Abstract
In previous work, we found that an anaerobic sludge efficiently degraded hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), but the role of isolates in the degradation process was unknown. Recently, we isolated a facultatively anaerobic bacterium, identified as Klebsiella pneumoniae strain SCZ-1, using MIDI and the 16S rRNA method from this sludge and employed it to degrade RDX. Strain SCZ-1 degraded RDX to formaldehyde (HCHO), methanol (CH3OH) (12% of total C), carbon dioxide (CO2) (72% of total C), and nitrous oxide (N2O) (60% of total N) through intermediary formation of methylenedinitramine (O2NNHCH2NHNO2). Likewise, hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine (MNX) was degraded to HCHO, CH3OH, and N2O (16.5%) with a removal rate (0.39 μmol · h−1 · g [dry weight] of cells−1) similar to that of RDX (0.41 μmol · h−1 · g [dry weight] of cells−1) (biomass, 0.91 g [dry weight] of cells · liter−1). These findings suggested the possible involvement of a common initial reaction, possibly denitration, followed by ring cleavage and decomposition in water. The trace amounts of MNX detected during RDX degradation and the trace amounts of hexahydro-1,3-dinitroso-5-nitro-1,3,5-triazine detected during MNX degradation suggested that another minor degradation pathway was also present that reduced —NO2 groups to the corresponding —NO groups.
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) is a widely used explosive that severely contaminates soil and groundwater (13, 22). RDX has been found to be toxic in various terrestrial and aquatic environments (26, 29). The toxicity of cyclic nitramines means that contaminated soil and groundwater must be remediated, preferably by bioremediation. Biodegradation of RDX with anaerobic sludge was extensively studied by McCormick et al. (21) and Kaplan (19), who proposed a pathway based on the sequential reduction of RDX to hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine (MNX), hexahydro-1,3-dinitroso-5-nitro-1,3,5-triazine (DNX), and hexahydro-1,3,5-trinitroso-1,3,5-triazine (TNX). It was proposed that the nitroso compounds undergo further transformation to hydroxylamino derivatives (HOHN-RDX), which are subsequently cleaved to produce formaldehyde (HCHO), methanol (CH3OH), hydrazine (NH2NH2), and dimethyl hydrazine [(H3C)2NNH2].
Other research groups have employed mixed anaerobic microbial cultures that have included methanogens (1, 2), acetogens (4), nitrate reducers (12), and individual isolates such as isolates of Desulfovibrio sp. (5), Clostridium bifermentans (25) Providencia rettgeri (20), Citrobacter freundii (20), Morganella morganii (20), and Serratia marcescens (31) to biotransform RDX, but in most cases no clear details concerning ring cleavage products or mineralization were provided in the descriptions. Subsequent work in our laboratory (14, 15, 16) showed that RDX could be degraded in anaerobic sludge via intermediary formation of methylenedinitramine (O2NNHCH2NHNO2), which decomposes in water to nitrous oxide (N2O) and HCHO. Recently, Oh et al. (23) confirmed the formation of O2NNHCH2NHNO2 during RDX degradation with anaerobic sludge. Despite these previous efforts, there is still insufficient information regarding the roles of individual anaerobic isolates and their products during RDX degradation. Without these details, particularly details concerning specific initial enzymatic reactions and their products, it will be difficult to understand degradation pathways and to optimize degradation towards mineralization.
The objective of the present study was to investigate degradation of RDX by an RDX-mineralizing strain, Klebsiella pneumoniae strain SCZ-1, which was isolated from the anaerobic sludge that was successfully used previously to degrade this energetic chemical (14, 16). We also used this strain to degrade MNX, an RDX degradation metabolite, under the same conditions that were used for the RDX degradation in an attempt to provide new insight into the anaerobic degradation pathway of cyclic nitramine explosives.
MATERIALS AND METHODS
Chemicals.
RDX (99% pure) and l-[U-14C]RDX (chemical purity, >98%; radiochemical purity, 96%; specific radioactivity, 28.7 μCi · mmol−1) were provided by Defense Research and Development Canada, Quebec, Canada (3). TNX (99% pure) was synthesized by the method described by Brockman et al. (7). H14CHO (specific activity, 53 mCi · mmol−1) was obtained from Aldrich, Oakville, Ontario, Canada. MNX (98% pure) was provided by R. J. Spanggord of SRI International (Menlo Park, Calif.). Methylenedinitramine was purchased from the rare chemical department of Aldrich. All other chemicals used were reagent grade.
Media.
The medium used for isolation of anaerobic bacteria was 5.8% Difco anaerobic agar (Becton Dickinson, Sparks, Md.). The YPG medium (pH 7.3) used for fermentative bacterial growth and RDX degradation contained (per liter of basic salts medium) 3.0 g of yeast extract, 0.6 g of Bacto Peptone (Difco), and 1.0 g of glucose. The basic salts medium was prepared as described previously (30), except that no (NH4)2SO4 was added and (NH4)6Mo7O24 · 4H2O was replaced with NaMoO4 · 2H2O. YPS agar was prepared as described by Zhao et al. (32). Liquid media were sterilized either by autoclaving at 120°C for 20 min or by filtration with sterile filters (pore size, 0.22 μm; Millex GP; Millipore, Bedford, Mass.) for trace metals and the glucose stock solution. The dry serum bottles used were sterilized by autoclaving them at 120°C for 60 min.
Bacterial isolation and characterization.
A methanogenic industrial sludge was obtained from Sensient Flavor Canada (Cornwall, Ontario, Canada) and was incubated on Difco anaerobic agar plates at 37°C under an atmosphere containing a mixture of H2 and CO2 (GasPak Plus; BBL, Sparks, Md.). Anaerobic conditions were monitored with a BBL indicator (VWR Canlab, Mississauga, Ontario, Canada). One strain, SCZ-1, was selected for subsequent characterization due to its high growth rate under both aerobic and anaerobic conditions and its capacity to mineralize RDX anaerobically. As determined by the Sherlock microbial identification system (MIDI, Newark, Del.), strain SCZ-1 was closely related to Klebsiella pneumoniae, with a similarity index of 0.9. As determined by the 16S rRNA identification method (18), the gene sequence of a 0.7-kb fragment (representing one-half of the total length of the gene) of the 16S rRNA gene from the isolate was 99% similar to the K. pneumoniae sequence.
Bacterial growth and biodegradation tests.
By using a procedure described previously (16), serum bottles (60 ml), each containing 18.5 ml of YPG medium and 106 μM RDX, were made anaerobic by repeated degassing under a vacuum and charging with filter-sterilized oxygen-free nitrogen or argon. The sealed bottles were then inoculated with 1.5 ml of an aerobic liquid culture (initial optical density at 600 nm [OD600], 0.15) of strain SCZ-1 and incubated in a rotatory shaker (200 rpm) at 37°C. After 3 h of anaerobic incubation, the bacterial growth reached a maximum OD600 of 0.7 (0.91 g [dry weight] of cells per liter). We measured the redox potential (Eh) with a Pt/Ag/AgCl electrode (Fisher Scientific, Montreal, Canada) and found that it decreased from 200 to −300 mV during this incubation period (meanwhile the pH decreased from 7.3 to 6.3). MNX (100 μM) and TNX (100 μM) biodegradation tests were conducted under the same conditions that were used for the RDX degradation tests. Separate bottles were used for monitoring production of either aqueous metabolites or N2O. For sampling, sterile syringes and needles washed with reduced buffer were used. Some microcosms were spiked with l-[U-14C]RDX (0.038 μCi) to measure mineralization (liberated 14CO2) with a Tri-Carb 4530 liquid scintillation counter (model 2100 TR; Packard Instrument Company, Meriden, Conn.) (16). When 14CO2 ceased to form in a microcosm, the microcosm was sacrificed to measure the remaining radioactivity in the culture supernatant and in the biomass. A similar procedure was used to mineralize HCHO (166 μM) supplemented with H14CHO (0.030 μCi).
RDX and metabolite analyses.
The concentrations of RDX and its nitroso products MNX, DNX, and TNX (in the aqueous phase obtained by centrifugation at 9,000 × g for 3 min) were analyzed at 230 nm by a high-performance liquid chromatography-UV method as described previously (16). The methods used for analyses of methylenedinitramine, (methyl)hydrazines, N2O (14, 16), and HCHO (11, 28) have also been described previously. CH3OH was measured by gas chromatography with a flame ionization detector (HP 6890) by using a Hayesep Q micropacked column (2 m by 0.03 mm; Supelco, Bellefonte, Pa.) (detection limit, 0.25 ppm). Nitrite was analyzed by U.S. Environmental Protection Agency method 345.1 (10), which has a detection limit of 10 ppb. Ammonia was analyzed by an enzymatic assay in which l-glutamate dehydrogenase and NADPH (Sigma, St. Louis, Mo.) were used. All tests were performed in triplicate.
RESULTS AND DISCUSSION
Isolation of strain SCZ-1.
An anaerobic RDX-mineralizing bacterium, strain SCZ-1, was isolated from the anaerobic sludge that was previously used to degrade RDX (14, 16). Strain SCZ-1 is a nonmotile, non-spore-forming, gram-negative, rod-shaped, facultatively anaerobic bacterium. Under aerobic and anaerobic conditions, strain SCZ-1 grew on glucose and peptone, but it did not grow when RDX was used as a sole source of carbon or nitrogen. On the basis of MIDI and 16S rRNA data, we identified strain SCZ-1 as K. pneumoniae.
RDX degradation with strain SCZ-1.
We found that strain SCZ-1 grew better aerobically but degraded RDX only under anaerobic conditions in the presence of glucose and peptone. K. pneumoniae strain SCZ-1 degraded RDX at a rate of 0.41 μmol · h−1 · g (dry weight) of cells−1, which was lower than the rate observed with the mixed anaerobic culture (10 μmol · h−1 · g [dry weight] of cells−1) (16). On the other hand, the rate of removal of RDX with strain SCZ-1 (9.0 μM · day−1) (biomass OD600, 0.7) is comparable to removal rates previously reported for other facultatively anaerobic organisms, such as M. morganii (12.1 μM · day−1), P. rettgeri (12.1 μM · day−1), C. freundii (7.3 μM · day−1; biomass OD580, 1.6 to 1.8) (20), and S. marcescens (20.4 μM · day−1; biomass unknown) (31). The previously described pure anaerobic cultures exhibited only low RDX mineralization activity (Table 1). In contrast, strain SCZ-1 could mineralize 72% of RDX (Fig. 1A).
TABLE 1.
Comparison of RDX mineralization values and maximal nitroso metabolite yields obtained with pure anaerobic bacterial isolates
| Bacterial strain | % Mineralization, as 14CO2 | Nitroso metabolite yields (%)
|
Reference | ||
|---|---|---|---|---|---|
| MNX | DNX | TNX | |||
| Klebsiella pneumoniae SCZ-1a | 72.3 (0.8) | 2 (0.5) | 0 | 0 | This study |
| Morganella morganii B2b | 5 (1) | 25 | 2.5 | 0 | 20 |
| Providencia rettgeri B1b | 8 (2) | 35 | 15 | 0 | 20 |
| Citrobacter freundii NS2b | 9 (2) | 50 | 15 | 5 | 20 |
| Clostridium bifermentans | NDd | ND | ND | ND | 25 |
| Desulfovibrio sp. | ND | ND | ND | ND | 5 |
| Serratia marcescensc | ND | 50 | 30 | 1 | 31 |
The values are averages for triplicate samples; standard deviations are indicated in parentheses. The maximal bacterial OD600 was 0.70.
The mineralization values are averages for duplicate samples; standard deviations are indicated in parentheses. The maximal bacterial OD560 for strains B2, B1, and NS2 were 1.8, 1.8, and 1.6, respectively. The yields of metabolites were calculated from data provided in reference 20.
Biomass was not given. The yields of nitroso metabolites were estimated from values provided in reference 31.
ND, no data available.
FIG. 1.
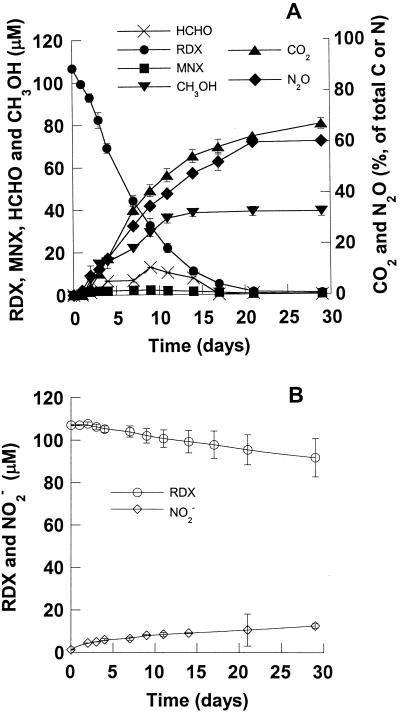
Anaerobic degradation of 106 μM RDX with K. pneumoniae strain SCZ-1. (A) Live cells (initial biomass, 0.15 OD600 unit; maximal growth, 0.7 OD600 unit or 0.9 g [dry weight] of cells · liter−1). (B) Killed cells (initial biomass, 0.15 OD600 unit).
Removal of RDX by strain SCZ-1 was accompanied by formation of several products, including N2O (60%), HCHO, CO2 (72%), and CH3OH (12%) (Fig. 1A). In controls containing RDX and killed cells, approximately 10% of the RDX was degraded (hydrolyzed) and nitrite accumulated (Fig. 1B). Nitrite was not seen in RDX degradation experiments when live strain SCZ-1 was used, and this was attributed to the bacterial conversion of nitrite to ammonia. For instance, when we incubated a standard solution of sodium nitrite (130 μM) with strain SCZ-1, the anion disappeared at a rate of 128 μmol · h−1 · g (dry weight) of cells−1, which was more than 300 times higher than the rate of RDX removal (0.41 μmol · h−1 · g [dry weight] of cells−1). Unfortunately, we were unable to quantify ammonia because relatively large amounts of it were released from YPG medium. Strain SCZ-1 did not degrade RDX in basic salts medium with RDX added as a carbon or nitrogen source; thus, we were unable to quantify ammonia production from RDX degradation. Other potential N-containing products, such as hydrazine and dimethyl hydrazines, which were reported by McCormick et al. (21), were not detected in the present study.
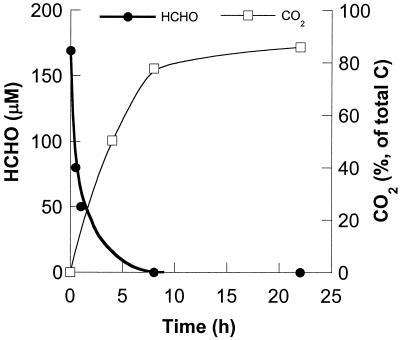
HCHO did not accumulate and was mineralized. For instance, when we incubated a reference standard of HCHO spiked with H14CHO with strain SCZ-1, we observed more than 85% mineralization (14CO2) in less than 20 h (Fig. 2). Figure 2 clearly shows that long after the removal of the aldehyde CO2 continued to form slowly.
FIG. 2.
Anaerobic mineralization of 169 μM HCHO with K. pneumoniae strain SCZ-1 culture supplemented with 0.03 μCi of H14CHO. The biomass was the same as that described in the legend to Fig. 1.
Using liquid chromatography-mass spectrometry with negative electron spray ionization, we were able to identify methylenedinitramine as a key ring cleavage intermediate, and spontaneous decomposition of this compound gave N2O and HCHO, as reported previously (14). Furthermore, we detected trace amounts of MNX, but we were unable to detect DNX or TNX. The product distribution described above is strikingly similar to that obtained during RDX incubation with the anaerobic sludge, indicating that there are similarities between the degradation pathways in the two cases (see below).
Stoichiometry of RDX degradation.
Conversion of 2.1 μmol of RDX (106 μM) produced 3.8 μmol of N2O, representing 60% of the total N content of RDX (4 N atoms) (Fig. 1A). Methylenedinitramine (O2NNHCH2NHNO2), a key intermediate during degradation of RDX by strain SCZ-1, is the main source of N2O. It has been demonstrated previously that 1 mol of O2NNHCH2NHNO2 decomposes quantitatively in water at pH 7 and 30°C to produce 2 mol of N2O and 1 mol of HCHO (14). The stoichiometry of N2O formation supported our belief that methylenedinitramine, which has two —NH-NO2 groups, was responsible for the quantitative formation of N2O from RDX in strain SCZ-1 (14).
Conversion of 106 μM RDX also produced 39 μM CH3OH, representing 12% of the total carbon in transformed RDX (Fig. 1A). Using l-[U-14C]RDX as the substrate, we found that strain SCZ-1 mineralized 72% of the total carbon in RDX (liberated 14CO2) over 36 days (Fig. 1A). Most of the remaining radioactivity (27%) was in the aqueous phase of the culture medium, and less than 0.5% of the RDX carbon was in biomass, which left roughly 15% of the carbon content of RDX unaccounted for. However, slow release of CO2 was still occurring long after the complete disappearance of RDX (Fig. 1A).
MNX and TNX degradation.
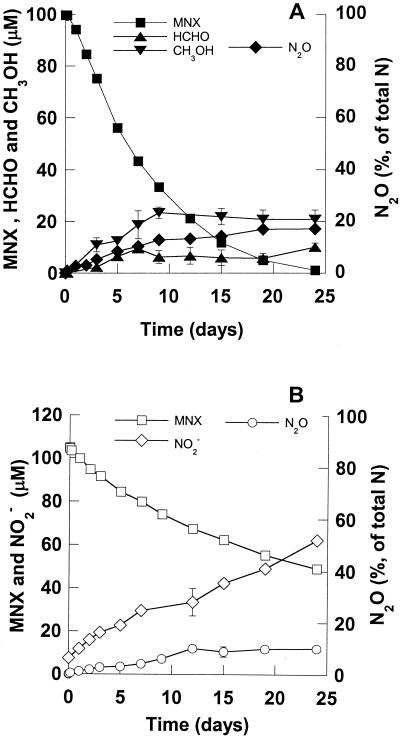
When we incubated MNX (100 μM) with live cells of strain SCZ-1, the compound was degraded at an initial rate of 0.39 μmol · h−1 · g (dry weight) of cells−1 (Fig. 3A), which was close to the rate of RDX degradation under the same conditions (0.41 μmol · h−1 · g [dry weight] of cells−1). The MNX transformation products were N2O, CH3OH, and HCHO (Fig. 3A) and thus were similar to those of RDX. However, removal of 2 μmol of MNX produced only 1.0 μmol of N2O (17% of the total N content of MNX) (Fig. 3A), compared to production of 3.8 μmol of N2O (60% of the total N content of RDX) from 2.1 μmol of RDX (Fig. 1A). Another notable observation is the detection of a small amount of DNX after MNX degradation; however, no TNX was detected. During MNX degradation we did not observe formation of O2NHNCH2NHNO2, the key intermediate during RDX degradation by strain SCZ-1.
FIG. 3.
Anaerobic degradation of MNX (2.0 μmol in 20 ml) with K. pneumoniae strain SCZ-1. (A) Live cells. (B) Killed cells. The biomass was the same as that described in the legend to Fig. 1.
In controls containing killed cells (Fig. 3B), MNX decomposed at a rate of 0.16 μmol · h−1 · g (dry weight) of cells−1, which was almost one-half its rate of removal in the presence of live cells (0.39 μmol · h−1 · g [dry weight] of cells−1), indicating that MNX was not a stable compound and underwent hydrolysis in water (Fig. 3B). Abiotic degradation of each 1 mol of MNX released approximately 1 mol of nitrite.
When we incubated the trinitroso derivative TNX (100 μM) under the same conditions, we found that the compound was converted much more slowly than either RDX or MNX. The rate of removal of TNX (0.07 μmol · h−1 · g [dry weight] of cells−1) was six times lower than that of MNX (0.39 μmol · h−1 · g [dry weight] of cells−1) or that of RDX (0.41 μmol · h−1 · g [dry weight] of cells−1). In controls with killed cells, TNX was not degraded. It has been reported frequently that TNX tends to accumulate during RDX biodegradation (20, 21, 23, 27, 31).
Pathway(s) of RDX degradation: initial denitration versus nitroso formation.
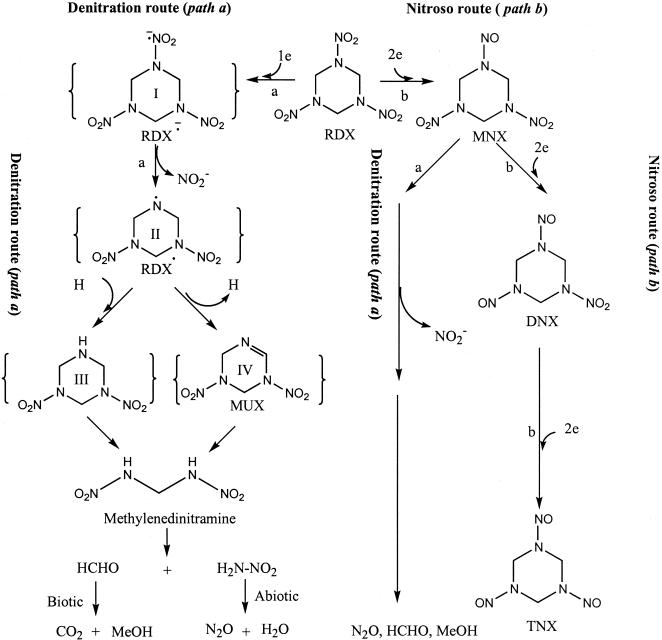
Based on the product distribution described above, we propose an RDX degradation pathway in K. pneumoniae strain SCZ-1 (Fig. 4). The RDX degradation pathway in this isolate is similar to the pathway(s) described previously for anaerobic sludge in that both involve methylenedinitramine as a ring cleavage intermediate (14, 16). Here we suggest that there is an important initial denitration step during RDX degradation that leads to ring cleavage and decomposition (Fig. 4, path a). The fact that oxygen completely quenched RDX removal by strain SCZ-1 supports the belief that an O2-sensitive reaction involving transfer of one electron to an NO2 group in RDX might initiate degradation. Such a process would produce an unstable radical anion, RDX·− (compound I), whose denitration could produce the unstable radical RDX (compound II). Subsequent reaction of compound II via abstraction would give the amine (compound III), or subsequent reaction of compound II via elimination of an H atom would give the monounsaturated intermediate (compound IV) (6, 17, 24). Methylenedinitramine is one of the expected products of decomposition of compound III or IV. The N denitration mechanism suggested in the present RDX study is similar to the mechanism reported previously for N denitration of nitramines by dihydronicotinamide (8). In the latter case, cleavage of an N-NO2 bond proceeds via a one-electron transfer process (8). Also, N denitration of RDX to compound IV has frequently been reported during alkaline hydrolysis (9, 17), in which the latter compound is found to decompose with a rate constant (k) that is 105 times larger than that of RDX (17).
FIG. 4.
Postulated routes for anaerobic degradation of RDX and MNX by K. pneumoniae strain SCZ-1. Compounds in brackets were not detected in this study. MeOH, methanol.
Sequential reduction of the nitro group(s) (—NO2) in RDX to the corresponding nitroso (—NO) derivative(s) did not seem to be a major degradation route for strain SCZ-1. If the sequential reduction of —NO2 to —NO is an important degradation pathway for RDX, then one might expect the accumulation of TNX (Fig. 4, path a), which we did not observe in the present study. As discussed above, TNX was degraded at a rate much lower than the rates of degradation of MNX and RDX with strain SCZ-1. In addition, we observed only traces of MNX during RDX degradation and traces of DNX during MNX degradation. Likewise, we suggest that MNX was also degraded mainly by initial denitration prior to ring cleavage (Fig. 4, path b). The absence of methylenedinitramine during MNX degradation supports the hypothesis that one of the two nitro functional groups (—NO2) in MNX might be removed prior to ring cleavage, leaving the compound unable to act as a precursor to methylenedinitramine.
In conclusion, we found that a facultatively anaerobic bacterium isolated from an anaerobic sludge mineralized RDX predominantly via initial denitration, whereas reduction of the nitro group of RDX to the corresponding nitroso products was a minor secondary reaction. Although the isolated strain improved our understanding of the pathways involved in the degradation of cyclic nitramines, use of this organism in remedial applications might not be practical because of a low rate of degradation. However, the high level of mineralization observed during RDX degradation should encourage us to optimize various physiological parameters to enhance the rate of degradation of RDX.
Acknowledgments
We thank Kevin Orouke of Sensient Flavor Canada for providing the anaerobic sludge. We are grateful to Sonia Thiboutot and Guy Ampleman of the Defense Research Establishment Valcartier, Quebec, Canada, for providing the energetic chemicals. We thank Jim Spain for valuable discussions, and we thank Carl Groom, Alain Corriveau, Stéphane Deschamps, and Philomena D'Cruz for technical assistance.
We thank the Natural Sciences and Engineering Research Council (NSERC) and the National Research Council (NRC) of Canada for granting a visiting fellowship to J.-S. Zhao. Funding was provided by the U.S. DoD/DoE/EPA Strategic Environmental Research and Development Program (grant SERDP CU1213).
REFERENCES
- 1.Adrian, N. R., and T. Chow. 2001. Identification of hydroxylaminodinitroso-1,3,5-triazine as a transient intermediate formed during the anaerobic biodegradation of RDX. Environ. Toxicol. Chem. 20:1874-1877. [PubMed] [Google Scholar]
- 2.Adrian, N. R., and A. Lowder. 1999. Biodegradation of RDX and HMX by a methanogenic enrichment culture, p. 1-6. In. B. C. Alleman and A. Leeson (ed.), Bioremediation of nitroaromatic and haloaromatic compounds, vol. 7. Battelle Press, Columbus, Ohio.
- 3.Ampleman, G., S. Thiboutot, J. Lavigne, A. Marois, J. Hawari, A. M. Jones, and D. Rho. 1995. Synthesis of 14C-labelled hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), 2,4,6-trinitrotoluene (TNT), nitrocellulose (NC) and glycidylazide polymer (GAP) for use in assessing the biodegradation potential of these energetic compounds. J. Labelled Compd. Radiopharm. 36:559-577. [Google Scholar]
- 4.Beller, H. R. 2002. Anaerobic biotransformation of RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) by aquifer bacteria using hydrogen as the sole electron donor. Water Res. 36:2533-2540. [DOI] [PubMed] [Google Scholar]
- 5.Boopathy, R. B., M. Gurgas, J. Ullian, and J. F. Manning. 1998. Metabolism of explosive compounds by sulfate-reducing bacteria. Curr. Microbiol. 37:127-131. [DOI] [PubMed] [Google Scholar]
- 6.Bose, P., W. H. Glaze, and S. Maddox. 1998. Degradation of RDX by various advanced oxidation processes. II. Organic byproducts. Water Res. 32:1005-1018. [Google Scholar]
- 7.Brockman, F. J., D. C. Downing, and G. F. Wright. 1949. Nitrolysis of hexamethylenetetramine. Can. J. Res. Sect. B 27:469-474. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, R. D., R. A. O'Brien, and P. A. Kondracki. 1996. N-denitration of nitramines by dihydronicotinamides. Tetrahedron 52:9655-9664. [Google Scholar]
- 9.Croce, M., and Y. Okamoto. 1979. Cationic micellar catalysis of the aqueous alkaline hydrolyses of 1,3,5-triaza-1,3,5-trinitrocyclohexane and 1,3,5,7-tetraza-1,3,5,7-tetranitrocyclooctane. J. Org. Chem. 44:2100-2103. [Google Scholar]
- 10.Environmental Protection Agency Environmental Monitoring and Support Laboratory Office of Research and Development. 1979. USA EPA method 345.1. Methods for chemical analysis of water and wastes. Environmental Protection Agency, Cincinnati, Ohio.
- 11.Fournier, D., A. Halasz, J. Spain, P. Fiurasek, and J. Hawari. 2002. Determination of key metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine with Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 68:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman, D. L., and K. W. Sutherland. 1998. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) under nitrate-reducing conditions. Water Sci. Technol. 38:33-40. [Google Scholar]
- 13.Haas, R., I. Schreiber, E. von Löw, and G. Stork. 1990. Conception for the investigation of contaminated munitions plants. 2. Investigation of former RDX-plants and filling stations. Fresenius' J. Anal. Chem. 338:41-45. [Google Scholar]
- 14.Halasz, A., J. Spain, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Insights into the formation and degradation mechanisms of methylenedinitramine during the incubation of RDX with anaerobic sludge. Environ. Sci. Technol. 36:633-638. [DOI] [PubMed] [Google Scholar]
- 15.Hawari, J. 2000. Biodegradation of RDX and HMX: from basic research to field application, p. 277-310. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. Lewis Publishers, Boca Raton, Fla.
- 16.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal sludge. Appl. Environ. Microbiol. 66:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffsommer, J. C., D. A. Kubose, and D. J. Glover. 1977. Kinetic isotope effects and intermediate formation for the aqueous alkaline homogeneous hydrolysis of 1,3,5-triaza-1,3,5-trinitrocyclohexane (RDX). J. Phys. Chem. 81:380-385. [Google Scholar]
- 18.Johnson, J. L. 1994. Similarity analysis of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murry, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 19.Kaplan, D. L. 1998. Biotransformation and bioremediation of munitions and explosives, p. 549-575. In S. K. Sikdar and R. L. Irvine (ed.), Bioremediation: principles and practice, vol. II. Biodegradation technology developments. Technomic Publishing Inc., Lancaster, Pa.
- 20.Kitts, C. L., D. P. Cunningham, and P. J. Unkefer. 1994. Isolation of three hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading species of the family Enterobacteriaceae from nitramine explosive-contaminated soil. Appl. Environ. Microbiol. 60:4608-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick, N. G., J. H. Cornell, and A. M. Kaplan. 1981. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine. Appl. Environ. Microbiol. 42:817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myler, C. A., and W. Sisk. 1991. Bioremediation of explosives contaminated soils: scientific questions/engineering realities, p. 137-146. In G. S. Sayler, R. Fox, and J. W. Blackburn (ed.), Environmental bio/technology for waste treatment. Plenum Press, New York, N.Y.
- 23.Oh, B.-T., C. L. Just, and P. J. J. Alvarez. 2001. Hexahydro-1,3,5-trinitro-1,3,5-triazine mineralization by zerovalent ion and mixed anaerobic cultures. Environ. Sci. Technol. 35:4341-4346. [DOI] [PubMed] [Google Scholar]
- 24.Peyton, G. R., M. H. LeFaivre, and S. W. Maloney. 1999. Verification of RDX photolysis mechanisms. Report 99/93. Environmental Processes Branch, Installations Division, Construction Engineering Research Laboratory, Champaign, Ill.
- 25.Regan, K. M., and R. L. Crawford. 1994. Characterization of Clostridium bifermentans and its biotransformation of 2,4,6-trinitrotoluene (TNT) and 1,3,5-triaza-1,3,5-trinitrocyclohexane (RDX). Biotechnol. Lett. 16:1081-1086. [Google Scholar]
- 26.Robidoux, P. Y., C. Svendsen, J. Caumartin, J. Hawari, G. Ampleman, S. Thiboutot, J. M. Weeks, and G. I. Sunahara. 2000. Chronic toxicity of energetic compounds in soil determined using the earthworm (Eisenia andrei) reproduction test. Environ. Toxicol. Chem. 19:1764-1773. [Google Scholar]
- 27.Sheremata, T. W., A. Halasz, L. Paquet, S. Thiboutot, G. Ampleman, and J. Hawari. 2001. The fate of the cyclic nitramine explosive RDX in natural soil. Environ. Sci. Technol. 35:1037-1040. [DOI] [PubMed] [Google Scholar]
- 28.Summers, W. R. 1990. Characterization of formaldehyde and formaldehyde releasing preservatives by combined reversed phase cation-exchange high performance liquid chromatography with postcolumn derivatization using Nash's reagent. Anal. Chem. 62:1397-1402. [Google Scholar]
- 29.Talmage, S. S., D. M. Opresko, C. J. Maxwell, C. J. E. Welsh, F. M. Cretella, P. H. Reno, and F. B. Daniel. 1999. Nitroaromatic munition compounds: environmental effects and screening values. Rev. Environ. Contam. Toxicol. 161:1-156. [DOI] [PubMed] [Google Scholar]
- 30.Weimer, P. J. 1984. Control of product formation during glucose fermentation by Bacillus macerans. J. Gen. Microbiol. 130:103-111. [Google Scholar]
- 31.Young, D. M., P. J. Unkefer, and K. L. Ogden. 1997. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a prospective consortium and its most effective isolate, Serratia marcescens. Biotechnol. Bioeng. 53:515-522. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, J. S., A. Singh, X. D. Huang, and O. P. Ward. 2000. Biotransformation of hydroxylaminobenzene and aminophenol by Pseudomonas putida 2NP8 cells grown in the presence of 3-nitrophenol. Appl. Environ. Microbiol. 66:2336-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]