Abstract
The Drosophila Polycomb group protein E(z) is a histone methyltransferase (HMTase) that is essential for maintaining HOX gene silencing during development. E(z) exists in a multiprotein complex called Polycomb repressive complex 2 (PRC2) that also contains Su(z)12, Esc and Nurf55. Reconstituted recombinant PRC2 methylates nucleosomes in vitro, but recombinant E(z) on its own shows only poor HMTase activity on nucleosomes. Here, we investigate the function of the PRC2 subunits. We show that PRC2 binds to nucleosomes in vitro but that individual PRC2 subunits alone do not bind to nucleosomes. By analysing PRC2 subcomplexes, we show that Su(z)12–Nurf55 is the minimal nucleosome-binding module of PRC2 and that Esc contributes to high-affinity binding of PRC2 nucleosomes. We find that nucleosome binding of PRC2 is not sufficient for histone methylation and that only complexes that contain Esc protein show robust HMTase activity. These observations suggest that different subunits provide mechanistically distinct functions within the PRC2 HMTase: the nucleosome-binding subunits Su(z)12 and Nurf55 anchor the E(z) enzyme on chromatin substrates, whereas Esc is needed to boost enzymatic activity.
Keywords: E(z)/EZH2, histone methylation, Polycomb repression, chromatin proteins
Introduction
The compaction of eukaryotic DNA into arrays of nucleosomes creates constraints for diverse processes that have to occur on the DNA template, but it also generates a level of control for regulating these processes. Eukaryotic cells use two principal classes of enzymes to modify the structure of chromatin at the nucleosome level: nucleosome-remodelling factors that hydrolyse ATP to catalyse the mobilization of core histones on DNA, and enzymes that covalently modify amino-acid side chains in core or linker histones. Typically, these enzymes exist in multiprotein complexes together with other proteins but, in most cases, the molecular function of these non-catalytic subunits is poorly understood.
The control of gene expression by Polycomb group (PcG) and trithorax group (trxG) proteins provides a paradigm for studying transcriptional regulation by chromatin-modifying protein complexes. Genetic studies originally identified PcG and trxG proteins as antagonistic regulators that maintain transcriptional OFF and ON states of HOX genes in Drosophila (reviewed by Ringrose & Paro, 2004). Studies over the past decade have shown that the PcG/trxG system is widely conserved and regulates HOX and other developmental control genes in both animals and plants. Biochemical purification and characterization of PcG and trxG proteins showed that they exist in distinct multiprotein complexes that contain different PcG and trxG proteins, and that some of these proteins are chromatin-modifying enzymes. In particular, the PcG protein E(z) and the trxG proteins Ash1 and ALL1/MLL/hTRX are histone methyltransferases (HMTases) that methylate distinct lysine residues in the amino-terminal tails of core histones (Beisel et al, 2002; Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Milne et al, 2002; Müller et al, 2002; Nakamura et al, 2002).
E(z) protein is the catalytic subunit of Drosophila Polycomb repressive complex 2 (PRC2), a protein complex that contains the PcG proteins Su(z)12 and Esc as well as Nurf55/CAF1, a protein that is present in many different chromatin complexes (Czermin et al, 2002; Müller et al, 2002). Mammalian PRC2 has a similar composition; it contains EZH2, SU(Z)12, EED, RbAp46 and RbAp48, the last two being mammalian homologues of Nurf55 (Cao et al, 2002; Kuzmichev et al, 2002). Drosophila PRC2 methylates K27 in histone H3 (H3-K27) in nucleosome templates in vitro (Müller et al, 2002). Mammalian PRC2 also methylates H3-K27 in nucleosome templates and, depending on the EED isoform present in the complex, it also methylates K26 in histone H1 (Cao et al, 2002; Kuzmichev et al, 2002, 2004; Cao & Zhang, 2004). Several lines of evidence support the idea that H3-K27 methylation by E(z) has an important role in the transcriptional repression of PRC2 target genes. First, the enzymatic activity of E(z) protein is strictly required for maintenance of HOX gene silencing in Drosophila (Müller et al, 2002). Second, H3-K27 methylation of chromatin in the Drosophila HOX gene Ubx depends on E(z) function (Cao et al, 2002). Moreover, H3-K27 methylation in mammals correlates with the presence of the mammalian E(Z), SU(Z)12 and EED proteins at target genes (Cao & Zhang, 2004; Kirmizis et al, 2004). Finally, Drosophila Polycomb (Pc), a core subunit of PRC1, selectively binds to a histone H3 tail peptide trimethylated at K27, suggesting that H3 K27 methylation may contribute to targeting of PRC1 to HOX genes (Fischle et al, 2003; Min et al, 2003).
The HMTase activity of recombinant E(z) protein on nucleosome substrates in vitro is about 1,000-fold lower than the HMTase activity of recombinant tetrameric PRC2, implying that non-catalytic subunits are important for methylation of chromatin (Müller et al, 2002). Recent studies on mammalian PRC2 showed that SU(Z)12 is crucial to HMTase activity of PRC2 on nucleosomes in vitro, but the molecular basis for this requirement is still unknown (Cao & Zhang, 2004; Pasini et al, 2004). Here, we report the biochemical characterization and dissection of Drosophila PRC2. We show that different PRC2 subunits make mechanistically distinct contributions to chromatin binding and activation of the E(z) HMTase.
Results And Discussion
Reconstitution of PRC2 subcomplexes
We previously reported the use of baculovirus expression vectors to reconstitute and purify recombinant tetrameric PRC2 from Sf9 cells (Müller et al, 2002). To identify intermolecular interactions within this complex, we used the same approach and tested for reconstitution of PRC2 subcomplexes on coexpression of two or more subunits in Sf9 cells. In each case, we used the Flag-epitope tag present on one of the subunits for affinity purification from Sf9 cell extracts. We could thus purify the following stable dimeric complexes: Esc–E(z), E(z)–Su(z)12, E(z)–Nurf55 and Su(z)12–Nurf55 (Fig 1, lanes 8–11). In contrast, purification either from cells expressing Flag–Esc and Nurf55 or from cells expressing Flag–Esc and Su(z)12 resulted in the isolation of Flag–Esc protein only, suggesting that these proteins do not bind directly to each other (data not shown). Purification from Sf9 cells expressing three subunits allowed the isolation of trimeric Esc–E(z)–Su(z)12 and E(z)–Su(z)12–Nurf55 complexes (Fig 1, lanes 12,13). Finally, we also reconstituted tetrameric PRC2, using either Flag–E(z) or Flag–Su(z)12 for affinity purification (Fig 1, lanes 6,7). Importantly, each of these complexes was stable in buffers containing up to 1 M KCl. Taken together, these data suggest that E(z) binds tightly to Su(z)12, Esc and Nurf55 and that Su(z)12 also binds to Nurf55. The failure to isolate dimeric complexes that contain Esc and Su(z)12 or those that contain Esc and Nurf55 indicates that E(z), Su(z)12 and Nurf55 form a trimeric core complex to which Esc binds through interaction with E(z) (Fig 1). Our observation that E(z) forms stable dimeric complexes with either Esc or Nurf55 in this reconstitution assay is consistent with earlier studies that reported physical interactions between these proteins in glutathione-S-transferase (GST) pull-down assays (Jones et al, 1998; Tie et al, 2001).
Figure 1.
Reconstitution of PRC2 subcomplexes. Molecular weight marker (lane 1), PRC2 subunits (lanes 2–5) and (sub)complexes (lanes 6–13) purified from Sf9 cells were separated by SDS–polyacrylamide gel electrophoresis and visualized by Coomassie staining; in each case, the Flag-tagged subunit is indicated by a red ‘+'. Note that most complexes seem to contain stoichiometric amounts of subunits; tetrameric PRC2 (lanes 6,7) contains substoichiometric amounts of Esc, but the Esc protein band is not well resolved on this 6–12% gradient gel (see supplementary Fig S1 online for details on stoichiometry). Right: schematic representation of molecular interactions between PRC2 subunits.
We note that the molecular architecture of mammalian PRC2 is unclear at present; conflicting data on intermolecular interactions between subunits have been reported (Cao & Zhang, 2004; Pasini et al, 2004; Yamamoto et al, 2004). Specifically, Cao & Zhang (2004) reported that human EZH2, SU(Z)12 and RbAp48 all bind to EED, the Esc homologue, and that EZH2 does not interact with SU(Z)12 or RbAp48 in GST pull-down assays; the authors proposed that EED is the core component of the complex and EZH2 associates with other components through EED. Yamamoto et al (2004) reported that EZH2 binds to SUZ12 in GST pull-down assays, which is consistent with our finding that Drosophila E(z) and Su(z)12 form a stable complex.
Nucleosome binding requires Su(z)12 and Nurf55
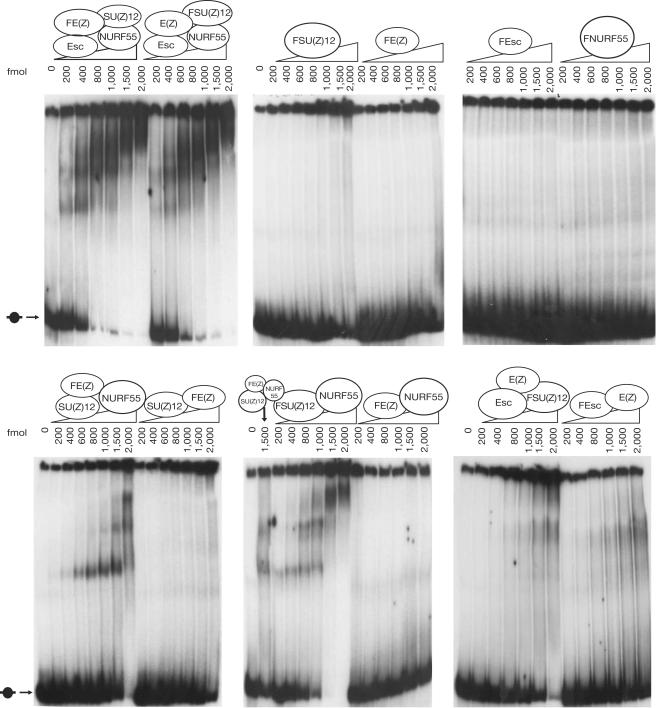
The HMTase activity of recombinant E(z) protein is significantly lower than the activity observed with recombinant tetrameric PRC2 (Müller et al, 2002). A simple mechanistic explanation would be that one or several PRC2 subunits are needed for nucleosome binding to facilitate interaction of the E(z) HMTase with its substrate, the histone H3 tail. As it is not known whether any of the PRC2 subunits binds to nucleosomes, we tested whether complex components alone or in combination could form stable complexes with mononucleosomes, in a bandshift assay. To this end, we reconstituted mononucleosomes with recombinant core histones that we expressed in Escherichia coli and a 201 base pairs (bp) long radioactively labelled DNA template that contained a strong nucleosome-binding sequence called ‘601' (Thaström et al, 1999). When we incubated recombinant tetrameric PRC2 with such mononucleosomes and resolved the reaction mixture on a polyacrylamide gel, we observed distinct, slowly migrating complexes that appeared in a concentration-dependent manner (Fig 2). In contrast, when we incubated PRC2 with naked 601 DNA template, we were unable to resolve specific protein–DNA complexes (data not shown). Together, these observations suggest that PRC2 binds to mononucleosomes and that these protein–nucleosome complexes remain stably associated under electrophoretic conditions. We next tested individual PRC2 subunits for nucleosome binding, but did not detect formation of protein–nucleosome complexes with any of the four proteins (Fig 2). This suggests that more than one subunit is needed for nucleosome binding and we therefore tested the different di- and trimeric PRC2 subcomplexes. Among the different subcomplexes, only incubation with the Su(z)12–Nurf55 or E(z)–Su(z)12–Nurf55 complexes results in the appearance of distinct, slowly migrating protein–nucleosome complexes (Fig 2). These protein–nucleosome complexes migrate similarly to the complexes observed with tetrameric PRC2, but two- to threefold higher concentrations of the Su(z)12–Nurf55 or E(z)–Su(z)12–Nurf55 complex are needed to shift all of the nucleosome probe (Fig 2; compare the lanes with 800 fmol PRC2 and 2,000 fmol E(z)–Su(z)12–Nurf55). Thus, the presence of Esc in PRC2 increases the affinity of the complex for nucleosomes or allows the complex to bind more stably under our experimental conditions compared with PRC2 subcomplexes that lack Esc. In contrast to the distinct protein–nucleosome complexes observed with the Su(z)12–Nurf55 or E(z)–Su(z)12–Nurf55 complex, no specific protein–nucleosome complexes are formed if nucleosomes are incubated with the E(z)–Su(z)12, E(z)–Nurf55, Esc–E(z) or Esc–E(z)–Su(z)12 complexes (Fig 2). However, incubation with high concentrations of the trimeric Esc–E(z)–Su(z)12 complex also shifts almost all of the nucleosome probe, and much of the probe is retained in the well of the gel (Fig 2, 2,000 fmol lane). Thus, it seems that the Esc–E(z)–Su(z)12 complex also binds to nucleosomes but that it binds in a manner distinct from the other PRC2 (sub)complexes. Taken together, these binding assays suggest that several subunits need to cooperate for nucleosome binding of PRC2, as follows. First, Su(z)12 is essential for nucleosome binding because only complexes containing Su(z)12 bind. Second, the minimal nucleosome-binding complex contains Su(z)12 and Nurf55, and only complexes that contain these two proteins give rise to distinct, slowly migrating protein–nucleosome complexes; Su(z)12 and Nurf55 together thus form the minimal nucleosome-binding module of PRC2. Third, as discussed above, Esc also contributes to nucleosome binding because (i) tetrameric PRC2 binds more strongly than the E(z)–Su(z)12–Nurf55 complex and (ii) in the absence of Nurf55, that is, in the Esc–E(z)–Su(z)12 complex, Esc seems to cooperate with Su(z)12 to cause retention of the nucleosome probe. E(z) is thus the only subunit for which we have not been able to detect a contribution to nucleosome binding. Note that comparable concentrations of the Su(z)12–Nurf55 or E(z)–Su(z)12–Nurf55 complex are required to shift 50% of the nucleosome probe (Fig 2).
Figure 2.
Nucleosome binding by PRC2 requires Su(z)12 and Nurf55 or Esc. Binding reactions containing 32P-labelled recombinant mononucleosomes (250 fmol) and the indicated recombinant PRC2 subunits/(sub)complexes (200–2,000 fmol; Flag-tagged subunit indicated by ‘F') were resolved on native polyacrylamide gels; position of free mononucleosome is indicated by an arrow. Note that tetrameric PRC2 purified through Flag–Su(z)12 or Flag–E(z) binds with indistinguishable affinity; for further details, see text. A binding reaction containing Flag–E(z)–Su(z)12–Nurf55 was loaded as a reference on some of the gels.
Nurf55 and Esc are both WD40 repeat proteins. RbAp46 and RbAp48, the mammalian homologues of Nurf55, have been reported to bind directly to helix 1 of histone H4, a portion of H4 that is thought to be inaccessible within the nucleosome, and, consistent with this, RbAp46 and RbAp48 are unable to bind to H4 in nucleosomal templates (Verreault et al, 1997). As shown here, Drosophila Nurf55 or Esc alone are not able to bind to mononucleosomes but they bind in combination with Su(z)12, which, by itself, also does not bind to nucleosomes. It is possible that the combination of Su(z)12 and Nurf55 or Esc is needed to create the necessary surface for stable nucleosome binding. Alternatively, it could be that these proteins act in a cooperative manner to disrupt histone–DNA contacts locally to expose the histone core (i.e. H4) for binding by Nurf55 or Esc.
HMTase activity requires Esc
The results described above suggest that Su(z)12 together with Nurf55 or Esc tethers the complex to nucleosomes, whereas E(z), the catalytic subunit of the complex, contributes little to nucleosome binding. We next analysed the HMTase activity of the different PRC2 (sub)complexes. As substrates for these reactions, we used non-radiolabelled mononucleosomes identical to those used in the bandshift assay. Recombinant tetrameric PRC2 methylates H3 in mononucleosomes (Fig 3, lane 9). In contrast, E(z) protein alone, the dimeric Esc–E(z), and the Su(z)12–E(z) or E(z)–Nurf55 complexes do not detectably methylate mononucleosomes (Fig 3, lanes 3–6). Strikingly, the Su(z)12–E(z)–Nurf55 complex also shows no detectable HMTase activity, whereas the Esc–E(z)–Su(z)12 complex methylates H3 in mononucleosomes with efficacy similar to tetrameric PRC2 (Fig 3, lanes 7,8). Thus, we do not observe a straightforward correlation between nucleosome binding in bandshift assays and HMTase activity. In particular, the Su(z)12–E(z)–Nurf55 complex seems to bind to nucleosomes with only two- to threefold lower affinity than tetrameric PRC2 (see above), but shows markedly reduced HMTase activity. In contrast, the Esc–E(z)–Su(z)12 complex, which shows almost no nucleosome binding at the concentration used in the HMTase assay (i.e. 1,000 fmol lane in Fig 2), shows HMTase activity comparable with PRC2. Together, these data suggest that nucleosome binding is not sufficient for HMTase activity and that Esc has a crucial role in boosting the enzymatic activity of E(z). It is possible that Esc is required to dock the complex in a specific orientation on the nucleosome that presents the H3 tail in a particularly favourable position to the E(z) enzyme. Alternatively, Esc could directly increase the catalytic activity of E(z) by inducing a conformational change in the enzyme. However, it is important to note that the Esc–E(z) complex shows no detectable HMTase activity on mononucleosomes and the presence of Su(z)12 is thus essential for HMTase activity of the Esc–E(z)–Su(z)12 complex. Our bandshift data show that Su(z)12 is strictly needed for nucleosome binding of PRC2. At the low complex concentrations used in the HMTase assay, there is probably very little nucleosome binding of the Esc–E(z)–Su(z)12 complex; nevertheless, it seems probable that the nucleosome interactions that Su(z)12 shows in cooperation with Esc (see above) contribute to the HMTase activity of the Esc–E(z)–Su(z)12 complex. Finally, it is puzzling that the absence of Nurf55 from the complex (i.e. in the Esc–E(z)–Su(z)12 complex) does not seem to diminish HMTase activity because Nurf55 is important for the formation of stable PRC2–nucleosome complexes in bandshift assays. It is possible that Nurf55 is not needed for HMTase activity under our assay conditions but that it is important for methylation of chromatin in vivo.
Figure 3.
HMTase activity of PRC2 depends on Esc protein. Molecular weight marker (lane 1) and HMTase reactions (lanes 2–9) that contained the indicated complexes, [14C]sAM and mononucleosomes were resolved by SDS–polyacrylamide gel electrophoresis; the gel was stained with Coomassie (top) and exposed for autoradiography (bottom). Note that only incubation with tetrameric PRC2 or Flag–Su(z)12–E(z)–Esc complex (lanes 7,9) results in detectable methylation of H3. E(z) and Su(z)12 proteins also become methylated in this assay (compare with Müller et al, 2002). H4 protein is present in stoichiometric quantities (see supplementary Fig S1 online) but did not run as a sharp band on this gel.
Concluding remarks
Recent studies have shown that SU(Z)12 is crucial for HMTase activity of mammalian PRC2 in vitro, but the molecular basis for this requirement has remained unclear (Cao & Zhang, 2004; Pasini et al, 2004). This study reports two main findings. First, we have shown that Su(z)12 has a crucial role in nucleosome binding of Drosophila PRC2. Second, we have shown that PRC2 subcomplexes that bind to nucleosomes but lack Esc are poorly active; Esc thus has an important role in boosting HMTase activity of the complex.
The requirement of Su(z)12 for HMTase activity can for the most part be explained by its ability to tether PRC2 to nucleosomes, and our data suggest that nucleosome binding requires Su(z)12 in combination with either Nurf55 or Esc. Although both Nurf55 and Esc contribute to nucleosome binding of the complex, only inclusion of Esc leads to an increase in HMTase activity. The contribution of Esc to HMTase activity thus goes beyond the activity that one would expect if Esc were required only for nucleosome binding. In summary, our data suggest that the Su(z)12, Nurf55 and Esc subunits all contribute to nucleosome binding of PRC2 but that these three subunits make distinct contributions to the activation of the E(z) HMTase.
The findings reported here imply that E(z) HMTase activity in vivo could be regulated at the level of chromatin binding and/or enzyme activity by modulating the abundance or activity of different PRC2 subunits. It is important to discuss the results reported here in the context of the in vivo requirement for different PRC2 subunits. Genetic studies have shown that Su(z)12 and E(z) are required throughout development to maintain silencing of HOX genes in Drosophila and that this process requires the enzymatic activity of E(z) (Birve et al, 2001; Cao et al, 2002; Müller et al, 2002). In contrast, Esc protein is required early in development and then becomes to a large extent, although not completely, dispensable for maintenance of HOX gene silencing during postembryonic development (Struhl & Brower, 1982). There are two possible explanations for the paradoxical observation that Esc is required for strong HMTase activity in vitro but that the protein seems to be largely dispensable for the HMTase activity of E(z) that is needed to maintain HOX gene silencing during larval development. First, it is possible that strong HMTase activity of PRC2 is required primarily early in embryogenesis and that, once it is established, H3-K27 methylation can be maintained with a catalytically less active form of the complex (i.e. lacking Esc). Second, it is possible that another protein substitutes for Esc at later developmental stages. We noted that the Drosophila genome encodes a second Esc-like protein (CG5202) but, at present, it is not known whether this protein is required for HOX gene silencing.
Methods
Protein expression and purification. Baculovirus production and Flag affinity purification of complexes was carried out as described (Müller et al, 2002). Viruses expressing Flag–Esc, Flag–E(z), Flag–Su(z)12, Flag–Nurf55, Ha–Esc, E(z), Su(z)12 and Myc–Nurf55 have been described previously (Müller et al, 2002) or were generated for this study (plasmid maps are available on request).
Recombinant Xenopus octamers were prepared as described (Luger et al, 1999) and mononucleosomes were assembled onto a (radiolabelled) 201 bp ‘601' DNA template (Thaström et al, 1999). For assembly, 1.4 μg octamer was mixed with 1 μg DNA template in a volume of 10 μl (2 M NaCl, 10 mM Tris (pH 8.0), 0.1 mM EDTA, 10 mM β-mercaptoethanol) followed by stepwise reduction of the salt concentration by addition of Tris–EDTA to obtain a solution containing 0.2 pmol nucleosome/μl (200 mM NaCl/Tris–EDTA). Assembly reactions for bandshift probes contained 2 mg/ml BSA.
Bandshift assays. 0.25 pmol of radiolabelled mononucleosome was incubated with 0.2–2 pmol PRC2 subunit or (sub)complex in 25 μl binding reactions (10 mM Tris (pH 8.0), 200 mM NaCl, 50 μM ZnCl2, 10% glycerol) for 40 min at 25°C. Binding reactions were analysed on 4% 0.5 × Tris–borate 60:1 polyacrylamide gels. Gels were fixed, dried and exposed for autoradiography.
HMTase assays. Reactions were carried out in a volume of 50 μl and contained 2.8 pmol nucleosome, 2 pmol PRC2 (sub)complex, 10 mM Hepes (pH 7.9), 0.25 mM EDTA, 200 mM NaCl, 10% glycerol, 2 mM dithiothreitol, 2.5 mM MgCl2 and 4 μM S-adenosyl-L-[methyl-14C]methionine. Reactions were incubated for 40 min at 30°C and resolved by 20% SDS–polyacrylamide gel electrophoresis; after Coomassie staining and fluorography, the gel was dried and exposed for autoradiography.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400376s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank D. Rhodes and A. Akhtar for reagents and advice. Comments from E. Izaurralde and A. Akhtar helped to improve the manuscript.
References
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F (2002) Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857–862 [DOI] [PubMed] [Google Scholar]
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Müller J (2001) Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128: 3371–3379 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol Cell 15: 57–67 [DOI] [PubMed] [Google Scholar]
- Cao R et al. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS (1998) The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol Cell Biol 13: 2825–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A et al. (2004) Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 18: 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D (2004) Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 14: 183–193 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Milne TA et al. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nakamura T et al. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 10: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Struhl G, Brower D (1982) Early role of the esc+ gene product in the determination of segments in Drosophila. Cell 31: 285–292 [DOI] [PubMed] [Google Scholar]
- Thaström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J (1999) Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol 288: 213–229 [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasadsinha J, Jane E, Harte PJ (2001) The Drosophila polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275–286 [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B (1997) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltranferase. Curr Biol 8: 96–108 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T (2004) Polycomb group suppressor of zeste 12 links heterochromatin protein 1α and enhancer of zeste 2. J Biol Chem 279: 401–416 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information