Abstract
Telomerase is the ribonucleoprotein reverse transcriptase that adds telomeric DNA repeats to the ends of chromosomes. This involves annealing of the telomerase RNA template to the 3′ end of the chromosome, reverse transcription of the RNA template by the telomerase reverse transcriptase polypeptide and translocation. Here, we overexpress and partially purify the catalytically active yeast telomerase core in its natural host and probe telomerase RNA base methylation accessibility with dimethyl sulphate in the presence and absence of a DNA substrate and after substrate elongation. The length of the RNA–DNA hybrid is kept constant at seven base pairs after primer binding and elongation. Thus, new base-pair formation at the 3′ end of the substrate during elongation coincides with disruption of base-pair interactions at the other side of the template. Presumably, this circumvents the generation of an exceedingly high energy barrier for translocation and dissociation. Our analysis also corroborates recently proposed yeast telomerase RNA secondary structure models.
Keywords: telomerase reaction cycle, telomerase RNA structure, dimethyl sulphate, reverse transcriptase
Introduction
The telomerase catalytic core is provided by the telomerase reverse transcriptase (TERT) polypeptide, which reverse transcribes a short stretch of the telomerase RNA moiety (TER) to extend the chromosome 3′ ends. TERT is thought to function similarly to reverse transcriptases from retroviruses and retroelements because they share highly conserved sequence motifs (Kelleher et al, 2002). However, reverse transcription by telomerase involves resetting the relative positions of the RNA template and telomeric substrate in the active site after the addition of each telomeric repeat. Thus, in addition to the RT motifs, TERT contains one or two amino-terminal RNA-binding domains that bind TER outside the template and that may enable stable association with telomerase RNA while allowing the template to move through the active site (Bryan et al, 2000). Other telomerase-associated proteins have a role in telomerase assembly and its regulation at chromosome ends in vivo, but they are not required for enzymatic activity in vitro (Evans & Lundblad, 2000).
Single-stranded telomeric DNA is bound by telomerase through base pairing with the RNA template and through protein–DNA interactions at the so-called anchor site. Measurement of the dissociation rates of different telomeric oligonucleotide substrates with telomerase from Euplotes aediculatus showed that substrate affinity was largely independent of the extent of base complementarity between primer and template (Hammond & Cech, 1998). This suggested that either the protein–DNA interaction could compensate for different binding energies stemming from a variable number of base pairs or that telomerase maintains a more or less constant RNA–DNA hybrid length during the reaction cycle. In a study with Tetrahymena telomerase with mutant telomerase RNA templates and various DNA substrates, it was concluded that the requirement for potential pairing between primer 3′ end and template increases as catalysis moves along the template (Wang et al, 1998).
The comparison of the telomerase RNA sequences from different phyla demonstrates that this RNA diverged quickly in evolution. The sizes range from 150–200 nucleotides (nt) in ciliates to about 450 nt in vertebrates and about 1,200 nt in Saccharomyces. The telomerase RNA sequences can be aligned with confidence only among closely related organisms, and secondary structure models were derived for the different groups by phylogenetic comparison (Romero & Blackburn, 1991; Lingner et al, 1994; Chen et al, 2000; Dandjinou et al, 2004; Zappulla & Cech, 2004). In addition, chemical probes and structurespecific RNases have been instrumental in evaluating the secondary structure models (Bhattacharyya & Blackburn, 1994; Zaug & Cech, 1995; Sperger & Cech, 2001; Antal et al, 2002; Dandjinou et al, 2004). Despite the divergence in sequence and size, the template of all telomerases is single stranded, and all telomerase RNAs have been proposed to contain a pseudoknot domain. The potential of the telomerase RNA to form a pseudoknot structure is well supported through nucleotide covariation in ciliates (ten Dam et al, 1991; Lingner et al, 1994) and vertebrates (Chen et al, 2000), and alternative pseudoknot structures have also been proposed for telomerase RNAs from Saccharomyces (Chappell & Lundblad, 2004; Dandjinou et al, 2004; Lin et al, 2004) and Kluyveromyces (Tzfati et al, 2003).
Here, we overexpress the TERT (Est2) and TER (Tlc1) subunits from yeast and probe the telomerase RNA structure with dimethyl sulphate (DMS) in the absence and presence of DNA substrate primers and after substrate elongation. Our results indicate that telomerase maintains a constant number of base pairs between template RNA and telomeric DNA substrate. In addition, our results corroborate and refine the telomerase RNA secondary structure models that were recently proposed for Saccharomyces cerevisiae.
Results and Discussion
To obtain sufficient amounts of telomerase for structural probing, we co-overexpressed the telomerase reverse transcriptase Est2 and the RNA moiety Tlc1 with the Gal1 promoter in S. cerevisiae. These two components form the catalytic core of telomerase, and their overexpression leads to increased in vitro telomerase activity without strongly affecting telomere length (Teixeira et al, 2002). Overexpressed telomerase was enriched through purification over a DEAE column (see Methods). The telomerase RNA in this fraction was probed for accessibility by the methylating agent DMS in the template region (Fig 1) and other parts of the RNA (Fig 2; supplementary Fig 1 online). Our analysis covers the essential core region described previously (Livengood et al, 2002). DMS methylation at position N-1 of A and N-3 of C in RNA occurs readily in singlestranded RNA, but the bases become protected after base-pair formation or protein binding. The modified positions are detected as stop sites during primer extension by reverse transcriptase.
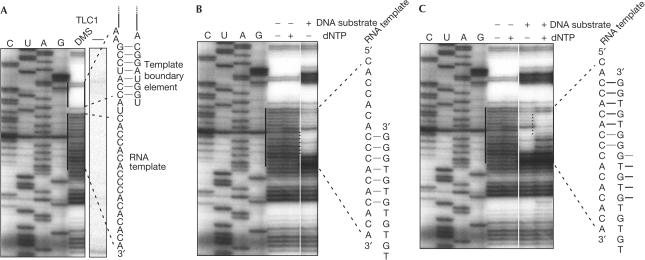
Figure 1.
DMS accessibility of the telomerase RNA template region analysed by primer extension with reverse transcriptase. (A) Lanes C, U, A and G denote dideoxynucleotide sequencing reactions using TLC1-containing plasmid DNA as a template. DMS denotes the lane in which enriched telomerase fraction was treated with DMS, RNA was extracted and TLC1 RNA was reverse transcribed in vitro. Termination at methylated sites occurs one nucleotide before the corresponding dideoxy termination product. The right panel shows a control experiment from a gel that was exposed under identical conditions in which the sample was not treated with DMS before reverse transcription. The RNA template region and the template boundary element are highlighted. (B) Preincubation of a DNA oligonucleotide substrate with telomerase before DMS modification in the absence of deoxynucleotide triphosphates (dNTPs). Base formation is indicated and inferred from the protection of seven bases of the telomerase RNA template. Note the particularly high accessibility of the base in the active site at the +1 position relative to the DNA substrate. (C) Primer extension on addition of dNTPs. The bases that are protected after primer extension are indicated.
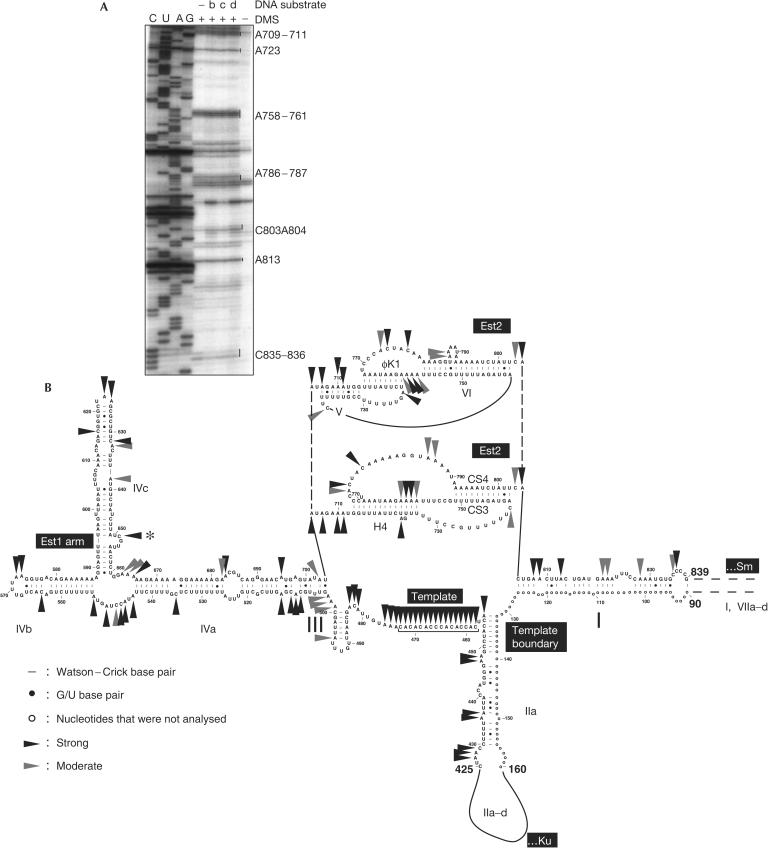
Figure 2.
DMS accessibility of the essential region of the telomerase RNA. (A) Est2-binding region. Some of the modified adenines and cytosines are highlighted on the right. DNA oligonucleotide substrates that were included in some of the reactions are b: 5′-GTGTGTGTGTGTGT-3′, c: 5′-GTGTGTGTGGG-3′ and d: 5′-TGTGTGTGGGTGTGGTG-3′. (B) Sites of DMS modification of the telomerase RNA superimposed on the Saccharomyces cerevisiae telomerase RNA secondary structure (Dandjinou et al, 2004; Zappulla & Cech, 2004). The upper pseudoknot structure is according to Dandjinou et al (2004) and the lower pseudoknot structure is according to Lin et al (2004). Only those bases of the RNA that were analysed in this study are shown. Strong modifications are indicated by black arrowheads and moderate modifications by grey arrowheads. Positions at which reverse transcription stopped also in the absence of DMS modification are ambiguous and are therefore not labelled. C651 (asterisk) in the bulge also shows a strong pause in the reverse transcriptase reaction of unmethylated RNA. Termination at this position may therefore reflect an in vivo-modified RNA base.
The template region of telomerase RNA
Telomerase from S. cerevisiae has the peculiarity of not dissociating from bound telomeric DNA oligonucleotides in vitro (Prescott & Blackburn, 1997). We took advantage of this stability to ‘lock' the enzyme in different states of the reaction cycle. We found that in the absence of DNA substrate, the entire template region of TLC1 was accessible to modification by DMS (Fig 1A), whereas in the absence of DMS, no stops of the reverse transcription reaction were observed in this region (Fig 1A, right panel). The accessibility to modification by DMS is consistent with the ability of the template to bind chromosome end substrates and is in accordance with the results obtained for Tetrahymena telomerase (Zaug & Cech, 1995) and recently proposed models for S. cerevisiae telomerase RNA (Dandjinou et al, 2004; Zappulla & Cech, 2004). Starting at three nucleotides 5′ of the template, seven bases in a row are completely protected. Strikingly, these bases have been proposed to form one strand of a stem–loop structure involved in defining the 5′ boundary of the template (Seto et al, 2003). On addition of a telomerase substrate before modification with DMS, the central portion of the template became protected as expected from the base complementarity of DNA substrate and RNA (Fig 1B). However, the protected region was only 7 bp long, starting at the DNA 3′ end. Closer to the 3′ boundary of the RNA template, the remaining three bases were readily modified by DMS even though they were complementary to the substrate. Thus, although these bases were accessible to DMS, they did not engage in base pairing. We also noted that the presence of substrate primer increased pausing or abortion of the reverse transcription reaction at G446 and G449. However, as G is not protected from DMS modification due to base pairing, the interpretation of this result is not straightforward.
To determine the protection pattern after substrate elongation, dNTPs were added to the telomerase–telomeric primer complex before DMS modification. The shift in the DMS protection pattern indicates that most of the bound substrates became elongated by five nucleotides (Fig 1C), as expected from analysis of product length obtained under similar reaction conditions (Prescott & Blackburn, 1997; Forstemann & Lingner, 2001; Teixeira et al, 2002; supplementary Fig 2 online). This result also indicates that most of the telomerase RNA molecules were assembled with Est2 in a catalytically active enzyme. Again, only seven bases of the telomerase RNA engaged in base pairing with the extended substrate, starting at the new DNA 3′ end. Thus, base-pair formation at the 3′ end of the substrate during elongation coincided with base-pair disruption at the 5′ end. The measured Km of the same substrate was about 0.8 μM (supplementary Fig 2 online), which is consistent with the free energy calculated for the formation of a 7–8 bp RNA–DNA hybrid (see legend to supplementary Fig 2 online for details). This assumes that the interaction of the telomerase protein with the DNA substrate does not contribute substantially to the binding energy. The short substrate oligonucleotides used in our study are too short to interact with the presumed anchor site in S. cerevisiae telomerase (Lue & Peng, 1998).
The putative pseudoknot domain
Two alternative pseudoknot structures have been proposed for the telomerase RNA from S. cerevisiae (Dandjinou et al, 2004; Lin et al, 2004; Fig 2B). Significantly, both pseudoknot structures include one stem, corresponding to CS3/CS4 in the lower pseudoknot structure in Fig 2B or to the lower part of stem VI in the upper structure, the base-pair potential of which has been demonstrated to be important for binding the yeast TERT Est2 (Chappell & Lundblad, 2004; Lin et al, 2004). Our DMS modification analysis (Fig 2A) is consistent with the existence of the CS3/CS4 stem or the extended bulged stem VI, as all adenines and cytosines in these stems are protected from modification. Also, the DMS protection is consistent with the existence of stem φK1 of the upper pseudoknot, whereas A758–762 in the extended H4 helix were accessible to DMS, indicating that stem H4 may be shorter than indicated, or that it may not be stably formed. Three adenines in stem V (A707, A709 and A710) of the upper structure were also strongly modified by DMS, indicating that this stem does not stably form in the Tlc1–Est2 catalytic core complex analysed here.
The Est1 binding bulged stem
The DMS modification data are in excellent agreement with the proposed secondary structures for stems IIa, III and IVa–c. Stem IVc corresponds to the previously described Est1 binding bulged stem (Seto et al, 2002). C651 in the bulge of IVc (Fig 2B, asterisk) also shows a strong pause in the reverse transcriptase reaction of unmethylated RNA (supplementary Fig 1 online). Termination at this position may therefore reflect an in vivo-modified RNA base.
Speculation
Here, we assess the telomerase RNA structure in the catalytically active yeast Tlc1–Est2 core complex. The accessibility of Tlc1 to DMS supports the recently proposed secondary structure models (Dandjinou et al, 2004; Zappulla & Cech, 2004). The discrepancy between DMS accessibility and predicted secondary structure in the pseudoknot domain might be reconciled with structural changes that may occur during substrate translocation, which was not resolved here.
Our results demonstrate that the analysed telomerase core allows the formation of only 7 bp for primer binding and after primer extension. Thus, our work provides direct proof for previous proposals that the TERT may limit the number of base pairs during the reaction cycle (Hammond & Cech, 1998). Product DNA disengagement from template RNA bases 3′ of the active site during elongation should facilitate telomeric DNA substrate dissociation and/or translocation (Fig 3). Although yeast telomerase only inefficiently dissociates or translocates from elongated primers in vitro, primer dissociation or translocation clearly occurs in vivo where many telomeric repeats can be added in a single cell cycle (Teixeira et al, 2004).
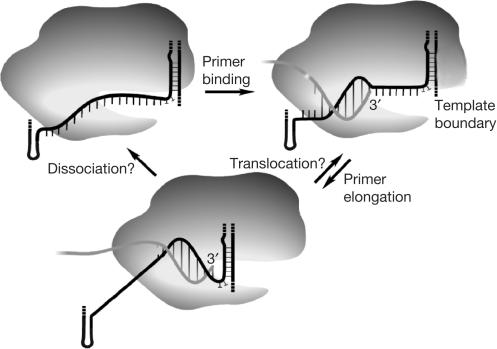
Figure 3.
Model for the telomerase reaction cycle. On substrate binding, a limited number of base pairs (seven in Saccharomyces cerevisiae) are formed between the 3′ end of the primer and the template region of telomerase RNA. During substrate elongation, base-pair formation at the DNA 3′ end coincides with base-pair disruption further 5′ on the DNA. Substrate elongation stops at or before the template boundary, which is imposed through a paired element in S. cerevisiae (Seto et al, 2003). Limitation of RNA–DNA duplex length should facilitate primer dissociation or translocation. Processive repeat addition is observed in vitro for telomerases from ciliates and vertebrates, but not from S. cerevisiae. The RNA template is modelled to be sterically constrained and under tension during primer binding and extension, allowing the formation of only three-quarters of a helical turn.
RNA–DNA helix unwinding seems to be a unique feature of the TERT when compared with the related retroviral reverse transcriptases, which are known to form several hundred nucleotide-long RNA–DNA hybrid molecules during reverse transcription. Conversely, RNA polymerase II also limits the length of the RNA–DNA hybrid to 9–10 residues by interaction with a set of protein loops (Westover et al, 2004). Whether the TERT polypeptide functions in a similar way remains to be seen. A concomitant accessibility of bases to DMS and inaccessibility to base pairing (Fig 1) could also be explained by a model in which a sterically constrained RNA template is under tension after primer binding, allowing the formation of only three-quarters of a helical turn (Fig 3).
Methods
Strain construction. Est2 was overexpressed from the Gal promoter as N-terminal His-tagged fusion protein. A plasmid coding for a PGAL1–His6–TEV cassette was constructed, by PCR-amplifying DNA fragments from pFA6a–TRP1–PGAL1 and pFA6a–HIS3MX6–PGAL1 (Longtine et al, 1998) with oligonucleotides His6–TEV–R2 (5′-GCGAATTCTCCTCCGGATTGGAAGTACAGGTT CTCACCAGAACCATGGTGATGGTGATGGTGCGATC CTCTCATTTTGAGATCCGGGTTTT-3′) and F4short (5′-CGGAATTCGAGCTCGTTTAAAC-3′). The products were cloned into pBluescript and sequenced. The plasmids, named pKF13 (His3MX marker) and pKF14 (TRP1 marker), can be used to construct PCR-based PGal–His–TEV integration cassettes analogous to previously described plasmids (Longtine et al, 1998). The amino-acid sequence appended to the N terminus of the tagged protein is MRGSHHHHHHGSGENLYFQSGG. The primer sequence for amplification is 5′-(genespecific sequence)-TCCTCCGGATTGGAAGTACAG-3′ (Longtine et al, 1998). Here, the EST2 gene was tagged by amplifying pKF13 with DNA oligonucleotides F4-Est2 (5′-CACAAGTGAAATAGAAAAGTGAAAAATTAAAA AAAAAAAAAAAAAAAAAAAAACTGATTTATACTCG AATTCGAGCTCGTTTAAAC-3′) and R6-Est2 (5′-TGTAGATCAATGTCAAGCTTGTCTTGAATGAA CTCGAATAAGATTTTCATTCCTCCGGATTGGAAGT ACAG-3′) and by transforming FYBL1-4D (MAT a ura3-Δ851 trp1Δ63 leu2Δ1 his3Δ200 lys2Δ202).
The overexpression construct for the TLC1 RNA was obtained by subcloning the PGAL1–TLC1 cassette from pRS314–PGAL1–TLC1 (Forstemann & Lingner, 2001) into pRS306, yielding pRS306–PGAL1–TLC1. The plasmid was linearized with HpaI and integrated at the TLC1 locus in the strain above, leading to duplication of the TLC1 gene with one TLC1 gene under the control of the endogenous promoter and one under the control of the GAL1 promoter. This strain is referred to as YKF24 (MAT a ura3-Δ851 trp1Δ63 leu2Δ1 his3Δ200 lys2Δ202 HIS3MX6 PGAL1-HIS/TEV-EST2 TLC1:PGAL1-TLC1 URA3).
Yeast culture and telomerase preparation. YKF24 was precultured in galactose medium lacking uracil for 48 h at 25°C to inoculate 2 l of rich galactose medium (2% peptone, 1% yeast extract, 3% galactose). Cells were grown at 25°C to an OD600 of 1, collected by centrifugation, washed once with H2O and resuspended in lysis buffer (150 mM Tris–HCl (pH 8.0), 150 mM NaAc (pH 8.0), 1 mM MgCl2, 1 mM dithiothreitol, 0.2% Triton X-100, 10% glycerol) supplemented with 2 × protease inhibitors without EDTA (Roche, Rotkreuz, Switzerland). Cells were lysed and telomerase was prepared by DEAE adsorption as described previously (Cohn & Blackburn, 1995; Prescott & Blackburn, 1997; Forstemann & Lingner, 2001). Most of the overexpressed Tlc1 RNA was associated with Est2 as determined by co-immunoprecipitation with a Myc-tagged version of Est2 (data not shown).
Telomerase RNA methylation. DMS modification was performed as described previously (Zaug & Cech, 1995; Antal et al, 2002). The reactions were carried out in 1 × telomerase reaction buffer (Forstemann & Lingner, 2001) in a volume of 20 μl containing DEAE-enriched telomerase fraction (20 μg of total protein). Substrate DNA oligonucleotides were included at a concentration of 25 μM. Substrate extension by the telomerase enzyme was initiated by addition of dNTPs (50 μM each) and incubation for 5 min at 23°C. For RNA methylation, 1.5 μl of 10% DMS (diluted in 100% ethanol) was added to 20 μl reaction mixture, yielding a final concentration of 0.75% DMS. The samples were incubated for 5 min at 23°C. The reactions were quenched by the addition of half volume of 2 M β-mercaptoethanol and incubated for 10 min at 23°C. To digest the proteins in the methylated RNA sample, 200 μl of proteinase K solution (200 μg/ml proteinase K in reaction buffer) was added and the samples were incubated for 30 min at 23°C. To acidify the solution, 25 μl of 3 M sodium acetate (pH 5.3) was added and the RNA was extracted with 150 μl of phenol/chloroform/isoamyl alcohol (25:25:1). The acid–phenol extraction partitioned DNA contaminations into the phenol phase. The aqueous phase containing RNA was transferred to a fresh tube containing 1 μl of glycogen solution (20 μg/ml) and precipitated with 600 μl of 100% ethanol. The RNA pellet was washed twice with 70% ethanol and resuspended in 10 μl of water. Dried gels were exposed for 1–3 days and signal strength was classified as strong or moderate visually, by both authors independently and discrepancies were re-evaluated by them together. All experiments were conducted at least twice, giving highly reproducible results.
Reverse transcriptase reactions and gel electrophoresis. The oligonucleotides used for primer extension by reverse transcriptase (tlcas600: 5′-GCAAATCTAACTTAAACTCC-3′ and tlcas900: 5′-TAAGAAAGGACACCCTTGCC-3′) were labelled with [32P-γ]ATP (equimolar amounts of ATP and oligonucleotide) and T4 DNA polynucleotide kinase (NEB) at their 5′ end. Labelled oligonucleotides were purified with the Nucleotide Removal Kit (Qiagen, Hombrechtikon, Switzerland). For reverse transcription, the entire 10 μl of methylated RNA was incubated with 1 pmol of radiolabelled oligonucleotide primer and 100 U of Superscript II reverse transcriptase (Invitrogen, Basel, Switzerland) according to the manufacturer's instructions. For parallel sequencing reactions, 1 pmol of the same radiolabelled oligonucleotide primer was used in standard dideoxynucleotide sequencing reactions (Amersham, Otelfingen, Switzerland) using pSD107 (Singer & Gottschling, 1994) as a template. After primer extension, all reaction products were precipitated with sodium acetate and ethanol according to standard protocols and resuspended in 5 μl of formamide loading buffer (90% formamide, 0.1 × TBE). The reaction products were separated on 6% acrylamide/urea gels according to standard procedures. The gels were dried and exposed to Kodak Biomax film for 2–14 days.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400374s1.pdf).
Supplementary Material
Legend to Supplementary Figures
Acknowledgments
This work was supported by the Swiss National Science Foundation.
References
- Antal M, Boros E, Solymosy F, Kiss T (2002) Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res 30: 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH (1994) Architecture of telomerase RNA. EMBO J 13: 5721–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR (2000) Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell 6: 493–499 [DOI] [PubMed] [Google Scholar]
- Chappell AS, Lundblad V (2004) Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol Cell Biol 24: 7720–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW (2000) Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH (1995) Telomerase in yeast. Science 269: 396–400 [DOI] [PubMed] [Google Scholar]
- Dandjinou AT, Levesque N, Larose S, Lucier JF, Elela SA, Wellinger RJ (2004) A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol 14: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (2000) Positive and negative regulation of telomerase access to the telomere. J Cell Sci 113: 3357–3364 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Lingner J (2001) Molecular basis for telomere repeat divergence in budding yeast. Mol Cell Biol 21: 7277–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond PW, Cech TR (1998) Euplotes telomerase—evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry 37: 5162–5172 [DOI] [PubMed] [Google Scholar]
- Kelleher C, Teixeira MT, Forstemann K, Lingner J (2002) Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem Sci 27: 572–579 [DOI] [PubMed] [Google Scholar]
- Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH (2004) A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci USA 101: 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR (1994) Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev 8: 1984–1998 [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR (2002) Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lue NF, Peng Y (1998) Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res 26: 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH (1997) Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev 11: 2790–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH (1991) A conserved secondary structure for telomerase RNA. Cell 67: 343–353 [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR (2002) A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR (2003) A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9: 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Sperger JM, Cech TR (2001) A stem–loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40: 7005–7016 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Forstemann K, Gasser SM, Lingner J (2002) Intracellular trafficking of yeast telomerase components. EMBO Rep 3: 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- ten Dam E, van Belkum A, Pleij K (1991) A conserved pseudoknot in telomerase RNA. Nucleic Acids Res 19: 6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Knight Z, Roy J, Blackburn EH (2003) A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev 17: 1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Guilley D, Backburn EH (1998) A novel specificity for the primer–template pairing requirement in Tetrahymena telomerase. EMBO J 17: 1152–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD (2004) Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303: 1014–1016 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR (2004) From the cover: yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci USA 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR (1995) Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the selfsplicing rRNA intron, and U2 snRNA. RNA 1: 363–374 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legend to Supplementary Figures