Abstract
Hypoxia-inducible factor (HIF)-1α is a short-lived protein and is ubiquitinated and degraded through the von Hippel–Lindau protein (pVHL)–E3 ubiquitin ligase pathway at normoxia. Deubiquitination, by reversing ubiquitination, has been recognized as an important regulatory step in ubiquitination-related processes. Here, we show that pVHL-interacting deubiquitinating enzyme 2, VDU2, but not VDU1, interacts with HIF-1α. VDU2 can specifically deubiquitinate and stabilize HIF-1α and, therefore, increase expression of HIF-1α targeted genes, such as vascular endothelial growth factor (VEGF). These findings suggest that ubiquitination of HIF-1α is a dynamic process and that ubiquitinated HIF-1α might be rescued from degradation by VDU2 through deubiquitination. Although pVHL functions as a master control for HIF-1α stabilization, as pVHL–E3 ligase mediates the ubiquitination of both HIF-1α and VDU2, the balance between the pVHL-mediated ubiquitination and VDU2-mediated deubiquitination of HIF-1α provides another level of control for HIF-1α stabilization.
Keywords: VDU1, VDU2, HIF-1α, VHL, ubiquitin
Introduction
Hypoxia-inducible factor (HIF)-1, consisting of an α-subunit and a β-subunit, is a sequence-specific DNA-binding transcriptional complex that regulates genes involved in angiogenesis, glucose metabolism, cell proliferation and invasion/metastasis (reviewed by Semenza, 2003). Whereas the HIF-1β level is kept stable in cells, the HIF-1α level is controlled by O2 tension as well as by growth factors. Under normoxic conditions, HIF-1α is regulated by O2-dependent prolyl hydroxylation, which facilitates HIF-1α polyubiquitination by E3 ubiquitin ligases and subsequent proteosomal degradation. In contrast, the O2-dependent prolyl hydroxylation is downregulated under hypoxia, resulting in HIF-1α accumulation and increased HIF-1 activity (Bruick & McKnight, 2001; Epstein et al, 2001; Ivan et al, 2001; Jaakkola et al, 2001). Growth factors can also increase HIF levels in their targeted cells through stimulation of HIF-1α synthesis. HIF-1α is overexpressed in many different human cancers as a result of intratumoral hypoxia as well as genetic alterations (for reviews, see Harris, 2001; Semenza, 2003). A unique example of the latter is von Hippel–Lindau (VHL) disease, in which affected individuals develop a variety of tumours, including haemangioblastomas of the central nervous system, clear-cell carcinomas of the kidneys and pheochromocytomas of the adrenal glands due to loss-of-function mutations of the VHL gene (reviewed by Kaelin, 2002). The von Hippel–Lindau tumour suppressor protein (pVHL) is the substrate recognition component of the E3 ligase that ubiquitinates prolyl hydroxylated HIF-1α. Without functional pVHL, polyubiquitination and degradation of HIF-1α become compromised, which leads to HIF-1α accumulation and overexpression of HIF-1 target genes (reviewed by Kaelin, 2002). Whereas ubiquitination is a crucial biochemical modification controlling diverse biological processes, deubiquitination has been recognized as an important regulatory step in ubiquitination-related processes (for reviews, see D'Andrea & Pellman, 1998; Wilkinson, 2000). In our previous studies, we have shown that pVHL-interacting deubiquitinating enzymes, VDU1 (USP33) and VDU2 (USP20), can be ubiquitinated by pVHL–E3 ubiquitin ligase for proteasomal degradation (Li et al, 2002a, 2002b). We suggested that the targeted degradation of VDU1 and VDU2 by pVHL–E3 ligase could be crucial for fine regulation of the ubiquitin–proteasome degradation pathway associated with pVHL-related physiological and pathological processes. In this report, we demonstrate that VDU2, but not VDU1, interacts with HIF-1α and has a role in HIF-1α stabilization.
Results and Discussion
VDU2, but not VDU1, interacts with HIF-1α
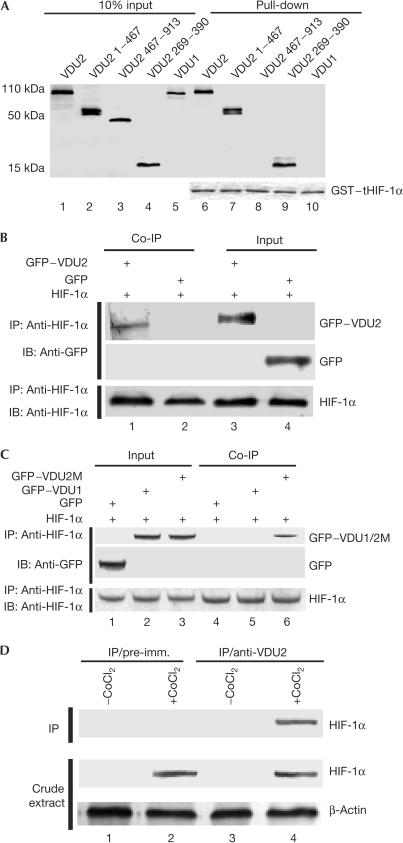
The fact that VDU1 and VDU2 are two substrates of the pVHL–E3 ligase for ubiquitination and both are deubiquitinating enzymes prompted us to examine whether VDU1 and VDU2 might be involved in deubiquitinating other downstream targets of pVHL–E3 ligase, salvaging them from proteasomal degradation. We first examined whether either of these two deubiquitinating enzymes could interact with HIF-1α. In a screening co-immunoprecipitation assay, the in vitro-translated T7–VDU2 fusion protein, but not T7–VDU1, was precipitated by anti-HIF-1α antibody in the presence of in vitro-translated HIF-1α (data not shown). On the basis of amino-acid sequences and domain structures of VDU1 and VDU2 (supplementary Fig S1 online), we proposed that the HIF-1α-interacting region might be located in the region between the UCH-1 and UCH-2 domains of VDU2, as this region shows only 25% amino-acid sequence identity with VDU1. We then performed direct binding assays using a glutathione-S-transferase (GST)-fused HIF-1α fragment (amino acids 530–826) to pull down the in vitro-translated VDU2 fragments and full-length VDU2 and VDU1. Our results showed that the full-length VDU2, VDU2 (1–467) and VDU2 (269–390), but neither VDU2 (467–913) nor the full-length VDU1, could bind to GST-fused HIF-1α (530–826) (Fig 1A), indicating that the HIF-1α-binding region is located within VDU2 amino acids 269–390. We then performed a mammalian two-hybrid assay in both COS-7 cells and 786-O cells (a VHL-null cell line) by coexpression of a GAL4-DBD-fused HIF-1α (amino acids 530–826) fragment and VP16-AD-fused VDU2, which demonstrated that these two proteins could interact with each other in the mammalian milieu (supplementary Fig S2A,B online). To demonstrate further that these two proteins interact with each other in mammalian cells, we performed immunoprecipitation assays, and our results showed that the GFP–VDU2 fusion protein was also readily immunoprecipitated from COS-7 cells transfected with the GFP–VDU2 fusion construct and the HIF-1α expression construct, but not GFP alone (Fig 1B) and GFP–VDU1 (Fig 1C). A VDU2 mutant (VDU2M-C154A), in which the key cysteine residue at amino acid 154 within the catalytic cysteine box was mutated to alanine, retained the ability to bind to HIF-1α (Fig 1C). Next, anti-VDU2 antisera were raised in rabbits against the purified 6 × His–VDU2 fragment, amino acids 269–390. This fragment was chosen owing to its minimal sequence identity to VDU1. Anti-VDU2 polyclonal antibodies were purified through an affinity column, and immunoblots using both in vitro-translated proteins and whole-cell lysates were performed. The results showed no crossreactivity of the purified antibodies to VDU1 (supplementary Fig S3A,B online). By using this antibody, we examined the interaction between endogenous HIF-1α and VDU2 proteins in HeLa cells cultured under normoxia or in those treated with hypoxia mimics (CoCl2). After immunoprecipitation, a western blot showed that HIF-1α could be immunoprecipitated by anti-VDU2 antibody but not by the pre-immune serum (Fig 1D). This interaction was detected only in cells subjected to hypoxia mimics, CoCl2 treatment, which stabilize HIF-1α in cells and make it detectable (Fig 1D). Together, these in vitro and in vivo assays demonstrate that VDU2, but not VDU1, can interact with HIF-1α. The fact that amino acids 269–390 in VDU2 have much less sequence identity to the same region of VDU1 explains why VDU1, which has high sequence identity with VDU2 in the zf-UBP, UCH1 and UCH2 domains, and carboxyl terminus, does not interact with HIF-1α.
Figure 1.
VDU2, but not VDU1, interacts with HIF-1α. (A) GST pull-down assay for mapping the HIF-1α–VDU2 interaction. In vitro-translated VDU2, VDU2 1–467, VDU2 467–913, VDU2 269–390 and VDU1 proteins were incubated with GST–tHIF-1α (530–826) and the bound proteins were eluted and analysed using SDS–polyacrylamide gel electrophoresis. Equal loading of GST fusion proteins is demonstrated in the lower panel. (B) In vivo co-immunoprecipitation (Co-IP) of HIF-1α with GFP–VDU2. Lysates prepared from COS-7 cells transfected with pCEP4–HIF-1α and pEGFP–C3 or pEGFP–C3–VDU2 were subjected to IP with anti-HIF-1α antibody followed by anti-GFP immunoblotting (IB). (C) In vivo Co-IP of HIF-1α with GFP–VDU1 or GFP–VDU2M-C154A. Lysates prepared from COS-7 cells transfected with pCEP4–HIF-1α plus pEGFP–C3, pEGFP–C3–VDU1 or pEGFP–C3–VDU2M-C154A were subjected to IP with anti-HIF-1α antibody followed by anti-GFP IB. (D) Interaction between endogenous HIF-1α and VDU2. The top panel shows a western blot of control IP with the pre-immune serum (lanes 1,2) and IP with VDU2 antibody (lanes 3,4) from HeLa cells untreated (lanes 1,3) or treated with CoCl2 (lanes 2,4). The middle and bottom panels show blots of cell extracts by anti-HIF-1α and anti-β-actin antibody, respectively.
VDU2 stabilizes HIF-1α
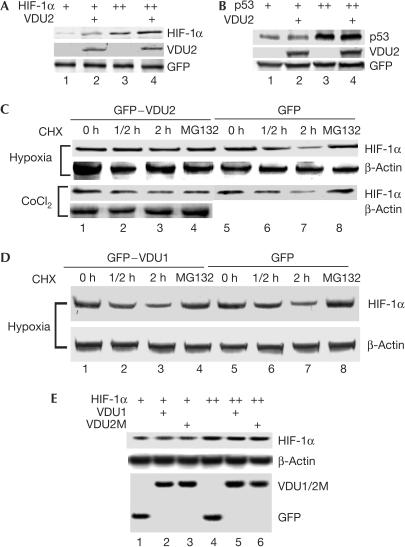
To determine the functional consequences of the VDU2–HIF-1α interaction, we asked whether VDU2 has a role in stabilizing HIF-1α. We first examined whether overexpression of VDU2 could increase the HIF-1α level. Our results showed that VDU2 overexpression significantly increased the steady-state cellular levels of HIF-1α under normoxia (Fig 2A). In contrast, VDU2 had no obvious effect on the levels of p53 (Fig 2B), which is another short-lived protein the stability of which is also regulated by the ubiquitination pathway (Li et al, 2002). To show directly the effect of VDU2 on HIF-1α stabilization, we examined the HIF-1α protein level in the presence of both pVHL and VDU2 in 786-O cells. Our results demonstrated that overexpression of pVHL significantly induced HIF-1α degradation, and this phenomenon could be inhibited on coexpression of VDU2 (supplementary Fig S4 online). Furthermore, we examined the effect of VDU2 overexpression on stabilization of endogenous HIF-1α. HEK293 cells stably transfected with pEGFP or pEGFP–VDU2/VDU1 were established and used in these experiments. Western blot analysis showed that the half-life of HIF-1α in cells stably transfected with VDU2 was significantly increased by expression of VDU2 after hypoxia and cycloheximide (CHX) treatment when compared with that in cells stably transfected with the empty pEGFP vector (Fig 2C). In a similar experiment, the half-life of HIF-1α in cells stably transfected with pEGFP–VDU1 showed no significant change when compared with that in cells stably transfected with the empty pEGFP vector (Fig 2D). MG132 also blocked the degradation of HIF-1α under these conditions (Fig 2C). To demonstrate further that the effect of VDU2 on the half-life of HIF-1α was related to the VDU2 deubiquitinating enzyme activity, VDU2M-C154A was tested on the half-life of HIF-1α. Although VDU2M-C154A was shown to still interact with HIF-1α (Fig 1C), its ability to increase the steadystate cellular levels of HIF-1α was abolished (Fig 2E) with the key cysteine changed to alanine. Additionally, VDU1 showed no effect on the steadystate cellular levels of HIF-1α under normoxia (Fig 2E). These data demonstrate that overexpression of VDU2 specifically stabilizes HIF-1α in cells under both normoxic and hypoxic conditions with CHX treatment. These results also raised the question of whether increased HIF-1α was caused by VDU2 rescuing HIF-1α or caused by VDU2 competing with HIF-1α to bind to pVHL.
Figure 2.
VDU2 stabilizes HIF-1α. (A) Enhancement of the steady levels of HIF-1α by VDU2. Western blot analysis of extracts from HEK293 cells transfected with HIF-1α alone or with HIF-1α and VDU2 together as indicated. (B) VDU2 has no effect on the level of p53. Western blot analysis of extracts from HEK293 cells transfected with p53 alone or with p53 and VDU2 together as indicated. (C) VDU2 increases the half-life of endogenous HIF-1α. Cell extracts from both the hypoxia- and CoCl2-treated HEK293 cells either stably transfected with pEGFP or pEGFP–VDU2 and collected at different time points as indicated after treatment with CHX or both CHX and MG132 (lane 4) were analysed for HIF-1α protein levels by western blot. (D) VDU1 does not increase the half-life of endogenous HIF-1α. Cell extracts prepared from hypoxia-treated HEK293 cells stably transfected with either pEGFP or pEGFP–VDU1 and collected at different time points as indicated after treatment with CHX or both CHX and MG132 (lane 4) were analysed for HIF-1α protein levels by western blot. (E) Neither VDU1 nor VDU2M-C154A enhances HIF-1α levels. Western blot analysis of extracts from HEK293 cells transfected with HIF-1α alone or with HIF-1α plus VDU2M-C154A or VDU1 was performed.
VDU2 deubiquitinates HIF-1α
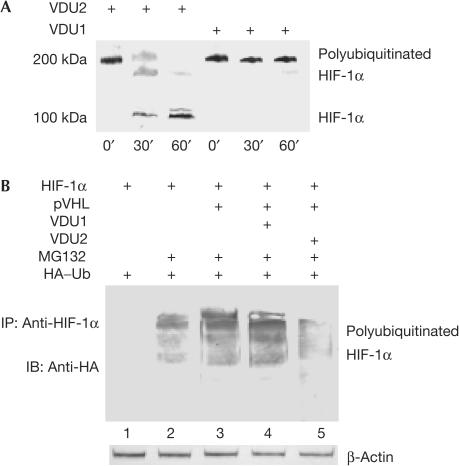
To explain further the molecular mechanism by which VDU2 stabilizes HIF-1α, we asked whether VDU2 could directly control the levels of HIF-1α ubiquitination. A highly purified in vitro test system was used in the in vitro assay to avoid possible contamination by either inhibitory factors or any enzymes involved in the ubiquitination and deubiquitination of HIF-1α. VDU2 and VDU1 were expressed in bacteria, which are known to have no ubiquitinating and deubiquitinating system, and purified to near homogeneity. HIF-1α was in vitro-translated and polyubiquitinated in an in vitro ubiquitination system. The ubiquitinated HIF-1α was purified by immunoprecipitation using anti-ubiquitin antibody. As shown in Fig 3A, the ubiquitinated HIF-1α was efficiently deubiquitinated on incubation with purified recombinant VDU2, but not VDU1, which also served as a negative control. Next, consistent with previous reports, we found a high level of ubiquitinated HIF-1α in cells transfected with HIF-1α and pVHL in the in vivo system. However, HIF-1α ubiquitination was significantly decreased by VDU2 expression, but not by VDU1 expression (Fig 3B). If this is caused by competition between VDU2 and HIF-1α to bind to pVHL–E3 ubiquitin ligase, one would expect that VDU1, similar to VDU2, could also decrease ubiquitinated HIF-1α and stabilize HIF-1α, as both VDU1 and VDU2 can interact with pVHL. These results demonstrate that VDU2 can specifically deubiquitinate HIF-1α both in vitro and in vivo. These results also demonstrated that, although their amino-acid sequences are similar, VDU1 and VDU2 are not redundant deubiquitinating enzymes. Interestingly, VDU1 and VDU2 genes are also regulated differently in response to external stress, such as hypothermia. Although both VDU1 and VDU2 bind to and deubiquitinate the type 2 iodothyronine deiodinase (D2), VDU1, but not VDU2, is markedly increased in brown adipocytes by norepinephrine or cold exposure, which results in the increase of D2 activity by prolonging the half-life of D2 through deubiquitination and, therefore, allows the animals to respond to the cold temperature quickly (Curcio-Morelli et al, 2003).
Figure 3.
VDU2 deubiquitinates HIF-1α. (A) Deubiquitination of HIF-1α by VDU2 in vitro. The purified polyubiquitinated HIF-1α was incubated with the purified recombinant VDU1 or VDU2 in a deubiquitination buffer for the durations indicated at 30°C followed by western blot with anti-HIF-1α antibody. (B) Regulation of HIF-1α ubiquitination levels in vivo. Western blot analysis of immunoprecipitates with anti-HIF-1α antibody from HEK293 cells transfected with HIF-1α and HA–ubiquitin (lanes 1–5), pVHL (lanes 3–5), VDU1 (lane 4), VDU2 (lane 5), and treated with MG132 (lanes 2–5). The blot was detected with anti-HA antibody.
VDU2 enhances HIF-1α-mediated activity
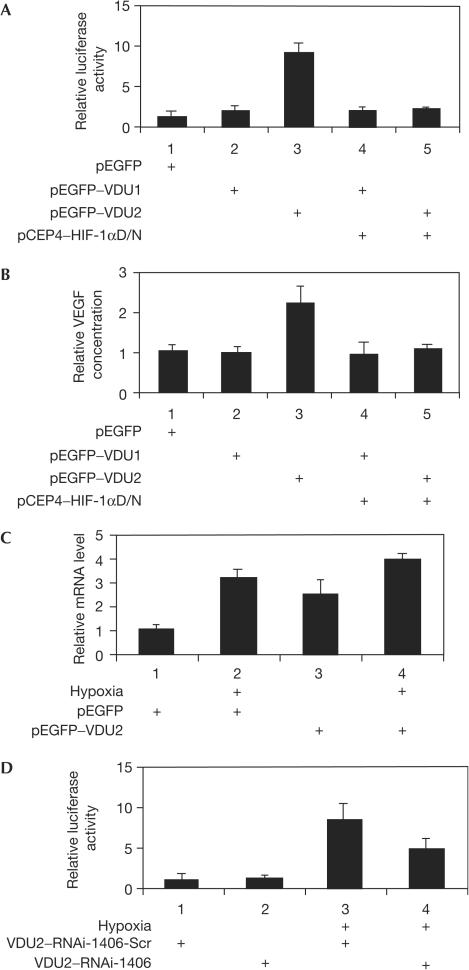
As VDU2 can deubiquitinate the ubiquitinated HIF-1α and salvage it from proteasomal degradation, we further examined whether VDU2 has any effect on the HIF-1α-induced hypoxia response element (HRE, hypoxia-inducible element) luciferase reporter gene activity and VEGF expression. Our results showed that in the HIF-1α- and pVHL-positive cells (HEK293), overexpression of VDU2 significantly increased HRE luciferase activity (Fig 4A) and VEGF protein secretion from HEK293 cells (Fig 4B), but not in a pVHL-null cell line (786-O) (supplementary Fig S7 online). The fact that coexpression of HIF-1αD/N (dominant negative) can abrogate the VDU2-induced HRE luciferase activity and VEGF protein secretion from these cells suggests that these effects are mediated through the HIF-1α pathway (Fig 4A,B). Notably, overexpression of VDU1 has no effects on HRE luciferase reporter gene activity and VEGF protein secretion (Fig 4A,B). We also examined VEGF messenger RNA levels in HEK293 cells stably transfected with GFP–VDU2 and their control counterparts, HEK293 cells stably transfected with GFP, cultured under normoxia or hypoxia (1% O2). We found that HEK293 cells with VDU2 overexpression showed a higher level of VEGF mRNA expression than those without VDU2 overexpression under both normoxic and hypoxic conditions (Fig 4C). A similar result was obtained when examining another HIF-1α-regulated gene, haeme oxygenase-1 (HO-1; supplementary Fig S5 online). We built two short interfering RNA (siRNA) constructs to suppress the overexpression of VDU2 in HEK293 cells stably transfected with GFP–VDU2. One of them, VDU2–RNAi-1406, was able to downregulate either the overexpressed GFP–VDU2 or endogenous VDU2 (supplementary Fig S6A,B online). A scrambled VDU2–RNAi-1406 served as a negative control. Using these siRNA constructs, we tested HRE luciferase reporter gene activity in HEK293 cells stably transfected with GFP–VDU2 under normoxic and hypoxic (1% O2) conditions. Our results showed that suppression of GFP–VDU2 decreased the level of HER luciferase reporter gene activity under hypoxia compared with the scrambled RNAi control (Fig 4D), suggesting that increased HIF-1 transcriptional activity in these cells was mediated through VDU2.
Figure 4.
VDU2 enhances HIF-1α-mediated HRE activity and VEGF expression. (A) Regulation of HIF-1α-mediated HRE luciferase reporter gene activity by VDU2. Various combinations of expression plasmids as indicated with 300 ng of pRE–tk–LUC and 10 ng of pRLSV40–LUC were transfected into HEK293 cells. At 24 h after transfection, cells were collected and measured in a dual-luciferase assay. (B) Regulation of HIF-1α-mediated VEGF protein production by VDU2. Various combinations of expression plasmids as indicated were transfected into HEK293 cells. At 36 h after transfection, aliquots of medium were collected for VEGF ELISA. Histograms represent the results of three independent experiments. (C) Effects of overexpression of VDU2 on VEGF mRNA levels. HEK293 cells stably transfected with GFP–VDU2 or GFP were cultured under normoxia or hypoxia (1% O2). VEGF mRNA levels were determined by real-time PCR. (D) Downregulation of HRE luciferase reporter gene activity in HEK293 cells stably transfected with VDU2 by VDU2–RNAi-1406 under hypoxia. Various combinations of expression plasmids as indicated with the dual reporter system were transfected into HEK293 cells stably transfected with GFP–VDU2. At 24 h after transfection, cells were cultured under normoxia or hypoxia (1% O2) overnight and collected for a dual-luciferase assay.
On the basis of the above findings, we conclude that deubiquitination is an important biochemical event in the regulation of HIF-1α stability. VDU2 may deubiquitinate ubiquitinated HIF-1α and rescue HIF-1α from degradation, therefore providing another level of control in HIF-1α stabilization. Conversely, with overexpression of VDU2, pVHL-mediated HIF-1α ubiquitination and subsequent proteosomal degradation may be compromised. Additionally, whether VDU2 has any role in O2-independent, growth-factor-stimulated increase of HIF-1α levels in certain types of cancer cell and certain physiological conditions requires further exploration.
Methods
Plasmid construction. Plasmids pCMX–VHL19, pCDNA4–VHL19, pEGFP–C3–VDU1, pEGFP–C3–VDU2, pET28c–VDU1, pET28c–VDU2, pCDNA4–VDU2, pVP16–VDU1, pVP16–VDU2 and pSG5–p53 have been described previously (Li et al, 2002a, 2002b). Plasmids pET28c–VDU2 1–467, pET28c–VDU2 467–913 and pET28c–VDU2 269–390, containing truncated VDU2 fragments of different lengths, were constructed by inserting the PCR-amplified human VDU2 fragments into pET28c vector (Novagen, Madison, WI, USA). pEGFP–C3–VDU2M-C154A was generated by site-directed PCR mutagenesis using overlapping primer pairs covering the intended mutation site and containing the appropriate nucleotide changes. pCEP4–HIF-1α and pCEP4–HIF-1αD/N plasmids, pRE–tk–LUC and HA–ubiquitin were kindly provided by Dr Gregg L. Semenza, Dr Steven L. McKnight and Dr Dirk Bohmann, respectively. pM–tHIF-1α (amino acids 530–826) and pGEX6p1–tHIF-1α (amino acids 530–826) were constructed by inserting PCR-amplified HIF-1α fragments into the corresponding vectors. pcDNA3–HIF-1α was constructed by inserting the NotI- and KpnI-digested HIF-1α cDNA from pCEP4–HIF-1α into pcDNA3 vector (Invitrogen, Carlsbad, CA, USA). Two DNA vector-based RNAi constructs were created to target VDU2. After a preliminary test, VDU2–RNAi-1406 showed downregulation of VDU2 and was used in our experiments. A negative control, VDU2–RNA-1406scr, containing scrambled targeting sequence, was also constructed (supplementary information online). All DNA fragments amplified by PCR were confirmed by complete sequencing.
Cell culture, transfection and luciferase activity assay. COS-7, HeLa and 786-O cells were maintained as described previously (Li et al, 2002a). Transient transfection was performed using SuperFect (Qiagen, Valencia, CA, USA) reagent according to the manufacturer's protocol. For in vivo co-immunoprecipitation assays, HEK293 cells were transfected with 5 μg of pCEP4–HIF-1α and/or VDU2 plasmids. At 36 h after transfection, cells were treated with MG132 (5 μM, Calbiochem, San Diego, CA, USA) for 6 h. To examine the degradation of the HIF-1α protein, HEK293 cells were plated in 60-mm dishes and transfected with different combinations of plasmids. At 36 h after transfection, cells were treated with or without MG132 for 6 h and collected for lysis. For stable transfection, HEK293 cells were transfected with pEGFP–VDU2, pEGFP–VDU1 or the empty pEGFP–C3 as a negative control with SuperFect reagent and selected with neomycin (Invitrogen). To examine the half-life of endogenous HIF-1α, HEK293 cells stably transfected with pEGFP, pEGFP–VDU2 or pEGFP–VDU1 were treated with CoCl2 or incubated under 1% O2 for 6 h. Cells were moved back to the normoxic condition and treated with CHX for different durations as indicated or with both CHX and MG132 for 2 h and then collected for lysis. For the luciferase assay, cells seeded in 12-well plates were transfected with a combination of plasmids as indicated in the figures, plus an internal control plasmid pRLSV40–LUC and a reporter plasmid pRE–tk–LUC, which contains three HREs and a herpes simplex virus thymidine kinase gene promoter. Analyses of luciferase activity were performed according to the protocol of the Dual Luciferase Assay System (Promega, Madison, WI, USA) and relative light units were measured using a luminometer.
Antibodies and immunoblotting. Anti-VDU2 antisera were raised in rabbits against the purified 6 × His–hVDU2 (amino acids 269–390; Covance Research Products, Inc., Denver, PA, USA). Anti-HA polyclonal antibody, anti-6 × His tag, anti-β-actin, anti-GFP (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), anti-T7 (Novagen), anti-VHL (BD Biosciences, Pharmingen, San José, CA, USA), anti-HIF-1α (Novus Biologicals, Littleton, CO, USA) and anti-p53 (Oncogene Research Products, San Diego, CA, USA) monoclonal antibodies were obtained from indicated commercial sources. Samples were resolved by SDS–polyacrylamide gel electrophoresis, transferred to PVDF membranes and immunoblotted as described previously (Li et al, 2002a).
Other methods. GST pull-down assay, in vivo and in vitro co-immunoprecipitation, mammalian two-hybrid assay, in vitro deubiquitination assay, RNA isolation, RT–PCR and determination of VEGF protein levels by ELISA were performed as described previously (Li et al, 2002a, 2002b, 2003) and are briefly summarized in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400377s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr D. Bohmann, Dr G.L. Semenza and Dr S.L. McKnight for plasmids.
References
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli C, Zavacki AM, Christofollete M, Gereben B, de Freitas BC, Harney JW, Li Z, Wu G, Bianco AC (2003) Deubiquitination of type 2 iodothyronine deiodinase by von Hippel–Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J Clin Invest 112: 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A, Pellman D (1998) Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol 33: 337–352 [DOI] [PubMed] [Google Scholar]
- Epstein AD et al. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Harris AL (2001) Hypoxia—a key regulatory factor in tumor growth. Nat Rev Cancer 2: 38–46 [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Jaakkola P et al. (2001) Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr (2002) Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer 2: 673–682 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–653 [DOI] [PubMed] [Google Scholar]
- Li Z, Na X, Wang D, Schoen SR, Messing EM, Wu G (2002a) Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel–Lindau tumor suppressor protein. J Biol Chem 277: 4656–4662 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Na X, Messing EM, Schoen SR, Wu G (2002b) Identification of a ubiquitin specific processing protease subfamily as the downstream substrates of pVHL E3 ubiquitin ligase. Biochem Biophys Res Commun 294: 700–709 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Na X, Messing EM, Schoen SR, Wu G (2003) The VHL protein recruits a novel KRAB-A domain protein to repress HIF-1α transcriptional activity. EMBO J 22: 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL (2003) Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3: 721–732 [DOI] [PubMed] [Google Scholar]
- Wilkinson KD (2000) Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol 11: 141–148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information