Abstract
The woodchuck post-transcriptional regulatory element (WPRE) can naturally accumulate hepatitis transcripts in the cytoplasm, and has been recently exploited as an enhancer of transgene expression. The retention of mutant myotonic dystrophy protein kinase (DMPK) transcripts in the nucleus of myotonic dystrophy (DM) cells has an important pathogenic role in the disease, resulting in pleiotropic effects including delayed myoblast differentiation. In this study, we report the first use of WPRE as a tool to enhance nuclear export of an aberrantly retained messenger RNA. Stable cell lines expressing the normal and mutant DMPK 3′ UTR (3′ untranslated region) complementary DNA, with or without WPRE, were produced. It is noteworthy that WPRE stimulated extensive transport of mutant transcripts to the cytoplasm. This was associated with repair of the defective cellular MyoD levels and a subsequent increase in myoblast differentiation. These results provide the basis for a cellular model that can be exploited in DM and in the study of RNA transport mechanisms.
Keywords: myotonic dystrophy, nuclear retention, WPRE
Introduction
The woodchuck post-transcriptional regulatory element (WPRE) is required for the cytoplasmic accumulation of hepatitis virus RNAs (Donello et al, 1998) and has been used recently in numerous cases to enhance transgene expression (Ramezani et al, 2000; Moreau-Gaudry et al, 2001; Johansen et al, 2003). When placed downstream of the complementary DNA to be expressed, it can cause a post-transcriptional increase in transgene expression (Loeb et al, 1999; Zufferey et al, 1999; Glover et al, 2002; Popa et al, 2002). Such transport elements can also be beneficial for enhancing cytoplasmic transport of nuclear-retained RNA molecules. Myotonic dystrophy type 1 (DM1) is an autosomal dominant disorder characterized by a plethora of symptoms such as myotonia, progressive muscle weakness and facial changes. Clinical expression of the disorder is extremely variable, ranging from a severe congenital form that is often fatal just after birth to an asymptomatic condition (Harper, 1989). The DM mutation is a CTG trinucleotide repeat expansion (TRE) in the 3′ untranslated region (3′ UTR) of the myotonic dystrophy protein kinase (DMPK) gene (Davies et al, 1983; Brook et al, 1992; Buxton et al, 1992; Fu et al, 1992; Mahadevan et al, 1992; Shaw et al, 1993). The number of CTG repeats is in the range of 5–35 in the normal population and increases to between 50 and several thousand in DM1 patients. An important part of the molecular pathogenesis of the disease is the retention of mutant DMPK transcripts in the nucleus of affected cells and the ‘toxic' effects that take place there. A growing body of evidence suggests that pleiotropic effects of aberrant interactions between mutant DMPK transcripts and RNA-binding proteins alter the metabolism of ‘target' messenger RNAs. Members of the muscleblind family (MBNL, MBXL and MBLL), which usually regulate mRNA splicing (Kanadia et al, 2003; Ho et al, 2004), have been shown to colocalize with the ribonuclear inclusions (Fardaei et al, 2002). Another factor, the CUG repeat binding protein 1 (CUGBP1), has been implicated in mRNA splicing and translation defects in DM cells (Philips et al, 1998; Timchenko et al, 2001a). Significant advances have been made in identifying the targeted myogenic steps that are involved in DM (Amack & Mahadevan, 2004; Ranum & Day, 2004). Recently, it was discovered that DM patient myoblasts fail to permanently withdraw from the cell cycle when stimulated to differentiate and were unable to initiate events that mediate cell-cycle arrest (Timchenko et al, 2001b). Using the C2C12 myoblast model system, MyoD has been identified as a target of mutant DMPK 3 UTR RNA (Amack et al, 2002). The compromised levels of MyoD probably explain why C2C12 myoblasts expressing the mutant 3′ UTR RNA fail to initiate the differentiation programme.
In this report, we show that the addition of the WPRE sequence downstream of the DMPK 3′ UTR stimulates the transport of mutant transcripts to the cytoplasm and repairs myoblast differentiation.
Results and Discussion
WPRE enhances mutant DMPK 3′ UTR expression
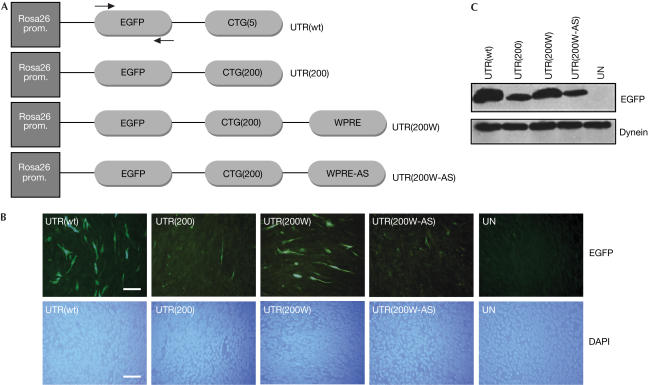
A series of constructs was created, containing the wild-type and mutant DMPK 3′ UTR cDNA with or without the WPRE sequence (Fig 1A). The first step was to transfect transiently the four constructs in C2C12 mouse myoblasts to evaluate the effect of WPRE on transgene expression, as assessed by enhanced green fluorescent protein (EGFP) fluorescence (Fig 1B). Although UTR(200) gave a low EGFP signal, owing to the retention of most of the transcripts in the nucleus, addition of the WPRE sequence downstream of the mutation (UTR(200W)) caused an increase in EGFP signal. The levels of EGFP expression of UTR(200W) approached those of UTR(wt) (Fig 1B,C). This is presumably due to the increased export of the transcripts to the cytoplasm. Cells transfected with the control UTR(200W-AS) showed no significant difference in the EGFP expression when compared with the mutant UTR(200), indicating that the antisense version of the WPRE sequence does not exert the same post-transcriptional effect on transgene expression as its sense counterpart (Fig 1B,C). PCR amplification from DNA extracts was carried out from all transfectants to ensure that an equal amount of plasmid was transfected in each case (data not shown). These results show that WPRE causes an increase of protein synthesis from transcripts that are known to be retained in the nucleus.
Figure 1.
Construction and transfection of plasmids in C2C12 myoblast cells. (A) Plasmids containing the wild type (UTR(wt)), mutant (UTR(200)), mutant with the WPRE sequence or its antisense version (UTR(200W) and UTR(200W-AS), respectively) were constructed. All plasmids coexpressed the EGFP gene under the Rosa26 promoter. Arrows indicate binding sites for PCR primers used in quantitative RT–PCR assays in Fig 3. (B) After transient transfections of the above constructs in C2C12 cells, low levels of EGFP were observed in cells transfected with the UTR(200) construct, whereas the addition of the WPRE sequence caused a significant increase in the EGFP expression (UTR(200W)). Normal EGFP levels were obtained with the UTR(wt) construct, whereas no EGFP was seen in untransfected cells (UN). The effect of WPRE was specific as no increase in EGFP levels was seen in cells transfected with its antisense version (UTR(200W-AS)). Nuclear staining by 4,6-diamidino-2-phenylindole (DAPI) shows the presence of cells. Scale bar, 0.1 mm. (C) Western blot analysis against EGFP showed results similar to (B), indicating that addition of the WPRE sequence increases transgene expression.
WPRE-mediated nuclear export of DM transcripts
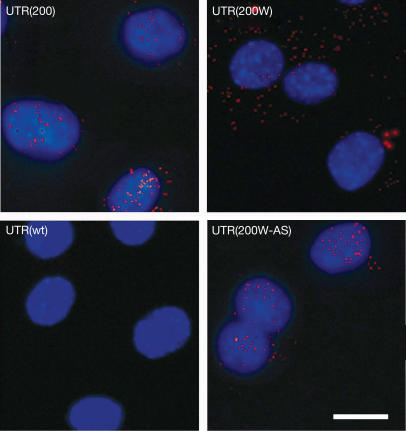
The mechanism by which WPRE exerts its effect was further investigated in stable clones. C2C12 stable cell lines, expressing the four different sequences described above (Fig 1A), were produced. RNA fluorescence in situ hybridization (FISH) was carried out to detect the cellular localization of mutant DMPK 3′ UTR transcripts in the presence or absence of WPRE (Fig 2). No RNA foci were detected in cells expressing the wild-type DMPK 3′ UTR sequence (UTR(wt)). As expected (Taneja et al, 1995; Amack & Mahadevan, 2001), most mutant DMPK 3′ UTR transcripts were retained in the nucleus of mouse myoblasts expressing this transcript (UTR(200); Fig 2). The presence of WPRE downstream of the mutant DMPK 3′ UTR (UTR(200W)) was associated mostly with cytoplasmic foci of mutant transcripts. These results indicate that WPRE acts as a post-transcriptional RNA transport enhancer, causing significant export of DMPK transcripts from the nucleus to the cytoplasm.
Figure 2.
WPRE-mediated release of mutant transcripts to the cytoplasm of myoblasts. Myoblasts expressing the mutant UTR(200) construct produced transcripts (shown as red foci), which were localized mainly in the nucleus after staining with DAPI. The presence of WPRE downstream of the mutant DMPK 3′ UTR (UTR(200W)) released most of the transcripts in the cytoplasm. No RNA foci were detected in cells expressing the wild-type UTR(wt) construct, whereas the presence of the antisense form of WPRE (UTR(200W-AS)) did not stimulate the export of the transcript to the cytoplasm. Scale bar, 0.25 mm.
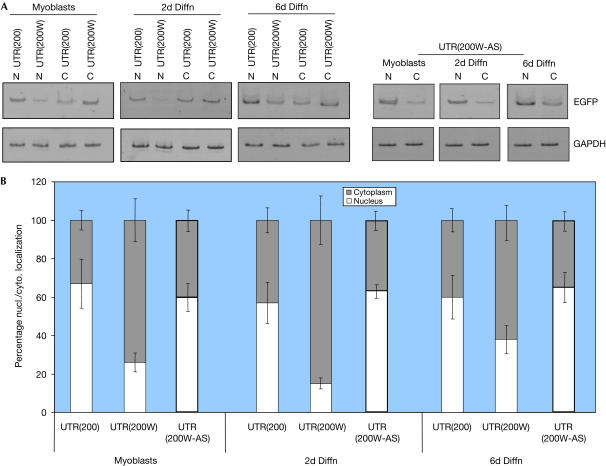
Further analysis was carried out by subcellular quantitative reverse transcription–PCR (RT–PCR) to quantify and characterize the transport of DM transcripts to the cytoplasm of myoblasts or differentiated cells (Fig 3). Mutant transcripts (UTR(200)) were predominantly localized in the nucleus of myoblasts and differentiated cells (Fig 3A). In contrast, WPRE-containing mutant transcripts were mostly present in the cytoplasm, confirming that the WPRE sequence causes a significant shift of mutant RNA molecules from the nucleus to the cytoplasm. For example, 2 days after the induction of differentiation, only 15% of the total levels of mutant transcripts remained in the nucleus (Fig 3B). The effect on RNA transport was specific for WPRE and probably not related to possible alterations of the 3′ UTR structure, as no increase of nuclear transport was observed when the antisense version of WPRE was included instead (Fig 3).
Figure 3.
Quantification of WPRE-mediated release of DMPK transcripts to the cytoplasm by RT–PCR. (A) Detection of nuclear (N) and cytoplasmic (C) mutant DMPK 3′ UTR transcripts (UTR(200)), in the presence of WPRE or its antisense version (UTR(200W) and UTR(200W-AS), respectively). RNA analysis was carried out from myoblasts or 2- and 6-day differentiated cells (2d Diffn and 6d Diffn, respectively) by amplifying the EGFP gene, which was connected to the mutant DMPK 3′ UTR. As a control, mouse GAPDH cDNA was also amplified. (B) Summary of mutant DMPK 3′ UTR RNA subcellular localization from myoblasts or differentiated cells. In all cases, WPRE (UTR(200W)) caused a significant increase of mutant DMPK 3′ UTR transcripts to the cytoplasm.
Defective muscle-cell differentiation is reversed by WPRE
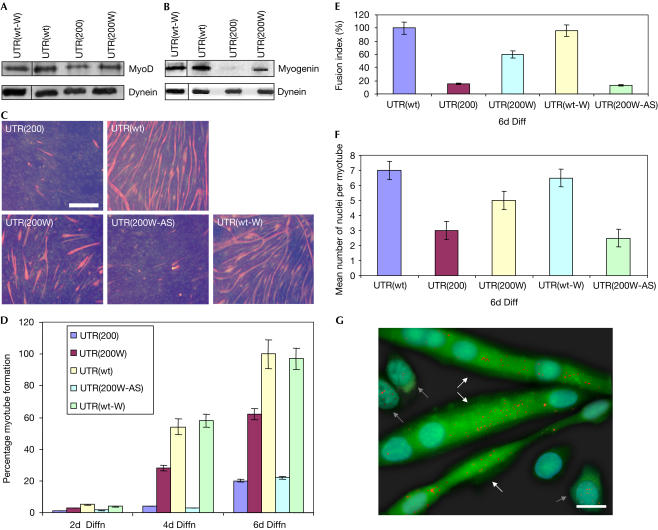
Previous evidence suggests that nuclear accumulation of mutant DMPK transcripts causes alterations associated with the cell cycle and cellular differentiation (Timchenko et al, 2001b; Amack et al, 2002). It is known that mutant 3′ UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD (Amack et al, 2002). Therefore, to determine the effect(s) on myoblasts of WPRE-mediated induction of mutant DMPK 3′ UTR transport to the cytoplasm, MyoD levels and myogenic differentiation were next investigated. Indeed, MyoD levels were reduced in the pool of cells expressing the mutant DMPK 3′ UTR (UTR(200W)), as compared with cells expressing the wild-type DMPK 3′ UTR (Fig 4A). However, MyoD levels in cells expressing the WPRE sequence, downstream of the mutant DMPK 3′ UTR (UTR(200W)), were twice those from UTR(200) and 70% of the average levels of the UTR(wt) pool. To investigate whether WPRE restored MyoD levels because of the cytoplasmic transport of mutant DMPK 3′ UTR and not because of other nonspecific effects, another C2C12 cell line—expressing the wild-type DMPK 3′ UTR and the WPRE sequence (UTR(wt-W))—was created. No alterations in MyoD levels were observed in UTR(wt-W) clones, as compared with the control UTR(wt), thereby indicating that WPRE repairs MyoD by transporting the mutant DMPK 3′ UTR transcripts to the cytoplasm (Fig 4A).
Figure 4.
Restoration of defective MyoD levels and muscle-cell differentiation. (A) Reduced MyoD levels were detected by western blotting in cells expressing the mutant DMPK 3′ UTR (UTR(200)), as compared with cells expressing the wild-type DMPK 3′ UTR (UTR(wt)) or the wild-type DMPK 3′ UTR and WPRE (UTR(wt-W)). WPRE-mediated transport of mutant DMPK 3′ UTR caused an increase in the cellular MyoD levels (UTR(200W)). (B) After 4 days of cell differentiation, myogenin levels were found to be increased, by western blotting, in cells expressing the UTR(200W) construct, as compared with cells expressing the UTR(200) construct. Normal myogenin was detected in cells that expressed UTR(wt) and UTR(wt-W). (C) Immunocytochemistry against myosin heavy chain (MHC) showed a marked reduction of myotube formation in cells expressing the mutant DMPK 3′ UTR (UTR(200)), as compared with cells expressing the wild-type DMPK 3′ UTR (UTR(wt)) or the wild-type DMPK 3′ UTR and WPRE (UTR(wt-W)), after 6 days of differentiation. Addition of WPRE downstream of the mutant DMPK 3′ UTR significantly increased myoblast differentiation (UTR(200W)) but not when it was present in its antisense form (UTR(200W-AS)). Scale bar, 0.5 mm. (D–F) Summary of MHC staining indicates that MHC levels are highly increased in differentiated UTR(200W) cells compared with UTR(200) cells, as detected by counting positively stained myotubes, the rate of fusion into myotubes and the number of nuclei per myotube. (G) RNA foci localization on differentiated cells expressing the WPRE sequence downstream of the mutant DMPK 3′ UTR, as detected by FISH, showed that transcripts/foci (shown as red dots) are mainly located in the cytoplasm in multinucleated myotubes (shown with white arrows), whereas undifferentiated myoblasts (shown with grey arrows) contain most of the RNA foci in the nucleus. Moreover, EGFP signal is significantly higher in differentiated myotubes than in undifferentiated myoblasts. Scale bar, 0.05 mm.
Next, the ability of cell pools to form myotubes was investigated by culturing the cells in differentiation medium and analysing them for expression of the myogenic factors myogenin and myosin heavy chain (MHC). As the differentiation response of C2C12 cells is known to be sensitive to cell confluency, each cell pool was tested in at least three independent experiments. Myogenin is known to be expressed early during normal muscle-cell differentiation and was almost undetectable in cells expressing the mutant UTR(200) (Fig 4B). The presence of WPRE caused a significant increase in myogenin levels 4 days after cell differentiation and approached 40% that of UTR(wt) or UTR(wt-W). Cells expressing UTR(200) showed little MHC (late marker of muscle-cell differentiation) immunostaining during differentiation, as compared with cells expressing the wild-type DMPK 3′ UTR (UTR(wt) and UTR(wt-W)), which showed strong formation of myotubes (Fig 4C). A high increase in the ability of myoblasts to be converted to multinucleated myotubes was shown in cells expressing the WPRE sequence downstream of the mutant DMPK 3′ UTR (UTR(200W)) (Fig 4D–F). Cell differentiation was, in some cases (for example, 6 days after differentiation), 60–70% of the wild-type levels, due to the presence of WPRE (Fig 4D,E). Finally, RNA localization experiments in these cells showed that differentiated myotubes contained cytoplasmic RNA foci, whereas undifferentiated myoblasts possessed nuclear-retained DMPK 3′ UTR transcripts (Fig 4G). This is consistent with the findings showing that most of the DMPK 3′ UTR transcripts were transported to the cytoplasm (Fig 3). As a result of the RNA cytoplasmic transport in myotubes, transgene (EGFP) expression was also higher than in myoblasts. This indicates that the transport of mutant DMPK 3′ UTR transcripts to the cytoplasm releases the cell from the inhibition of differentiation. These results show that WPRE can repair defective differentiation in cells expressing the DM mutation and show that the presence of mutant DMPK 3′ UTR mRNA in the cytoplasm of myoblasts does not result in inhibition of myogenic differentiation.
The retention of DMPK transcripts as ribonuclear inclusions in the nucleus of DM cells is considered to be an important pathogenic mechanism in DM. Any attempt to facilitate their export to the cytoplasm might provide insight about the importance of nuclear retention and RNA foci to the disease process and perhaps to the development of therapeutic approaches. Thus far, WPRE, which has been linked to the CRM1-dependent export pathway (Popa et al, 2002), has been used to enhance transgene expression when placed downstream of the cDNA to be expressed. Here, evidence is presented that shows the successful application of WPRE to enhance nuclear export of mutant DMPK 3′ UTR transcripts, accompanied by a repair of muscle-cell differentiation. This is the first demonstration of a means by which nuclear-retained transcripts can be liberated to the cytoplasm. It also provides a unique system for the study of RNA transport mechanisms in mammalian cells and the dynamics and kinetics of the RNA foci before and during differentiation, some of which are presented here. Moreover, the above-described system can be exploited to study the downstream mechanisms that lead to myoblast differentiation defects due to the expression of the mutant DMPK 3′ UTR.
In the future, the model described here may also prove useful to determine the capacity of WPRE as a trans-acting genetic tool against DM. Recently, group I intron ribozymes (Phylactou et al, 1998), hammerhead ribozymes (Langlois et al, 2003) and antisense RNA (Furling et al, 2003) have been used as potential gene therapy tools for DM. Further experiments will determine the ability of WPRE to act in trans on the DMPK transcripts. Post-transcriptional regulatory elements hold great promise in the field of genetic manipulation, and further research should exploit their value.
Methods
Preparation of constructs and transfections. UTR(wt) and UTR(200) constructs were prepared as described (Amack & Mahadevan, 2001). UTR(200W), UTR(200W-AS) and UTR(wt-W) were prepared by cloning the sense and antisense versions of WPRE sequence in the NruI site of the UTR(200) or UTR(wt) constructs. C2C12 cells (5 × 105) were transfected with 10 μg of each plasmid and 10 μl Lipofectamine 2000 reagent (Invitrogen, UK). After 48 h, transfections were either stopped and EGFP fluorescence captured under a NIKON TE 2000 fluorescence microscope, or transferred to cell-culture media containing 0.8 mg/ml G418 (Invitrogen, UK) for stable transfections as described by Amack & Mahadevan (2001). All clonal pools were composed of ten individual C2C12 clones. For differentiation studies, cells were grown to 80–90% confluence and then cultured in differentiation media containing Dulbecco's Modified Eagle Media (DMEM) plus 2% horse serum (Invitrogen, UK) for up to 6 days.
RNA fluorescence in situ hybridization. Myoblasts grown on coverslips or differentiated myotubes grown on chamber slides (covered in collagen) were fixed in 4% paraformaldehyde for 15 min at 37°C. A CY3-conjugated (CAG)10 oligonucleotide probe (Operon, Germany) was used for RNA FISH experiments and performed as described previously (Amack & Mahadevan, 2001). CY3 and 4,6-diamidino-2-phenylindole (DAPI) fluorescent signals were visualized using a NIKON Eclipse TE2000 microscope with epifluorescence. Images were captured with a NIKON digital camera and then assembled using Adobe Photoshop software.
cDNA synthesis and PCR amplification. Nuclear and cytoplasmic RNA was isolated (PARIS kit, Ambion, USA). A 500 ng portion of cytoplasmic or nuclear-extracted RNA was subjected to reverse transcription and PCR amplification was carried out to amplify EGFP (Fwd: 5′-GTGAGCAAGGGCGAGGAGC-3′; Rev: 5′-TTACTTGTACAGCTCGTCCA-3′) and the housekeeping gene GAPDH (Fwd: 5′-TCATCATCTCCGCCCCTTCC-3′; Rev: 5′-GAGGGGCCATCCACAGTCTT-3′). Gel bands were quantified using the Scion software, and the levels of EGFP were normalized with those of GAPDH.
Immunocytochemistry. After fixation, cells were incubated with a monoclonal MY32 antibody against MHC (Sigma, Germany) and a Texas-red-conjugated anti-mouse secondary antibody (Jackson Laboratories, USA) and nuclei stained with DAPI (Vysis, USA). Images were captured with a NIKON digital camera and then assembled using Adobe Photoshop software. Myotubes were counted at least three times from each pool of clones in ten different cellular areas.
Western blotting. Protein extracts (50 μg) were incubated with myogenin (Santa Cruz, Germany), dynein (Santa Cruz), MyoD (Pharmingen, USA) and EGFP (Santa Cruz) primary antibodies followed by secondary antibodies conjugated to horseradish peroxidase (Santa Cruz).
Acknowledgments
We thank E. Kyriacou for technical assistance. This work was supported by the A.G. Leventis Foundation and the Cyprus Research Promotion Foundation (grants to L.A.P.).
References
- Amack JD, Mahadevan MS (2001) The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum Mol Genet 10: 1879–1887 [DOI] [PubMed] [Google Scholar]
- Amack JD, Mahadevan MS (2004) Myogenic defects in myotonic dystrophy. Dev Biol 265: 294–301 [DOI] [PubMed] [Google Scholar]
- Amack JD, Reagan SR, Mahadevan MS (2002) Mutant DMPK 3′-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD. J Cell Biol 159: 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook JD et al. (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68: 799–808 [DOI] [PubMed] [Google Scholar]
- Buxton J et al. (1992) Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature 355: 547–548 [DOI] [PubMed] [Google Scholar]
- Davies KE, Jackson J, Williamson R, Harper PS, Ball S, Sarfarazi M, Meredith L, Fey G (1983) Linkage analysis of myotonic dystrophy and sequences on chromosome 19 using a cloned complement 3 gene probe. J Med Genet 20: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donello JE, Loeb JE, Hope TJ (1998) Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol 72: 5085–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD (2002) Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 11: 805–814 [DOI] [PubMed] [Google Scholar]
- Fu YH et al. (1992) An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Furling D, Doucet G, Langlois MA, Timchenko L, Belanger E, Cossette L, Puymirat J (2003) Viral vector producing antisense RNA restores myotonic dystrophy myoblast functions. Gene Ther 10: 795–802 [DOI] [PubMed] [Google Scholar]
- Glover CP, Bienemann AS, Heywood DJ, Cosgrave AS, Uney JB (2002) Adenoviral-mediated, high-level, cellspecific transgene expression: a SYN1-WPRE cassette mediates increased transgene expression with no loss of neuron specificity. Mol Ther 5: 509–516 [DOI] [PubMed] [Google Scholar]
- Harper PS (1989) Myotonic Dystrophy. Philadelphia, USA: WB Saunders [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA (2004) Muscleblind proteins regulate alternative splicing. EMBO J 23: 3103–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Tornoe J, Moller A, Johansen TE (2003) Increased in vitro and in vivo transgene expression levels mediated through cis-acting elements. J Gene Med 5: 1080–1089 [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS (2003) A muscleblind knockout model for myotonic dystrophy. Science 302: 1978–1980 [DOI] [PubMed] [Google Scholar]
- Langlois MA, Lee NS, Rossi JJ, Puymirat J (2003) Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol Ther 7: 670–680 [DOI] [PubMed] [Google Scholar]
- Loeb JE, Cordier WS, Harris ME, Weitzman MD, Hope TJ (1999) Enhanced expression of transgenes from adeno-associated virus vectors with the woodchuck hepatitis virus posttranscriptional regulatory element: implications for gene therapy. Hum Gene Ther 10: 2295–2305 [DOI] [PubMed] [Google Scholar]
- Mahadevan M et al. (1992) Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 255: 1253–1255 [DOI] [PubMed] [Google Scholar]
- Moreau-Gaudry F, Xia P, Jiang G, Perelman NP, Bauer G, Ellis J, Surinya KH, Mavilio F, Shen CK, Malik P (2001) High-level erythroidspecific gene expression in primary human and murine hematopoietic cells with self-inactivating lentiviral vectors. Blood 98: 2664–2672 [DOI] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280: 737–741 [DOI] [PubMed] [Google Scholar]
- Phylactou LA, Darrah C, Wood MJA (1998) Ribozyme-mediated transsplicing of a trinucleotide repeat. Nat Genet 18: 378–381 [DOI] [PubMed] [Google Scholar]
- Popa I, Harris ME, Donello JE, Hope TJ (2002) CRM1-dependent function of a cis-acting RNA export element. Mol Cell Biol 22: 2057–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS, Hawley RG (2000) Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther 2: 458–469 [DOI] [PubMed] [Google Scholar]
- Ranum LP, Day JW (2004) Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Shaw DJ et al. (1993) Genomic organization and transcriptional units at the myotonic dystrophy locus. Genomics 18: 673–679 [DOI] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH (1995) Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol 128: 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT (2001a) RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem 276: 7820–7826 [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT (2001b) Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol 21: 6927–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Donello JE, Trono D, Hope TJ (1999) Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol 73: 2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]