute to the bright fluorescence of KikGR (supplementary Table S1 online).
Similar to most GFP-like proteins, KikGR oligomerizes. This oligomerization will not limit the use of KikGR for labelling cells, but will preclude its use in many fusion protein applications. Recently, two monomeric photoconvertible fluorescent proteins have been developed from GFP-like proteins by site-directed and/or random mutagenesis approaches, and their ability to track protein movement with high contrast has been demonstrated (Chudakov et al, 2004; Miyawaki, 2004; Wiedenmann et al, 2004). Monomerization of KikGR is now under way, although thorough optimization is required to obtain a practically useful mutant.
Conclusions
We report the in vitro structure-aided evolution of a newly cloned fluorescent protein from the stony coral Favia favus into a photoconvertible tracer. The most evolved variant, KikGR, is more advantageous for cell labelling and optical marking than the previously discovered photoconvertible protein Kaede. KikGR shows more efficient photoconversion and several-fold greater brightness of green and red fluorescence when expressed in cells. In addition, KikGR fluorescence demonstrates a larger separation of green and red emission wavelengths, which enables more efficient detection of photoconversion. The photoconvertible properties of KikGR allowed us to mark individual cells with high spatial resolution by means of two-photon excitation. Our study demonstrates both the usefulness of this engineered highlighter for biological applications and the validity of structure-aided mutagenesis to generate fluorescent proteins as useful optical tools.
Methods
Complementary DNA cloning. A small piece of stony coral, F. favus, was collected with the aid of an aqualung in the shallow water off Sesoko Island (Okinawa, Japan). Total RNA was isolated with TRIZOL reagent (Gibco BRL, MD, USA). Reverse transcription of cDNA from total RNA, PCR-based amplification of the cDNA fragments of interest using degenerate primers and subsequent generation of a full-length cDNA by rapid amplification of cloned ends were performed as described previously (Matz et al, 1999; Karasawa et al, 2003). The degenerate primers used were 5′-GGIWSBGTIAAYGGVCAYDANTT-3′ and 5′-ACVGGDCCATYDGVAAGAAARTT-3′ (R=A,G; Y=C,T; V=A,C,G; D=A,G,T; W=A,T; S=C,G). For bacterial expression of the protein, a cDNA fragment containing the coding region was amplified using primers containing 5′-BamHI and 3′-XhoI sites, and was cloned in-frame into the BamHI/XhoI site of the modified pRSETb vector, which introduces a 6 × -His tag at the carboxyl terminus of the protein after the addition of two amino acids (-Leu-Glu-) corresponding to the XhoI site. For mammalian expression, the cDNA containing the coding region with a Kozak consensus sequence at the 5′ end was cloned into the BamHI/EcoRI site of the pCS2+ vector.
Mutagenesis. Site-directed random mutations were introduced as described previously (Sawano & Miyawaki, 2000). Several degenerative primers, more than ten in some cases, were used together for reactions. Additional random mutations were introduced using error-prone PCR (Cadwell & Joyce, 1994).
Protein expression, purification and spectroscopy. Protein expression in E. coli (JM109DE3) and purification by Ni2+ affinity chromatography were performed as described previously (Ando et al, 2002). Absorption/fluorescence spectroscopy and pH titration were performed at 20–25°C as previously described (Ando et al, 2002).
Cell imaging. HeLa and HEK293 cells were transfected with plasmids using Lipofectamine reagent (Invitrogen, CA, USA) and imaged from 6 to 24 h post-transfection. Cells were imaged on an inverted microscope (IX70 Olympus, Tokyo, Japan) with a 75 W xenon lamp. Excitation, dichroic and emission filters for imaging green and red fluorescence were selected as described (Ando et al, 2002). The excitation filters used were 400DF10 for photoconversion, and 475AF20 and 550DF30 for excitation of green and red chromophores, respectively. To examine the photoconversion kinetics, a drop of the protein solution (10 μM, ∼100 nl) was placed on a glass-bottomed dish under mineral oil. ΦPC was calculated from the development of red fluorescence obtained with direct excitation at 550 nm. Excitation and emission filtering were controlled by means of Lamda 10-2 (Sutter Instrument, CA, USA). Images were acquired using a cooled CCD camera (Cool Snap HQ, Roper Scientific, Tucson, AZ, USA), controlled using MetaFluor 4.5 software (Universal Imaging, Media, PA, USA). The power of the excitation light was measured with a power meter (OPHIR Optronics, Israel) on a home-made mount. Confocal imaging was performed using a laserscanning system (FV500 Olympus) equipped with a × 60 objective (NA 1.30) and Ar (488 nm) and He/Ne (543 nm) lasers. Two-photon excitation for KikGR photoconversion was performed using a Ti-Sapphire laser tuned at 760 nm (Tsunami, Spectra Physics).
Crystallization. After purification on a cation exchange column (MonoQ, Amersham, NJ, USA), the protein was concentrated and exchanged at ∼30 mg ml−1 into 10 mM NaCl and 10 mM Tris–HCl (pH 9.0) by filtration (Vivaspin, Vivascience). The protein solution, mixed with an equal volume of a crystallization mother liquid (4.0 M NaCl, 0.1 M Hepes–NaOH (pH 7.0)), was microseeded and crystallized by sitting-drop vapour diffusion. Crystals grew in a few days at 20°C. After soaking in a cryoprotectant (20% v/v glycerol, 4.0 M NaCl, 0.1 M Hepes–NaOH (pH 7.0)), samples were flash-frozen with a 100 K N2-gas stream before data collection.
Crystal structure determination. Diffraction data from a single crystal were collected at beamline 44B2 at the SPring-8 facility. Data were processed using Mosfilm packages. The crystals belong to space group C2 with two molecules in the asymmetrical unit. The unit cell dimensions were a=97, b=119, c=49 Å and β=120°. The statistics of the crystals analysed in this report are summarized in the supplementary information online section. Initial phasing was performed according to the molecular replacement method using the Molrep program in the CCP4 suite. The DsRed structure was used as a search probe (PDB# 1GGX; Wall et al, 2000). Refinement and graphical modelling were performed with CNS and TURBO software, respectively. After data collection, radiated crystals were re-dissolved to test their spectral properties, which were not altered from those of fresh protein solutions (data not shown).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400361s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Higa and the staff at Sesoko Marine Station of Ryukyu University for their support during our sample collection excursions. This work was partly supported by grants from the Special Postdoctoral Researcher Program of RIKEN to H.T., CREST of Japan Science and Technology, the Special Coordination Fund for the promotion of the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government, the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan, the New Energy and Industrial Technology Development Organization and the Human Frontier Science Program.
References
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A (2002) An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA 99: 12651–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF (1994) Mutagenic PCR. PCR Methods Appl 3: 136–140 [DOI] [PubMed] [Google Scholar]
- Chudakov DM, Belousov VV, Zaraisky AG, Novoselov VV, Staroverov DB, Zorov DB, Lukyanov S, Lukyanov K (2003) Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol 21: 191–194 [DOI] [PubMed] [Google Scholar]
- Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA (2004) Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol 22: 1435–1439 [DOI] [PubMed] [Google Scholar]
- Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A (2003) Green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem 278: 34167–34171 [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradokov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyyanov SA (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17: 969–973 [DOI] [PubMed] [Google Scholar]
- Miyawaki A (2004) Fluorescent proteins in a new light. Nat Biotechnol 22: 1374–1376 [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Sawano A, Kogure T (2003) Lighting up cells: labeling proteins with fluorophores. Nat Cell Biol 5(Suppl): S1–S7 [PubMed] [Google Scholar]
- Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A (2003) Photo-induced peptide cleavage in the green-to-red conversion of a fluorescence protein. Mol Cell 12: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Örmo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (1996) Crystal structure of the Aequorea victoria green fluorescent protein. Science 273: 1392–1395 [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincottschwartz J (2002) A photoactivatable GFP for selective labelling of proteins and cells. Science 273: 1873–1877 [DOI] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A (2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res 28: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67: 509–544 [DOI] [PubMed] [Google Scholar]
- Wall MA, Socolich M, Ranganathan R (2000) The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol 12: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU (2004) EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA 101: 15905–15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Moss LG, Phillips GN Jr (1996) The molecular structure of green fluorescent protein. Nat Biotechnol 14: 1246–1251 [DOI] [PubMed] [Google Scholar]
- Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ (2001) Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc Natl Acad Sci USA 98: 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Campbell RE, Ting AY, Tsien RY (2002) Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3: 906–918 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information