Abstract
It is well documented that single eukaryotic genes can give rise to proteins that are localized to several subcellular locations. This is achieved at the level of transcription, splicing and translation, and results in two or more translation products that either harbour or lack specific targeting signals. Nevertheless, the possibility of dual targeting of a single translation product has recently emerged. Here, we review cases of such dual targeting with emphasis on the mechanisms through which these phenomena occur. Proteins that harbour one signal, two separate signals or an overlapping ambiguous signal may follow dual distribution in the cell. The mechanism of dual targeting is driven by the competition or promiscuity of various molecular events. Protein folding, post-translational modification and protein–protein interaction are key players in this phenomenon.
Keywords: subcellular distribution, dual targeting, membrane translocation, isoenzymes, signal peptides
Introduction
Subcellular compartments and organelles contain specific proteins that determine their structure and catalytic capacity. In certain cases, identical proteins can be found in more than one compartment, which gives rise to isoprotein distribution. In this review, we discuss dual targeting of proteins in which at least one of the isoproteins undergoes transmembrane translocation into a membrane-sealed compartment such as mitochondria, chloroplasts, the endoplasmic reticulum (ER) or peroxisomes. The nucleus is not included among these membrane-sealed compartments because nucleocytoplasmic transport takes place through the huge nuclear pore complexes that do not inflict the same constraints on protein movement (into or out of the organelle) as does transmembrane translocation (Alberts et al, 2002).
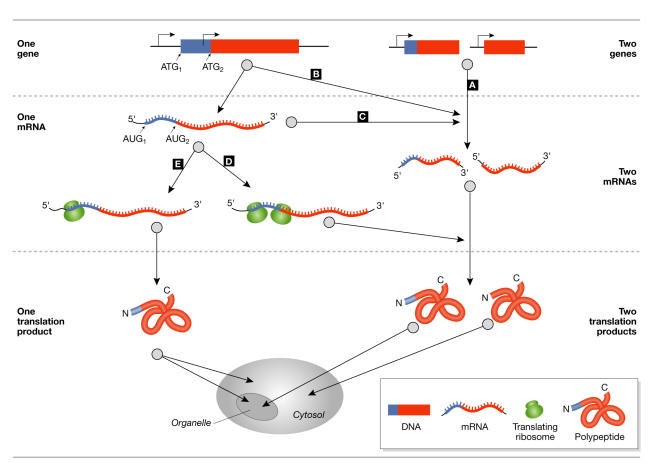
Dual distribution between subcellular compartments can be achieved by one of several routes that are based on two genes, two mRNAs from a single gene, or two translation initiations on a single mRNA (Fig 1A-D). In all of these situations, the end result is the creation of two translation products that differ by the presence or absence of a targeting signal (for reviews, see Danpure 1995; Small et al, 1998). More recently, the intriguing possibility of dual localization of membrane-targeted single-translation products (Fig 1E) has emerged. Here, we review cases of such dual targeting and, more importantly, emphasize the mechanisms by which these phenomena occur.
Figure 1.
Mechanisms by which proteins with identical activities are distributed in the eukaryotic cell. (A) mRNA molecules transcribed from two genes, of which only one carries an organelle targeting sequence (TS, shown in green). (B) Two mRNAs are transcribed from one gene, but only one encodes a TS. (C) A non-spliced mRNA and the spliced mRNA (TS lost) encode products in the organelle and in the rest of the cell, respectively. (D) A single message is translated from alternative initiation codons to produce TS-containing and TS-lacking products. (E) A single translation product containing the TS is nevertheless distributed between the organelle and the rest of the cell by mechanisms described in this review.
How can a single protein reach many locations?
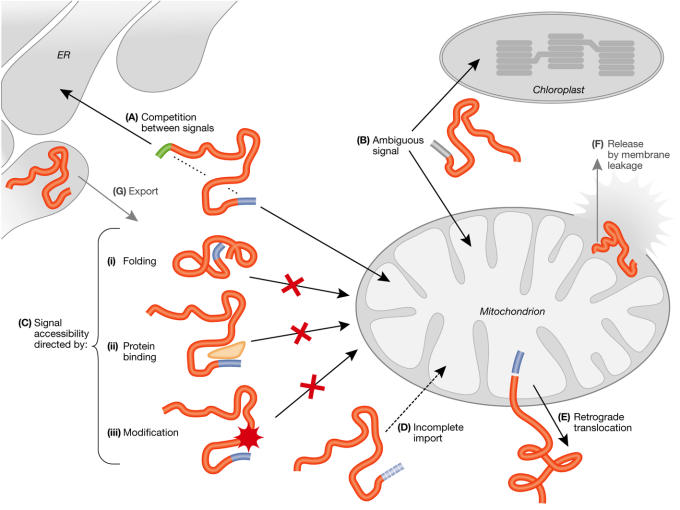
When dealing with dual or multiple distribution of proteins, a crucial issue is the identification of amino-acid sequences in these proteins that define specific topogenic information. Proteins with two separate targeting signals, a single ambiguous signal that is recognized by two receptors or a single signal can be distributed between separate locations in the cell. In the latter case, the cytosol may be considered the default compartment for proteins that did not succumb to their initial programmed destination. Sorting of a protein to a membrane-sealed organelle involves two main steps: targeting to the organelle and interaction with its surface receptors; and translocation through the organellar membranes by specific import machineries. Therefore, dual distribution of a single translation product must reflect competition and/or promiscuity in one or both of these sorting steps. In this section, we provide an overview of the 'tricks' or mechanisms that cells use to achieve dual localization (Fig 2).
Figure 2.
'Tricks' used for dual distribution: proteins can undergo dual distribution in the eukaryotic cell by using a combination of various processes described in detail in the text. (A) Competition between two signals on the same polypeptide. (B) An ambiguous signal that is recognized by two organelles. (C) Inaccessibility of a signal by (i) folding, (ii) protein binding, and (iii) modification of the distributed polypeptide. (D) Incomplete import and (E) retrograde translocation. Possible mechanisms based on either release by membrane leakage (F), or export of proteins from organelles (G) are indicated in grey.
Several proteins harbour two targeting signals, which leads to distribution among mitochondria, peroxisomes, the ER or one of these organelles and the nucleus. When two signals are present on the same polypeptide, the question is how the relative contribution of each signal is achieved. When both signals are accessible, distribution may be dictated by the relative affinities to their receptors (Fig 2A). Competition over target receptors also occurs in proteins that are destined for both mitochondria and chloroplasts. These proteins use a single targeting presequence that is termed 'ambiguous' as it is recognized by both import systems (Fig 2B).
Differential distribution may be achieved if, before import, the targeting signal in a subpopulation of the protein becomes inaccessible to its destination receptor (Fig 2C). The accessibility of a targeting signal may be controlled by folding, hindrance from other proteins, or post-translational modification (Fig 2C(i),(ii) and (iii), respectively). This may lead either to retention of the precursor in the cytosol, or to targeting to a different organelle through a second targeting signal. Proteins that are efficiently targeted to an organelle can nevertheless be distributed as a result of incomplete translocation of a fraction of the targeted molecules across the organellar membranes (Fig 2D). Alternatively, proteins can be retrieved from the organelle to the cytosol by retrograde movement through the translocon (Fig 2E). Finally, fully translocated proteins may be redistributed as a result of leakage out of an organelle that has lost its membrane integrity (Fig 2F), or by active export from the organelle (Fig 2G).
In the following sections, we review dual subcellular distribution through the routes outlined above. Most of these cases naturally include a combination rather than a single means for achieving dual targeting. Each of the sections was designed to include examples that most clearly demonstrate a specific theme, but also may include some 'tricks' that are relevant to other sections.
Competition between two targeting signals
Catalase A in yeast has two types of signal: (i) a mitochondrial targeting sequence (MTS) located at, or in the vicinity of, the amino terminus, and (ii) a peroxisomal-like targeting signal (PTS1) located at the carboxyl terminus (Petrova et al, 2004). Distribution of catalase A between mitochondria and peroxisomes is affected greatly by cell physiology and by the nature of the carbon source in the growth medium. Cells grown on oleate are enriched in peroxisomes, which contain most of the cellular catalase A, whereas targeting to mitochondria prevails when cells are grown on raffinose (Petrova et al, 2004). Possibly, enrichment of the target organelle competes for cellular catalase A and shifts the distribution between mitochondria and peroxisomes; however, unravelling the mechanism of distribution of catalase A awaits completion.
Several members of the cytochrome P450 family (CYP) are targeted to both the ER and mitochondrial compartments (Addya et al, 1997; Anandatheerthavarada et al, 1999; Robin et al, 2001). Dual targeting of CYP apoproteins to the ER and to mitochondria is modulated by an amino-terminal bipartite signal, which includes an ER-targeting sequence followed by a cryptic mitochondrial targeting sequence. Activation of the cryptic signal is mediated through post-translational modification. Here, the case of CYP2B1 is presented as a prototype for competition between two signals (Anandatheerthavarada et al, 1999). This elegant study examined the affinity of in vitro-translated CYP2B1 towards the targeting and translocation machineries of the ER and the mitochondria. Interaction of CYP2B1 with signal recognition particle (SRP) was examined by two approaches: an SRP-mediated inhibition of in vitro translation and direct chemical crosslinking. The interaction of a CYP2B1-dihydrofolate reductase (DHFR) hybrid with the mitochondrial translocase components Tom40, Tim44 and mtHsp70 was also determined by chemical crosslinking. The results show that under-phosphorylated CYP2B1 protein binds to SRP more efficiently than fully phosphorylated CYP2B1 and also preferentially associates with the ER membrane in a transmembrane orientation. Concomitantly, the same protein (and also a phosphorylation-defective mutant) is imported poorly into mitochondria, probably due to its inefficient interaction with mitochondrial translocase proteins. Phosphorylated CYP2B1, however, shows reduced affinity for SRP and higher affinity to mitochondrial translocase proteins (Anandatheerthavarada et al, 1999). The model implies that in CYP2B1, the relative affinities of the targeting signals to the ER versus mitochondria are regulated through cellular cAMP levels and phosphorylation of an internal site in the protein. It is also worth mentioning that the bipartite signal of the Alzheimer amyloid precursor protein includes an ER signal peptide followed by a cryptic MTS. The mechanism that governs this dual distribution remains to be elucidated (Anandatheerthavarada et al, 2003).
Competition between two organelles for one signal
Most chloroplast and mitochondrial proteins are imported from the cytosol through distinct routes that ensure specificity in targeting. However, several proteins are targeted to both organelles because they have an ambiguous signal that can be recognized by both import systems (Silva-Filho, 2003). The contribution of the receptors and translocation systems of the organelles to this sort of dual distribution is essentially unknown. However, several studies have analysed the structure and function of such ambiguous targeting signals. Comparison of sequences of known ambiguous signals shows an overall similarity to mitochondria- and chloroplastspecific signals. However, ambiguous signals contain additional leucine and phenylalanine residues, which result in an overall increase in hydrophobicity compared with either mitochondrial-specific or chloroplast-specific signals (Peeters & Small, 2001). The relative significance of particular amino acids in the ambiguous signal of pea glutathione reductase (GR) was examined by mutation analysis. The mutants were studied in vitro using import assays and in vivo by tracing the location of GRsignal-GFP (green fluorescent protein) fusions (Chew et al, 2003). The results suggest that hydrophobic residues are important for targeting both mitochondria and chloroplasts. Single positively charged residues seem to be more important for mitochondrial targeting, but when a basic residue was co-mutated with a hydrophobic one, the mutations also had a significant effect on import into chloroplasts (Chew et al, 2003). Thus, dual targeting by ambiguous signals appears to be achieved by combining distinct targeting instructions in a single N-terminal peptide. Nevertheless, as several amino-acid residues are crucial for targeting to both organelles, this is not a simple superimposition of two separate signals.
Accessibility of a signal governs targeting
The accessibility of a targeting sequence may be governed by several molecular events. Precursor folding might conceal the signal in the protein, or interaction with another protein might directly mask the signal or indirectly cause conformational changes in the precursor. Post-translational modification of the precursor might directly affect its targeting properties (that is, its affinity to a target), or affect them indirectly through protein folding or binding, as mentioned above.
Folding buries the targeting signal. The main adenylate kinase in yeast, Aky2, has a dual location, with the bulk of the protein residing in the cytoplasm and a minor fraction in the mitochondrial intermembrane space (IMS). Aky2 lacks a cleavable presequence, but its N-terminal 18-amino-acid stretch is able, although inefficiently, to drive a heterologous passenger protein into the IMS. This whole region has a greater preference for β-strand than α-helix conformations (Egner et al, 1987; Bandlow et al, 1998). Accordingly, targeting and mitochondrial localization of Aky2 can be improved markedly by exchanging the N-terminal peptide for efficient targeting sequences taken from Aky3 or cytochrome c1 (Bandlow et al, 1998; Strobel et al, 2002). Folding of Aky2 is spontaneous and rapid. After denaturation and renaturation, the protein acquires enzymatic activity within a few minutes and, once folded, is thermally and proteolytically stable. In a cell-free organellar import system, post-translational uptake of Aky2 is negligible, but can be improved by previous denaturation with urea or guanidinium isothiocyanate (Angermayr et al, 2001; Strobel et al, 2002). Bandlow and colleagues suggest that partitioning of Aky2 between the cytoplasm and mitochondria is derived from competition between rapid protein folding and inefficient mitochondrial targeting (Bandlow et al, 1998). Their model suggests that folding hinders the accessibility of the targeting signal to the receptor. It is also possible that the signal remains accessible to the import apparatus, but that the folded protein cannot be translocated. In both scenarios, the nascent Aky2 has two options: either it is synthesized to completion and folds into an enzymatically active import-incompetent conformation that remains in the cytosol, or, during synthesis and before formation of a significant tertiary structure, it reaches a mitochondrial surface receptor and is internalized.
Protein–protein interactions mask the targeting signal. Apurinic/apyrimidinic endonuclease 1 (Apn1) is a DNA-repair enzyme, which is a key enzyme in the base excision repair pathway in the nucleus and mitochondria. The C terminus of Apn1 has a nuclear localization signal (NLS), whereas its N terminus harbours a putative MTS (Vongsamphanh et al, 2001). Using a two-hybrid screen, Pir1 was shown to interact with the C terminus of Apn1. Deletion of Pir1 resulted in elevated nuclear targeting of Apn1 and a simultaneous reduction in mitochondrial targeting. This suggests that when Pir1 binds, it masks the NLS of Apn1, thus leading to mitochondrial targeting of the protein. However, Pir1 may also have an active role in mitochondrial targeting of Apn1: deletion of the C terminus of Apn1 (which is both an NLS and a binding site for Pir1) blocks not only nuclear import, but also transport to the mitochondria, which results in its cytosolic location. Thus, the predicted N-terminal MTS of Apn1 is not sufficient for mitochondrial targeting in the absence of Pir1 (Vongsamphanh et al, 2001).
Modification controls interaction with receptors. As mentioned above, the distribution of CYP2B1 between the ER and the mitochondria is modulated by the level of cAMP in the cell. Phosphorylation at an internal protein kinase A (PKA) signature sequence RRFSL of CYP2B1 results in preferential localization of the protein to mitochondria with a concomitant reduction in ER targeting (Anandatheerthavarada et al, 1999). How phosphorylation of an internal site activates a mitochondrial signal at the distant N terminus is not fully understood. Phosphorylation of CYP2B1 induces association with the cytosolic chaperone Hsp70, which is essential for mitochondrial import (Robin et al, 2002). The interaction with this chaperone leads to a conformational change in CYP2B1 as indicated by a shift in its circular dichroism (CD) spectra (Robin et al, 2001). This conformational change may activate the cryptic mitochondria targeting signal in CYP2B1.
Although the distribution of other proteins is governed by their phosphorylation, some of these show variations on the theme presented by CYP2B1 above. The glutathione S-transferase protein GSTA4-4 is distributed between mitochondria and the cytosol. This protein is modulated by not one but two kinases—PKA and protein kinase C (PKC)—which mediate efficient import into mitochondria (Robin et al, 2003). For the yeast nucleotide diphosphate kinase 1 (Ynk1) protein, phosphorylation inhibits rather than enhances mitochondrial import and, accordingly, decreases the affinity of Ynk1 for Tom40 (Amutha & Pain, 2003).
In CYP1A1, which also has an N-terminal bipartite signal, the cryptic mitochondrial signal is activated by a conformational change that is driven by a different post-translational modification; endoproteolytic cleavage by a cytosolic protease leads to removal of the ER-targeting signal (Addya et al, 1997). Recently, Colombo et al (2005) demonstrated that the distribution of mammalian NADH-cytochrome b5 reductase between the ER and the mitochondrial outer membrane is determined by N-myristoylation. This modification of the N terminus perturbs the interaction of the protein with SRP and in turn promotes mitochondrial targeting.
The distribution of superoxide dismutase (SOD1) between the cytosol and IMS has been proposed to involve a combination of protein modification, protein folding and protein–protein interaction. Binding of the copper chaperone to SOD1 (CCS) induces disulphide bond formation and a conformational change in the latter, which may prevent its translocation across the outer membrane into the IMS and prevent leakage of the mature SOD1 out of the IMS (Field et al, 2003).
Inefficient or aborted translocation
The Saccharomyces cerevisiae MCR1 gene encodes two mitochondrial isoforms of NADH-cytochrome b5 reductase: a 34-kDa isoform that is localized to the outer membrane of mitochondria, and a 32-kDa isoform that reaches the IMS. The small isoform results from cleavage of the larger isoform by the inner membrane protease. Large isoforms that accumulate in the presence of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) cannot be converted into the mature form on re-establishment of membrane potential. Thus, once fully arrested in the outer membrane, Mcr1 is impeded from further import (Hahne et al, 1994). The first 12 residues of Mcr1 function as a classical MTS, which is followed by a hydrophobic stop-transfer signal. A leaky stop-transfer model was proposed in which all translation products of Mcr1 are targeted to the mitochondrial outer membrane, but only one-third of the molecules move from the outer membrane translocon and integrate in the outer membrane. The rest of the molecules are further translocated through the import pathway, interact with the TIM complex, and are cleaved by the inner membrane protease (IMP) to produce the soluble mature form in the IMS (Hahne et al, 1994). Recently, a similar leaky stop-transfer model across the mitochondrial inner membrane was proposed to explain the alternative topogenesis of the yeast dynamin-like GTPase Mgm1 in mitochondria (Herlan et al, 2004).
Folding drives retrograde translocation
In S. cerevisiae, a single translation product of the FUM1 gene is distributed between the cytosol and the mitochondria (Stein et al, 1994). Fumarase precursors harbour a 24-amino-acid-long N-terminal presequence, which is removed by the mitochondrial matrix peptidase (MPP) on import into mitochondria. Mitochondrial and cytosolic isoenzymes of fumarase share identical N-termini that correspond to the mature processed form (Sass et al, 2001). In vivo, when processing is inhibited either by blocking import with the uncoupler CCCP or by inactivating MPP, all fumarase polypeptides accumulate as precursors (Stein et al, 1994). When fumarase is translated in vitro in the presence of mitochondria, correctly processed molecules are found not only within, but also outside, mitochondria (Knox et al, 1998). Thus, before distribution, all fumarase polypeptides are directed to mitochondria and processed by MPP. A subset of the processed fumarase molecules is then fully imported into the matrix, whereas the majority is released back into the cytosol. Folding of fumarase appears to be the driving force for its retrograde movement into the cytosol. Rapid folding impedes fumarase import into mitochondria post-translationally, both in vivo and in vitro (Stein et al, 1994; Knox et al, 1998). Mutations throughout the coding sequence of fumarase that alter its conformation do not impair targeting to mitochondria, but cause a loss of retrograde movement and distribution (Sass et al, 2003). Overexpression of a cytosolic Hsp70 (Ssa1) causes a shift of fumarase distribution to mitochondria, whereas partially impairing mitochondrial Hsp70 (Ssc1 conditional mutation) diminishes full import and allows more fumarase molecules to reside in the cytosol. Finally, mass spectrometry analysis revealed that mature cytosolic and mitochondrial fumarase molecules are identical, and contain no post-translational modifications. These results support the idea that folding per se is the driving force for the distribution of fumarase (Sass et al, 2003).
Perspectives and concluding remarks
There is ample knowledge with regard to the ways in which protein subcellular distribution is determined at the level of multiple genes, transcription initiation, splicing and initiation of translation (Fig 1). Conversely, the distribution of single translation products is less understood and documented, and it is only recently that information about specific cases has accumulated. In particular, there has been progress in our understanding of the mechanisms by which subcellular distribution of single translation products occurs (Fig 2). One underlying principle is that dynamic competition between molecular events determines the distribution of certain proteins between different compartments: target organelles can compete for separate or overlapping targeting signals; targeting signals may become inaccessible as a result of competitive folding, precursor modification or interaction with other proteins; and translocation may be incomplete or even reversed due to processes that compete with or block translocation.
Despite some advancement in the understanding of dual targeting of single gene products, many questions still remain; for example: Are there additional general regulators apart from cAMP that may control protein distribution under varying physiological conditions? Should dual targeting between mitochondria and chloroplasts be attributed solely to the nature of ambiguous signals or are there organelle-specific mediators that participate in distribution? What are the precise molecular events that lead to retrograde translocation out of mitochondria? The majority of cases of multiple distribution that are described in the literature result from multiple genes or multiple transcripts from a single gene. Cases of multiple translation initiation, splicing and distribution of single translation products are less abundant, with only about 50 known cases of the latter. It is interesting to speculate on why dual targeting of single versus multiple translation products developed in the course of evolution. Obviously, it is possible that this is an alternative and simple solution for placing identical activities in different compartments. Then again, such distribution mechanisms may provide unique regulation opportunities at a post-translational level, as in the case of cAMP and CYP2B1 phosphorylation. In this regard, dual targeting of proteins in eukaryotic cells is probably a much more abundant phenomenon than realized at present. Newly emerging examples include yeast Nsf1 (Nakai et al, 2001) and aconitase (N. Rudzki et al, in preparation). These proteins are mainly sorted to mitochondria, but sustain a minute, barely detectable, extra-mitochondrial population that is required for a specific cellular function. Nfs1 has an essential role in thio-modification of tRNAs in the nucleus (Muhlenhoff et al, 2004) and aconitase participates in the glyoxylate cycle in the cytosol (N. Rudzki et al, in preparation). Thus, our understanding of the function of eukaryotic cells may depend on our ability to identify even small amounts of isoproteins in different compartments and to analyse the mechanisms by which they are targeted and distributed.
In theory, one can also envisage additional multiple distribution mechanisms that are based on known processes in the cell such as apoptosis (programmed cell death), ER-associated degradation and autophagy, which might affect the subcellular location of proteins (for example, Fig 2F,G). At present, these processes are considered to be involved solely in protein degradation or cell death. However, components of these processes may well have a role in fully functional and growing cells, and provide an alternative (or additional) route for simultaneously establishing the activity of a protein in two locations.


Acknowledgments
We apologize to our colleagues whose work could not be cited, or was cited indirectly, due to space restrictions. We thank E. Bibi, D. Rapaport and N. Regev-Rudzki for critical reading of the manuscript. O.P. is supported by the Israel Science Foundation (ISF) and the German Israeli Project Cooperation (DIP) of the Bundesministerium für Bildung und Forschung.
References
- Addya S, Anandatheerthavarada HK, Biswas G, Bhagwat SV, Mullick J, Avadhani NG (1997) Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J Cell Biol 139: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson AJ, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular Biology of the Cell. New York, NY, USA: Garland Science [Google Scholar]
- Amutha B, Pain D (2003) Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochem J 370: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Biswas G, Mullick J, Sepuri NB, Otvos L, Pain D, Avadhani NG (1999) Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at ser128. EMBO J 18: 5494–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG (2003) Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol 161: 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr M, Strobel G, Zollner A, Korber D, Bandlow W (2001) Two parameters improve efficiency of mitochondrial uptake of adenylate kinase: decreased folding velocity and increased propensity of N-terminal alpha-helix formation. FEBS Lett 508: 427–432 [DOI] [PubMed] [Google Scholar]
- Bandlow W, Strobel G, Schricker R (1998) Influence of N-terminal sequence variation on the sorting of major adenylate kinase to the mitochondrial intermembrane space in yeast. Biochem J 329: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O, Rudhe C, Glaser E, Whelan J (2003) Characterization of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol Biol 53: 341–356 [DOI] [PubMed] [Google Scholar]
- Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, Borgese N (2005) N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J Cell Biol 168: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danpure CJ (1995) How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol 5: 230–238 [DOI] [PubMed] [Google Scholar]
- Egner U, Tomasselli AG, Schulz GE (1987) Structure of the complex of yeast adenylate kinase with the inhibitor P1,P5-di(adenosine-5′-) pentaphosphate at 2.6 Å resolution. J Mol Biol 195: 649–658 [DOI] [PubMed] [Google Scholar]
- Field LS, Furukawa Y, O'Halloran TV, Culotta VC (2003) Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J Biol Chem 278: 28052–28059 [DOI] [PubMed] [Google Scholar]
- Hahne K, Haucke V, Ramage L, Schatz G (1994) Incomplete arrest in the outer membrane sorts NADH-cytochrome b5 reductase to two different submitochondrial compartments. Cell 79: 829–839 [DOI] [PubMed] [Google Scholar]
- Herlan M, Bornhovd C, Hell K, Neupert W, Reichert AS (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol 165: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, Sass E, Neupert W, Pines O (1998) Import into mitochondria, folding and retrograde movement of fumarase in yeast. J Biol Chem 273: 25587–25593 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Balk J, Richhardt N, Kaiser JT, Sipos K, Kispal G, Lill R (2004) Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J Biol Chem 279: 36906–36915 [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Hayashi H, Kagamiyama H (2001) Nuclear localization of yeast Nfs1p is required for cell survival. J Biol Chem 276: 8314–8320 [DOI] [PubMed] [Google Scholar]
- Peeters N, Small I (2001) Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta 1541: 54–63 [DOI] [PubMed] [Google Scholar]
- Petrova VY, Drescher D, Kujumdzieva AV, Schmitt MJ (2004) Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem J 380: 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin MA, Anandatheerthavarada HK, Fang JK, Cudic M, Otvos L, Avadhani NG (2001) Mitochondrial targeted cytochrome P450 2E1 (P450 MT5) contains an intact N terminus and requires mitochondrial specific electron transfer proteins for activity. J Biol Chem 276: 24680–24689 [DOI] [PubMed] [Google Scholar]
- Robin MA, Anandatheerthavarada HK, Biswas G, Sepuri NB, Gordon DM, Pain D, Avadhani NG (2002) Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J Biol Chem 277: 40583–40593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin MA, Prabu SK, Raza H, Anandatheerthavarada HK, Avadhani NG (2003) Phosphorylation enhances mitochondrial targeting of GSTA4-4 through increased affinity for binding to cytoplasmic Hsp70. J Biol Chem 278: 18960–18970 [DOI] [PubMed] [Google Scholar]
- Sass E, Blachinsky E, Karniely S, Pines O (2001) Mitochondrial and cytosolic isoforms of yeast fumarase are derivatives of a single translation product and have identical amino termini. J Biol Chem 276: 46111–46117 [DOI] [PubMed] [Google Scholar]
- Sass E, Karniely S, Pines O (2003) Folding of fumarase during mitochondrial import determines its dual targeting in yeast. J Biol Chem 278: 45109–45116 [DOI] [PubMed] [Google Scholar]
- Silva-Filho MC (2003) One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr Opin Plant Biol 6: 589–595 [DOI] [PubMed] [Google Scholar]
- Small I, Wintz H, Akashi K, Mireau H (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol Biol 38: 265–277 [PubMed] [Google Scholar]
- Stein I, Peleg Y, Even-Ram S, Pines O (1994) The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol Cell Biol 14: 4770–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G, Zollner A, Angermayr M, Bandlow W (2002) Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol Biol Cell 13: 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongsamphanh R, Fortier PK, Ramotar D (2001) Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol Cell Biol 21: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]