Abstract
Pfiesteria piscicida is a heterotrophic dinoflagellate widely distributed along the middle Atlantic shore of the United States and associated with fish kills in the Neuse River (North Carolina) and the Chesapeake Bay (Maryland and Virginia). We constructed a genomic DNA library from clonally cultured P. piscicida and characterized the nontranscribed spacer (NTS), small subunit, internal transcribed spacer 1 (ITS1), 5.8S region, ITS2, and large subunit of the rRNA gene cluster. Based on the P. piscicida ribosomal DNA sequence, we developed a PCR-based detection assay that targets the NTS. The assay specificity was assessed by testing clonal P. piscicida and Pfiesteria shumwayae, 35 additional dinoflagellate species, and algal prey (Rhodomonas sp.). Only P. piscicida and nine presumptive P. piscicida isolates tested positive. All PCR-positive products yielded identical sequences for P. piscicida, suggesting that the PCR-based assay is species specific. The assay can detect a single P. piscicida zoospore in 1 ml of water, 10 resting cysts in 1 g of sediment, or 10 fg of P. piscicida DNA in 1 μg of heterologous DNA. An internal standard for the PCR assay was constructed to identify potential false-negative results in testing of environmental sediment and water samples and as a competitor for the development of a quantitative competitive PCR assay format. The specificities of both qualitative and quantitative PCR assay formats were validated with >200 environmental samples, and the assays provide simple, rapid, and accurate methods for the assessment of P. piscicida in water and sediments.
Pfiesteria piscicida is a heterotrophic dinoflagellate associated with skin lesions and massive kills in several estuarine and marine fish species and with detrimental effects on human health (7, 8, 10, 20, 62). In the summer of 1997, fish kills were recorded in the Chesapeake Bay tributaries and attributed to P. piscicida blooms (29, 36). Concurrent human health problems, such as skin lesions, respiratory problems, and neurological complications primarily involving short-term memory loss, were attributed to associated toxic effects (5, 21, 27, 32, 33, 57). Because very little information is available on the environmental factors that may trigger Pfiesteria blooms, they have been difficult—if not impossible—to anticipate. One approach that may significantly contribute to the understanding and prediction of Pfiesteria blooms is to examine the distribution of P. piscicida in the environment and to analyze this information in the context of bloom events (44). Until recently, the environmental detection of P. piscicida was based on in vitro propagation, morphological identification using scanning electron microscopy (61, 63), and confirmation of toxicity in a fish mortality bioassay (9, 10). Unfortunately, these identification methods are lengthy and laborious, and in most cases the interpretation of the results is subjective. More recently, and based on approaches implemented for other harmful algal bloom species (34, 47, 58), molecular detection methods have been developed for P. piscicida and related species (6, 45, 53, 67).
PCR assays based on genes or intergenic regions of the rRNA locus have been very useful for the environmental detection of both prokaryotic and eukaryotic microorganisms. Because the rRNA genes are tandemly repeated in high copy numbers (41, 64) and the small subunit (SSU) is highly conserved (38, 39), this region has been the target region of choice for the development of molecular assays. The relatively high conservation of SSU regions within the Dinozoa has enabled the design of phylum-selective PCR primers and their use in a heteroduplex mobility assay (45). The same region has been targeted for the development of quantitative assay formats based on real-time PCR (6). In addition to SSU sequences (26, 58), other regions of the rRNA cluster, such as 5.8S and the large subunit (LSU), have been selected as targets for PCR amplification. This has been based mostly on the level of variability of each region within a particular species of interest and requirements of assay specificity and sensitivity (13, 34, 47). For example, the high conservation of the SSU among closely related species, subspecies, and strains may represent a drawback for the accurate intraspecific identification of environmental isolates. Therefore, alternative detection assays that target intergenic regions such as the nontranscribed spacer (NTS), a region known in other Alveolata to have a higher intra- and interspecies variability than the SSU (4, 12), have been successfully developed. These include PCR assays for species- and strain-specific detection of parasites of the genus Perkinsus (15, 18, 35, 49, 50). Until now, the sequence of the rRNA gene cluster of P. piscicida had not been completed, and this has hindered a comprehensive analysis and rational selection of the most suitable target sequence for assay development. Furthermore, although a quantitative assay for P. piscicida (6) has been developed, the costly probes, reagents, and equipment required may not render this method accessible to most laboratories. The development of quantitative assays, such as the quantitative competitive PCR (QC-PCR), that only require the construction of an internal standard for the competitive amplification of the experimental DNA mixture provides inexpensive yet useful and reliable alternatives (43, 59).
Although PCR-based assays are sensitive and specific, the presence of interfering substances in the sample may yield false-negative results. In the case of soil or sediment samples, the ubiquitous presence of DNA polymerase inhibitor(s), such as humic acid, has been well documented as the cause of false-negative results in PCR assays (40, 42). This problem may not be as critical for the detection of P. piscicida in the water column, in which the dinoflagellate is present as flagellated forms from spring to fall. However, during the winter months, and depending on environmental conditions, P. piscicida may also be present in sediment beds as a resting nonmotile form (2, 55, 65). Therefore, in order to implement a “robust” environmental assessment strategy for the detection of P. piscicida, it is essential to develop a system that allows the accurate detection of the dinoflagellate in both the water column and sediments and minimizes the possibilities of false-negative results. Here, we report the nucleotide sequence of the P. piscicida rRNA gene cluster, including the NTS and the LSU, and the use of the NTS for the development of a reliable assay system. We developed and validated a species-specific PCR-based detection assay for P. piscicida targeting the newly characterized NTS sequence, constructed an internal standard for the implementation of this assay for the detection P. piscicida in sediment samples, and used this internal-standard plasmid to develop a QC-PCR assay for the assessment of P. piscicida in environmental samples.
MATERIALS AND METHODS
P. piscicida and other dinoflagellate isolates and cultures.
P. piscicida, used in this study and kindly provided by K. A. Steidinger (Florida Department of Environmental Protection, St. Petersburg, Fla.), was isolated in 1997 by Steidinger and J. M. Burkholder (Center for Applied Aquatic Ecology, North Carolina State University) from the Chicamacomico River, Maryland. Other cultures were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP; West Boothbay Harbor, Maine) (CCMP1830, CCMP1831, CCMP1834, CCMP1902, CCMP1921, and CCMP1928 [all presumptive P. piscicida], 24 previously unidentified Pfiesteria-like isolates [CCMP1832, CCMP1833, CCMP1835, CCMP1936, CCMP1838 to -1845, CCMP1872 to -1882, and CCMP1929], and CCMP768 [Rhodomonas sp.]). Pfiesteria shumwayae and Cryptoperidiniopsis brodii were gifts from R. W. Litaker (University of North Carolina, Chapel Hill). Karlodinium micrum and Prorocentrum minimum were provided by D. K. Stoecker (Center of Environmental Studies, University of Maryland, Cambridge) and D. W. Coats (Smithsonian Environmental Research Center, Edgewater, Md.), respectively. Isolates COMBH4, COMBH10 (Salisbury, Md.), COMBN1R2, and COMBN2F1 (Neuse River, N.C.) were collected and cultured at the Center of Marine Biotechnology (COMB) University of Maryland Biotechnology Institute, Baltimore. COMB4872, a presumptive Oxyrrhis sp., was isolated from a water sample (collection site unknown) provided by the Maryland Department of Natural Resources (DNR).
All dinoflagellate zoospore cultures used in this study were maintained in enriched f/2 medium (24) at a salinity of 15 ppt with Instant Ocean synthetic sea salt (Aquarium Systems Inc., Mentor, Ohio) under a standard light cycle (14 h light-10 h dark; white fluorescent light; 150 mE m−2 s−1 [milli-radiant flux per area]) at 23°C in a biosafety level 2 laboratory (COMB Dinoflagellate Culture Core Facility). Rhodomonas sp. was used as the algal prey for heterotrophic species (23, 25). P. piscicida cysts were obtained by incubating a clonal P. piscicida culture (5 × 104 cells ml−1; total, 20 ml of f/2 medium; salinity, 15 ppt) at 4°C in the dark without algal prey until no zoospores were observed in the water column (ca. >3 weeks). Flasks with encysted P. piscicida organisms were washed three times with f/2 medium (salinity, 7 ppt) to remove debris. The cysts were released from the flask surface by harsh shaking with artificial seawater (20 ml; salinity, 7 ppt) (three times), pelleted by centrifugation (15 min at 1,000 × g), and resuspended in artificial seawater (1 ml). The zoospores and cysts were counted in a hemocytometer under a microscope (magnification, ×100).
Environmental water and sediment samples.
Water (25) and sediment (43) samples were collected by COMB in Maryland and North Carolina from 1999 to 2001. Additional water samples (199) were provided by the Maryland DNR in 1999 and 2000 as part of the “Pfiesteria and fish health” field monitoring program (http://www.dnr.state.md.us/pfiesteria/).
P. piscicida genomic-library construction.
For DNA extraction, a P. piscicida culture (500 ml) was grown to 1 × 105 to 5 × 105 cells ml−1 and depleted of Rhodomonas sp. by maintaining it for 48 h without further addition of algal prey, and the zoospores were collected by centrifugation at 1,500 × g for 15 min. The cells were resuspended in TE-sodium dodecyl sulfate buffer (300 μl; 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 2% [wt/vol] sodium dodecyl sulfate) and transferred to a 10-ml glass tube. Tris-buffered phenol (300 μl) and an equal volume of acid-washed glass beads were added, and the tube was vortexed at full speed for 3 min at room temperature. The supernatant was collected, the beads were washed with 300 μl of TE (50 mM Tris-HCl [pH 7.4], 100 μM EDTA), the supernatant was recovered and pooled with the previous wash, and the total supernatant was transferred to a 1.5-ml Eppendorf tube and centrifuged at 11,000 × g for 5 min. The aqueous phase was transferred to a new tube, and the phenol phase was reextracted with 300 μl of TE and combined with the previous. After 20 min on ice, the pooled aqueous phases were mixed with an equal volume of isopropanol, and the precipitated DNA was separated by centrifugation (11,000 × g for 15 min) at 4°C. The pellet was washed once with 70% ethanol and centrifuged, the supernatant was discarded, and the pellet was air dried for 30 min at room temperature. The pellet was resuspended in 200 to 300 μl of TE, 1 μl of 10-μg μl−1 RNase A was added, the mixture was incubated at 37°C for 15 min, and the DNA was stored at −20°C.
P. piscicida genomic DNA (100 μg) was partially digested with 133 U of Sau3A (New England Biolabs Inc., Beverly, Mass.) at room temperature in a 6.6-ml reaction mixture for 9 min. After transfer of the mixture to 37°C, 900-μl aliquots were collected at 1-min intervals, pipetted into 600 μl of isopropanol, incubated on ice for 10 min, and centrifuged at 11,000 × g for 15 min at 4°C. The pellets were washed with 500 μl of 70% ethanol, centrifuged, air dried, and resuspended in 20 μl of TE. The DNA digests were pooled and separated on a 1% agarose gel and stained with ethidium bromide (EtBr). A DNA fraction of 8 to 25 kb was excised from the gel and extracted with 20 μl of QiaExpress II gel extraction resin (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. The DNA digest was eluted to a final volume of 40 μl of 10 mM Tris-HCl (pH 8.0) and ligated with BamHI/EcoRI-digested lambda DASH phage vector (Stratagene, La Jolla, Calif.) following the manufacturer's instructions. A total of 106 plaques were collected, and the library was amplified in four separate pools, each containing 2.5 × 105 independent clones; titrated; and stored in 7% dimethyl sulfoxide at −70°C.
Screening of P. piscicida genomic library and subcloning and sequencing of lambda clone 6132.
The DNA probe for screening the lambda DASH genomic library for clones containing the rRNA NTS region was generated from a P. piscicida Vectorette genomic library (Vectorette System, Sigma, Tex.) (51). Using a primer (PP2R) (Table 1) based on the P. piscicida SSU of ribosomal DNA (rDNA) (GenBank accession no. AF077055), we obtained 50 bp of NTS sequence upstream from the SSU rDNA gene. Subsequently, based on this partial NTS and the contiguous SSU sequence, we designed a forward primer, NTS/SSU1F, and a reverse primer, NTS/SSU2R, that amplify a 429-bp fragment containing the 50 bp of the P. piscicida NTS and a stretch on the 5′ end of the SSU (51). This fragment was used as the probe for screening the P. piscicida lambda DASH genomic library described above. Prior to screening with this DNA probe, 4 μl of each of the P. piscicida genomic library pools (containing at least 106 PFU) was tested in a PCR with the primers NTS/SSU1F and NTS/SSU2R. All four pools gave an amplicon of 429 bp, which indicated that all of them contained at least one clone of the NTS-SSU DNA. The pool with the highest titer was selected for further screening. Approximately 30,000 PFU were plated in Luria-Bertani soft agarose in 150-mm-diameter plates, and plaque lifts were carried out using nylon membranes (Hybond-XL; Amersham Pharmacia Biotech, Piscataway, N.J.). The screening was performed as reported elsewhere (17) using 25 ng of the purified PCR product labeled with Redivue 5′-[α-32P]dCTP (rediprime II; Amersham Pharmacia Biotech). Positive clones were rescreened up to four times using the same protocol.
TABLE 1.
Synthetic oligonucleotides for P. piscicida
| Name | Tma | Sequence (5′ to 3′) | Application |
|---|---|---|---|
| PP2R | 56 | AAACATCCTTGGCAAATGCTTTCGC | Vectorette gene walker |
| NTS/SSU1F | 54 | TTCGGCGATTTCGTGCTTCG | PCR of NTS and SSU rDNA (forward) |
| NTS/SSU2R | 57 | TTCTCCGTTACCCGTCATTGCC | PCR of NTS and SSU rDNA (reverse) |
| PP12 | 55 | CGCAGGTCCTAAGTTACGTTAAG | PCR of NTS rDNA (forward) |
| PP5 | 55 | ACAAGCATATGACTACTGGCAGG | PCR of NTS rDNA (reverse) |
| PP19 | 53 | GTCACAGCAAGGAGCTTCC | Sequencing of NTS rDNA |
| PP20 | 58 | TGCATGCTGCGGTCTGCACC | Sequencing of NTS rDNA |
| NTS2F | 54 | CGCCGCCAAAAATGACAAGG | PCR for P. piscicida NTS (forward) |
| NTS3R | 50 | CAGACGGGTTTACACACC | PCR for P. piscicida NTS (reverse) |
| NTS4R | 55 | GCAAAAGCGAAGCACGAAATCG | PCR for P. piscicida NTS (reverse) |
Tm, melting temperature.
DNA was isolated from each of four positive lambda clones by liquid lysis (3). DNA from clone 6132 was digested with NotI, and a 6-kb fragment of the insert was subcloned into the NotI-digested and dephosphorylated pGEM-T Easy vector (Promega, Madison, Wis.), yielding the plasmid pPPG3-1. Similarly, lambda 6132 was digested with EcoRI and ligated with the EcoRI-digested and dephosphorylated plasmid pGEM3z to yield the plasmid pPPG4-3. pPPG3-1 was initially sequenced with primers T7 and SP6. New primers were designed in the insert sequences and used for further sequencing. This was repeated until both strands of the pPPG3-1 insert were fully sequenced. Because the pPPG4-3 insert overlaps with the pPPG3-1 insert, the primers designed on the NTS portion of the pPPG3-1 insert were used to obtain the initial sequence of the pPPG4-3 insert. Two additional primers were designed on the new pPPG4-3 sequence until a repetitive sequence was reached.
DNA extraction for PCR assay on cultures and environmental water and sediment samples.
Dinoflagellate cultures (1 to 10 ml) and environmental water (50 ml) and sediment (a 10-ml volume of sediment suspended in 50 ml of water) samples were centrifuged at 2,000 × g for 15 min at room temperature. The pellets were resuspended in 200 μl of TE, subjected to freeze (on dry ice)-thaw (at room temperature) cycles, incubated at 100°C for 10 min, and centrifuged at 11,000 × g for 10 min at room temperature. The DNA-containing supernatants were stored at −20°C, and 10 μl was routinely used in 20 μl of PCR mixture. DNAs from P. shumwayae and C. brodii were kindly provided by R. W. Litaker. Alternatively, DNA was extracted from sediment samples with the FastDNA spin kit for soil (Q · BIOgene, Inc., Carlsbad, Calif.) following the manufacturer's instructions.
Intraspecific variability in the P. piscicida NTS.
Based on the P. piscicida rDNA sequence described above, we designed PCR primers targeting those regions (NTS, internal transcribed spacer 1 [ITS1], and ITS2) considered to be the most variable within the rRNA gene cluster (4, 12). The NTS regions of various isolates of P. piscicida were amplified in a PCR with primers PP12 and PP5. A 650-bp amplicon was separated by agarose gel electrophoresis, and DNA was extracted from the gel slice with a QIAquick Gel Extraction kit (Qiagen) following the manufacturer's instructions. The purified amplicon (150 ng) was directly sequenced in an ABI 373 automatic sequencer (Applied Biosystems, Foster City, Calif.), using the PP5 and PP12 primers and additional sequencing primers (PP19 and PP20). The amplicon (523 bp) of NTS2 PCR was also subcloned and sequenced. Contigs were assembled with Sequencher 3.1 software (Gene Codes Co., Ann Arbor, Mich.).
Development and optimization of a PCR-based detection assay for P. piscicida: specificity and sensitivity.
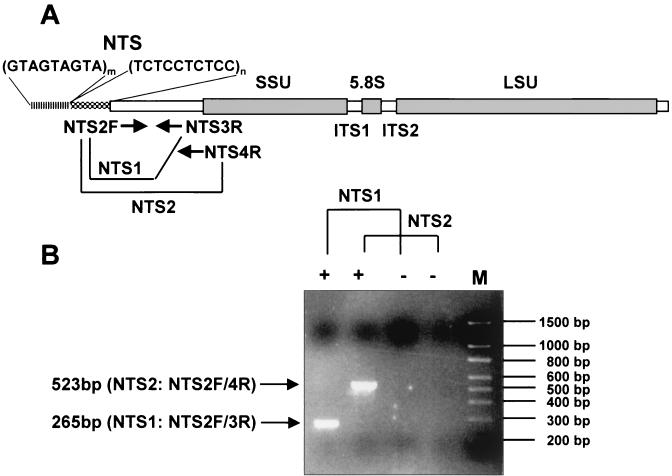
Oligonucleotide primers for PCR amplification were designed on the NTS region using the GeneJockey II program (Biosoft, Cambridge, United Kingdom). One forward primer, NTS2F, and two reverse primers, NTS3R and NTS4R, were used for the development of the PCR-based detection assay for P. piscicida (Fig. 1A).
FIG. 1.
Organization of P. piscicida rDNA and locations of primers for P. piscicida diagnostic PCR (A) and amplification of target DNA (B). One forward primer (NTS2F) and two reverse primers (NTS3R and NTS4R) amplify 265- (NTS2F-NTS3R) and 523-bp (NTS2F-NTS4R) amplicons localized on the NTS of rDNA. +, positive control; −, negative control; M, 100-bp ladder molecular size markers. The arrows indicate target amplicons separated by electrophoresis at 100 V for 1 h on 1.5% agarose gels stained by EtBr. Ten microliters of a 20-μl PCR mixture was applied per well.
Two PCR primer combinations, NTS2F-NTS3R (NTS1 PCR) and NTS2F-NTS4R (NTS2 PCR), amplify 265- and 523-bp amplicons, respectively. The PCR mixtures included a 0.3 mM concentration of each forward and reverse primer in a 20-μl reaction mixture; KCl PCR buffer (ENZYPOL, London, Ontario, Canada); 1.5 mM MgCl2; 0.25 mM tetramethyl-ammonium chloride; 150 μM (each) dATP, dTTP, dCTP, and dGTP; and 0.5 U of Taq DNA polymerase (ENZYPOL). The reaction conditions for NTS1 PCR were denaturation at 94°C for 4 min followed by 15 cycles of 94°C for 1 min, 60°C for 40 s, and 72°C for 40 s, and then 15 more cycles of 94°C for 1 min, 55°C for 40 s, and 72°C for 40 s with a final extension at 72°C for 5 min. The PCR protocol for NTS2 PCR was denaturation at 94°C for 4 min followed by 5 cycles of 94°C for 1 min, 68°C for 20 s, and 72°C for 40 s and 10 cycles of 94°C for 1 min, 65°C for 20 s, and 72°C for 40 s, followed by 15 additional cycles of 94°C for 1 min, 60°C for 20 s, and 72°C for 40 s with a final extension at 72°C for 5 min in a PTC-200 Peltier Thermal Cycler (MJ Research, Watertown, Mass.).
The specificity of the PCR assay was assessed by amplifying DNAs (100 ng each) of P. piscicida, P. shumwayae, C. brodii, CCMP768 (Rhodomonas sp.), K. micrum, CCMP isolates (1830, 1833, 1834, 1840, 1845, 1872, 1880, 1921, and 1929), and COMB4872 with the NTS2F-NTS3R and NTS2F-NTS4R primer sets. The integrity of the DNA templates was confirmed by PCR amplification with “universal” actin primers designed to amplify actin genes from lower eukaryotes to vertebrates (amplicons, ≅730 bp; primers, G-480 and G-482 [kindly provided by G. W. Warr, Medical University of South Carolina, Charleston]). P. piscicida DNA (100 pg) or target plasmid (106 copies, as described below) and distilled water were routinely used as positive and negative controls, respectively.
For assessment of the sensitivity of the PCR assay, 0.0000001 to 1 ng of P. piscicida DNA was spiked into 1 μg of the DNA of Rhodomonas sp. (CCMP768), K. micrum, or COMB4872, and the mixture was subjected to the P. piscicida PCR detection assay. The PCR products were resolved by electrophoresis in a 1.5% (wt/vol) agarose gel in the presence of EtBr (final concentration, 0.8 μg ml−1) in TAE (40 mM Tris-acetate [pH 7.4], 1 mM EDTA). The bands of interest were recovered from agarose gels with a QIAquick Gel Extraction kit for direct sequencing. To assess the detection limit of the PCR assay for P. piscicida cells, clonal cultures were diluted with f/2 medium to 103 cells ml−1 on a 24-well plate. Individual cells were isolated with a calibrated microcapillary with a stainless steel plunger (5 μl; Drummond Scientific Co., Broomall, Pa.), the cell numbers in the capillary tube were confirmed by light microscopy, and the cells were transferred into the PCR tubes. All samples were prepared in triplicate.
The potential inhibitory effect(s) of EDTA (a component of the TE DNA extraction buffer) on the PCR amplification from water and sediment samples was examined using the NTS2 PCR assay in the presence or absence of EDTA. Decreasing (≤50 μM) or constant (50 μM) concentrations of EDTA in the PCR mixture were tested by serially (twofold) diluting P. piscicida DNA (1 ng in sterile water or TE) in sterile water. Alternatively, P. piscicida DNA (1 ng) was serially diluted (twofold) with TE. To examine the effect(s) of EDTA on DNA extraction and PCR amplification, an extract of autoclaved sediment (0.1 g) was prepared in 200 μl of sterile water or TE (100 μM EDTA) by the freeze-thaw-boil procedure. The sediment extract was spiked with P. piscicida DNA (1 ng) and serially diluted (twofold) in sterile water for PCR.
For spike-recovery experiments, 10, 102, 103, or 104 P. piscicida zoospores or cysts were spiked into 0.1 g of autoclaved sediment. For DNA extraction from the spiked sediments, the FastDNA spin kit for soil (Q 158 BIOgene, Inc.) was used following the manufacturer's instructions with three times extra washing on the DNA binding matrix before DNA elution with 100 μl of sterile water. DNA extracts corresponding to 0.1, 1, 10, and 100 P. piscicida cells were amplified in the presence of 103 copies of pMNTS2 as an internal standard (described below).
Construction of an internal standard for the PCR-based detection assay and the QC-PCR assay format.
The PCR product (523 bp) resulting from NTS2 PCR amplification was cloned into pGEM-T vector (Promega) to obtain pTNTS2 for use as a positive control. A second plasmid, pMNTS2, containing P. piscicida-specific PCR priming site sequences for the NTS2F-NTS4R primer set, was constructed using the PCR MIMIC construction kit (Clontech Laboratories Inc., Palo Alto, Calif.). This plasmid was used as an internal standard for the PCR-based detection assay and as a competitor in the development of a QC-PCR and yields a 418-bp amplicon. The sensitivity of the PCR-based assay was assessed with a 10-fold serial dilution (each) of plasmids pTNTS2 and pMNTS2 from 108 copies in 20 μl of PCR mixture. To examine the amplification kinetics of both the target and competitor plasmids, pTNTS2 and pMNTS2 were amplified in a 20-μl PCR mixture by NTS2 PCR using the NTS2F and NTS4R primers. For use as an internal standard, the sample DNA was spiked with 2 × 104 copies of pMNTS2 in a 20-μl reaction mixture. For validation of competitive quantification, titration of pMNTS2 as the competitor and a constant amount of either pTNTS2 (105 copies), P. piscicida DNA (500 pg), or DNA extract from 770 cells of P. piscicida were coamplified. The PCR products were separated and visualized by electrophoresis on 1.5% agarose gels stained with EtBr. The gels were scanned (FluorImager 575; Molecular Dynamics, Sunnyvale, Calif.), and band intensity was measured with NIH Image version 1.62 software (Research Services Branch, National Institute of Mental Health-National Institutes of Health, Bethesda, Md.).
Nucleotide sequence accession number.
The complete rDNA sequence of P. piscicida has been deposited in GenBank (AY112746).
RESULTS
Sequence of the P. piscicida rRNA locus.
A genomic DNA library of clonally cultured P. piscicida was constructed and screened with a probe containing partial P. piscicida NTS and SSU sequences. Of four positive clones, one (clone 6132) was digested with NotI, and a 6-kb fragment was subcloned. The insert of the resulting plasmid pPPG3-1 yielded the almost-complete rRNA gene cluster of P. piscicida containing 650 bp of NTS sequence 5′ from the SSU; the complete SSU, ITS1, 5.8S region, and ITS2; and 2.8 kb of LSU sequence. A plasmid (pPPG4-3) containing a 9-kb EcoRI fragment of the 6132 clone revealed an additional 600 bp of NTS sequence. This sequence, however, consisted mostly of a repetitive unit (GTAGTAGTA). The sequences of the SSU, ITS1, 5.8S region, and ITS2 were almost identical to the available sequences of P. piscicida (complete SSU, ITS1, 5.8S region, and ITS2 and partial LSU sequences; GenBank accession no. AF330619 [CCMP1830] and AF330620 [CCMP1831], both isolated in Maryland), with differences in only two positions in the ITS1 sequence. The P. piscicida rDNA sequences were 97 (SSU), 68 (ITS1), 100 (5.8S), and 51% (ITS2) identical to those of P. shumwayae (GenBank accession no. AF218805 and AF352345). The 2.8-kb LSU sequence of P. piscicida was 87.4 and 79.6% identical to those of the Provocentrum micans and Gonyaulax polyedra LSUs (GenBank accession no. X16108 and AF377944, respectively).
Intraspecific variability of the P. piscicida rRNA locus.
To characterize the intraspecific variability within the rRNA locus of P. piscicida, DNA from putative P. piscicida cultures (CCMP1830, -1831, -1834, -1902, -1921, and -1928, COMBH4 [Maryland], and COMBN1SR2 [North Carolina]) were extracted and amplified, and the amplicons of the NTS, ITS1, and ITS2 were sequenced. All ITS1 amplicons were identical among seven clonal P. piscicida and two regional mixed cultures. Similarly, all ITS2 amplicons were identical among the cultures tested. Amplicons from the NTS, however, revealed differences in two positions. At 399 bp upstream from the SSU boundary, alternative bases (T or G) were present in the P. piscicida sequence, the T/G ratio at this position being variable among different isolates. The second site, 89 bp upstream from the SSU, had an insertion or deletion in approximately 50% of the P. piscicida cultures (Table 2).
TABLE 2.
Intraspecific variability in NTS of P. piscicida
| Source | Site 1 | Site 2a |
|---|---|---|
| P. piscicida (PCR) | T | Mixed (50/50) |
| P. piscicida (clone 6231) | G | −C |
| CCMP1830 (PCR) | Mixed | Mixed |
| CCMP1831 (PCR) | Mixed, T | −C |
| CCMP1834 (PCR) | T | Mixed |
| CCMP1902 (PCR) | Mixed | Mixed |
| CCMP1921 (PCR) | T | +C |
| CCMP1928 (PCR) | T | +C |
| COMBH4 (PCR) | T | Mixed |
| COMBN1R2 (PCR) | Mixed | Mixed |
−, deletion; +, insertion.
Development and validation of a PCR-based assay for P. piscicida: specificity and sensitivity.
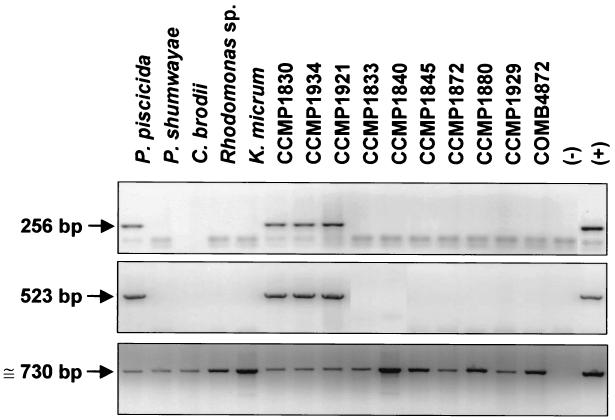
The primer sets NTS2F-NTS3R (NTS1 PCR) and NTS2F-NTS4R (NTS2 PCR) yielded the expected 265- and 523-bp amplicons, respectively (Fig. 1B). For assessment of the species specificities of both the NTS1 and NTS2 PCR assays, 37 dinoflagellate isolates were tested. Only 10 P. piscicida isolates tested positive with both NTS PCRs. Representative PCR results with 15 isolates are shown in Fig. 2, and together with those from additional PCR tests of 22 Pfiesteria-like dinoflagellates, are listed in Table 3.
FIG. 2.
Species specificity of PCR-based assay for P. piscicida. DNAs of dinoflagellate isolates were amplified with specific primers for P. piscicida and actin genes. (Top) NTS1 PCR for P. piscicida with NTS2F and NTS3R primers (256 bp). (Middle) NTS2 PCR for P. piscicida with NTS2F and NTS4R primers (523 bp). (Bottom) Actin-PCR using G-480 and G-482 universal primers (≅730 bp). (−), negative control; (+), positive control with P. piscicida DNA (100 pg) for NTS1 and NTS2 PCR and P. piscicida actin clone (1 ng) for actin-PCR.
TABLE 3.
Diagnosis by PCR assay based on NTS for P. piscicida with clonal culture of Pfiesteria complex and Pfiesteria-like dinoflagellates
| Isolate | Collection site | Result for P. piscicidaa |
|---|---|---|
| P. piscicida | Chicamacomico River, Md. | + |
| P. shumwayae | Neuse River, N.C. | − |
| C. brodii | Neuse River, N.C. | − |
| K. micrum | Fish farm, Salisbury, Md. | − |
| P. minimum | Unknown | − |
| CCMP768 (Rhodomonas sp.) | North Island, New Zealand | − |
| CCMP1830 (presumptive P. piscicida) | Chicamacomico River, Md. | + |
| CCMP1831 (presumptive P. piscicida) | As above | + |
| CCMP1832 | As above | − |
| CCMP1833 | As above | − |
| CCMP1834 (presumptive P. piscicida) | Pocomoke River, Md. | + |
| CCMP1835 | Pocomoke Sound, Va. | − |
| CCMP1836 | As above | − |
| CCMP1838 | Neuse River, N.C. | − |
| CCMP1839 | As above | − |
| CCMP1940 | As above | − |
| CCMP1841 | Neuse River, N.C. | − |
| CCMP1842 | As above | − |
| CCMP1843 | As above | − |
| CCMP1844 | As above | − |
| CCMP1845 | As above | − |
| CCMP1872 | As above | − |
| CCMP1873 | Wilmington River, Ga. | − |
| CCMP1874 | As above | − |
| CCMP1875 | As above | − |
| CCMP1876 | As above | − |
| CCMP1877 | As above | − |
| CCMP1878 | Neuse River, N.C. | − |
| CCMP1879 | Wilmington River, Ga. | − |
| CCMP1880 | As above | − |
| CCMP1881 | As above | − |
| CCMP1882 | As above | − |
| CCMP1902 (presumptive P. piscicida) | Chicamacomico River, Md. | + |
| CCMP1921 (presumptive P. piscicida) | As above | + |
| CCMP1928 (presumptive P. piscicida) | Wilmington River, Ga. | + |
| CCMP1929 | Middle River, Md. | − |
| COMB4872 | Unknown (DNR) | − |
| COMBH4 | Fish farm, Salisbury, Md. | + |
| COMBH10 | As above | + |
| COMBN1R2 | Neuse River, N.C. | + |
| COMBN2F1 | Neuse River, N.C. | − |
+, positive; −, negative (by NTS PCR for P. piscicida)
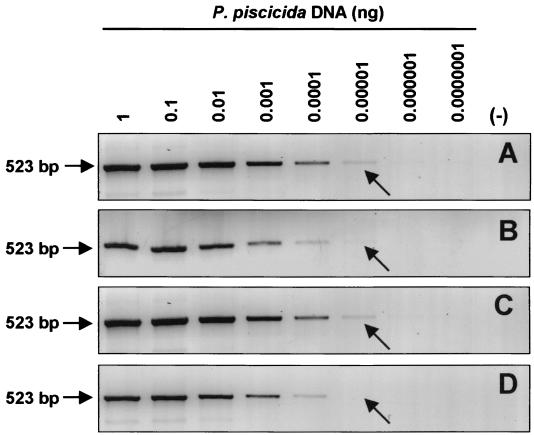
The detection limit of the PCR-based assay using the NTS2 PCR primers was 10 fg of P. piscicida DNA (Fig. 3A). The presence of up to 1 μg of background DNA from the algal-prey Rhodomonas sp. in the PCR mixture did not affect the sensitivity of the assay (Fig. 3C). The presence of 1 μg of background DNA from either K. micrum (Fig. 3B) or COMB4872 (Fig. 3D) decreased the detection sensitivity.
FIG. 3.
Sensitivity of PCR-based assay for P. piscicida. Shown are amplicons of 10-fold serial dilutions of P. piscicida DNA starting from 1 ng without background DNA (A), with 1 μg of Rhdomonas sp. DNA (B), with 1 μg of K. micrum DNA (C), and with 1 μg of COMB4872 DNA (D). The arrows indicate the limit of detection (10 fg of DNA). Although light bands can be observed at higher dilutions, the arrows indicate those that are clear under UV light with the naked eye. Fifteen microliters of a 20-μl PCR mixture was applied for electrophoresis. (−), negative control with sterile water.
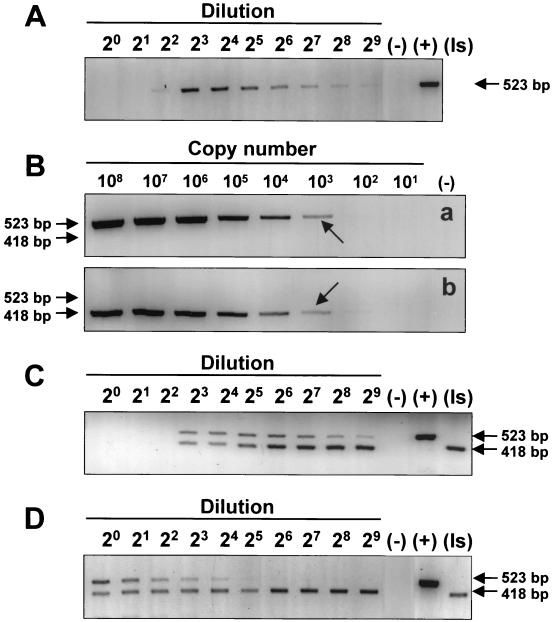
Construction of an internal standard for PCR diagnosis of environmental samples.
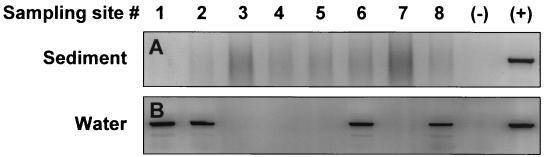
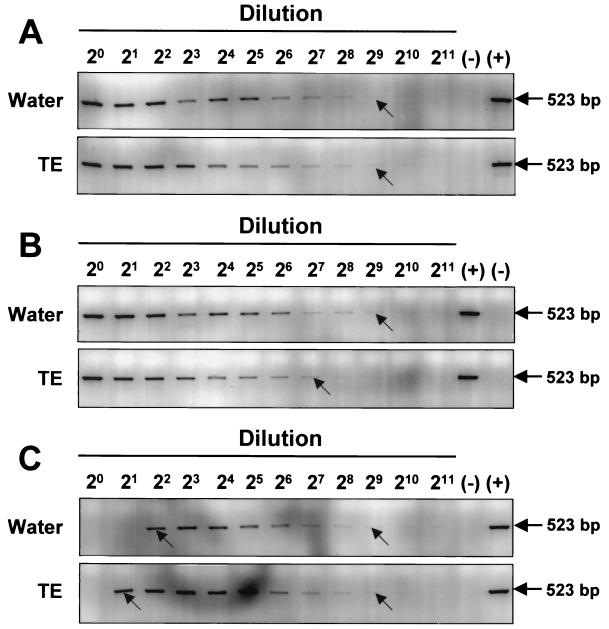
The application of the PCR assay to environmental samples revealed an unexpected complication when samples from dry ponds where P. piscicida had been detected earlier were tested. The freshly collected sediments failed to yield the expected positive results (Fig. 4A) that had been obtained with the water in which the sediment suspension had been incubated for 7 days (Fig. 4B). These positive results in water were corroborated by the observation of actively motile P. piscicida zoospores within 7 days, which could only have emerged from cysts or other life stages present in the sediment. However, a sediment sample that by incubation with water had previously yielded a supernatant that tested positive for P. piscicida required a 1:8 or greater dilution of the DNA extract for detection (Fig. 5A). This suggests that inhibitors for the PCR amplification were present in the sediment and were carried over through the DNA extraction. To address this potential problem of false-negative PCR results for sediment samples, an internal standard (pMNTS2) was constructed which has the same priming sites as the target sequence but yields a smaller amplicon (418 bp). The detection limit of the assay using either pTNTS2 or the internal standard, pMNTS2, was approximately 103 copies of P. piscicida NTS in the presence of Rhodomonas sp. DNA (1 μg) (Fig. 5B). When the internal standard was spiked (104 copies reaction−1) into the PCR mixture in which DNA from the sediment sample had been extracted with TE, inhibition of the PCR amplification was observed up to the 1:4 dilution (≥12.5 mM EDTA) for both P. piscicida DNA and the internal standard (Fig. 5C). When DNA was extracted from the sediment sample with the FastDNA spin kit for soil, however, no inhibition prozone was observed (Fig. 5D). To examine the possibility that the inhibition prozone is due to the extraction method used, particularly the presence of EDTA, PCR amplifications of P. piscicida DNA in the presence and absence of EDTA were carried out. The results indicate that the presence of EDTA at the highest concentration (50 μM) may slightly reduce the sensitivity of the assay, but no prozone was observed (Fig. 6A and B). In addition, DNA extractions of sediments in parallel with TE and water spiked with P. piscicida DNA were carried out and tested by PCR amplification. A higher dilution (1:4) of the water-extracted sample was required to obtain a positive result, whereas a 1:2 dilution was enough to avoid the inhibitory prozone in the TE-extracted sample (Fig. 6C).
FIG. 4.
Detection of P. piscicida in environmental sediment samples. DNA was extracted from sediment samples from eight individual locations and from water from a suspension of each sediment sample incubated for 7 days. P. piscicida was detected by NTS2 PCR. (+), positive with 100 pg of P. piscicida DNA; (−), negative control.
FIG. 5.
Construction and application of an internal-standard plasmid for NTS2 PCR assay. (A) Environmental sediment sample detection with twofold serial dilution. (B) Amplification of 10-fold dilution of pTNTS2 (a) and pMNTS2 (b) in 108 to 101 copies with 1 μg of Rhodomonas sp. DNA by NTS2 PCR. The 418- and 523-bp amplicons indicate the target and the internal standard PCR products, respectively. The arrows indicate the limit of detection (1 × 103 copies). (C) Application of the internal standard to the detection of P. piscicida with DNA extracted from environmental sediment samples; a twofold serial dilution of sample DNA was amplified with 2 × 104 spiked copies of pMNTS2. (D) Detection of P. piscicida with DNA of environmental sediment sample extracted by BIO101 kit for soil. (−), negative control; (+), positive control; (Is), positive control of an internal standard.
FIG. 6.
Effect(s) of the presence of EDTA in PCR. (A) Amplification of dilution of P. piscicida DNA (1 ng) in TE (10 μl) with sterile water in 20-μl PCR mixture. (B) PCR products of diluted P. piscicida DNA spiked into 10 μl of TE in a 20-μl reaction mixture on agarose gel. (C) Amplification of P. piscicida DNA extracted with sterile water or TE from cells spiked into sediment. (−), negative control; (+), positive control. The 523-bp PCR product indicates target amplification. The arrows indicate the limit of detection under UV light with the naked eye.
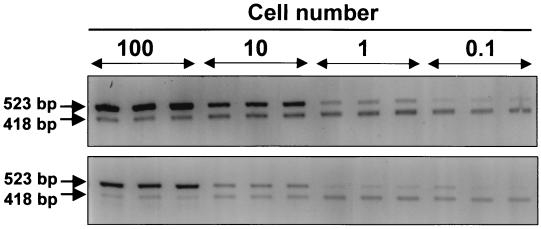
Because the DNA content per cell or the efficiency of DNA extraction may vary with the cell type, we spiked equal numbers of either P. piscicida zoospores or cysts in a sediment sample that had tested negative for P. piscicida. The lower detection limit was approximately one zoospore or 10 cysts per g of sediment (Fig. 7).
FIG. 7.
Detection sensitivity of P. piscicida PCR-based assay with DNA extracted with BIO101 kit for soil from P. piscicida cells spiked into autoclaved sediment (0.1 g). (Top) DNA from zoospores; (bottom) DNA from cysts. Amplicons corresponding to 102, 101, 1, and 0.1 P. piscicida cells were amplified with 103 copies of pMNTS2 and separated on agarose gels.
This optimized PCR assay was used for detection of P. piscicida in 267 environmental samples (Table 4). PCR-positive results were routinely confirmed by direct sequencing of the amplicons, and sequences identical to that of the P. piscicida NTS were obtained with all 49 PCR products analyzed.
TABLE 4.
Environmental sample diagnosis results in 1999-2001 by NTS2 PCR assay
| State and sample | No. positive | No. negative |
|---|---|---|
| Md. | ||
| Water | 22 | 196 |
| Sediment | 18 | 15 |
| N.C. | ||
| Water | 3 | 3 |
| Sediment | 6 | 4 |
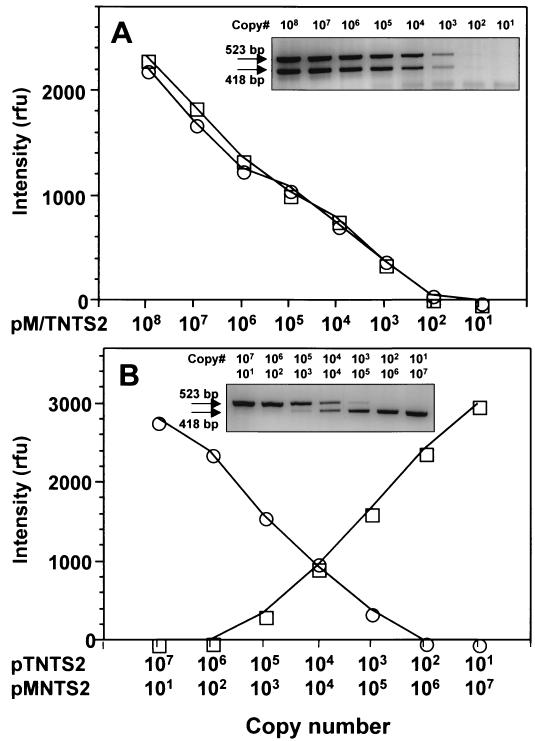
Development of a QC-PCR assay: assessment of amplification kinetics with target and competitor DNAs.
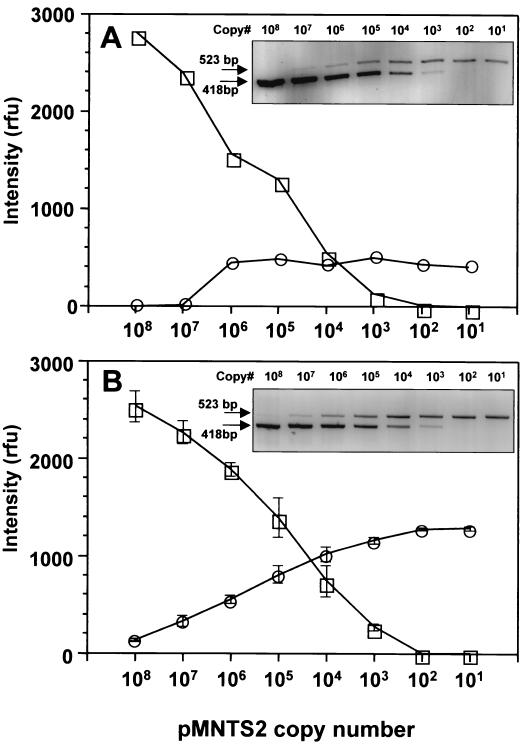
The internal standard, pMNTS2, was used as a competitor for the development of a QC-PCR assay. A prerequisite for the use of a competitor is an assessment of the amplification kinetics of target and competitor DNA templates, which have to be similar. When pTNTS2 was coamplified with pMNTS2 using the NTS2F and NTS4R primers, the band intensities of PCR products generated from equal copy numbers of pTNTS2 and pMNTS2 were comparable at every dilution tested (Fig. 8A). When reciprocal dilutions of pTNTS2 and pMNTS2 were coamplified (Fig. 8B), the band intensities of target and competitor amplicons confirmed that the amplification kinetics of both pTNTS2 and pMNTS2 are similar, with equal amplicon band intensities at the equivalence point.
FIG. 8.
Amplification kinetics of pTNTS2 and pMNTS2 with NTS2F and NTS4R primers. pTNTS2 and pMNTS2 were amplified in the same reaction mixture. PCR products of the same copy numbers of both plasmids in 10-fold dilutions starting from 108 copies (A) and various numbers of each plasmid (B) were separated by electrophoresis. Fifteen microliters of a 20-μl PCR mixture was applied to each well, and the band intensities of target and competitor amplicons are plotted. ○, target amplicon (523 bp); □, pMNTS2 amplicon (418 bp); rfu, relative fluorescence units.
Application of QC-PCR for assessing copy numbers of target DNA in P. piscicida.
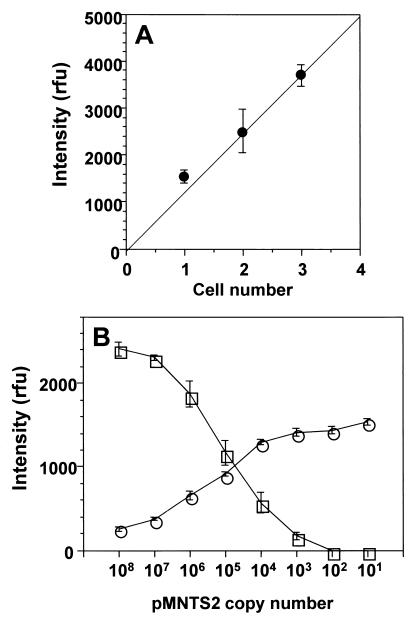
By coamplifying decreasing copy numbers of the competitor plasmid (pMNTS2) with a constant copy number (104) of the target plasmid (pTNTS2), it could be confirmed that the point of equal band intensities corresponds to the reaction mixture containing equal copy numbers of target and competitor (Fig. 9A). By replacing pTNTS2 with P. piscicida DNA (500 pg) as the target in a similar assay format (Fig. 9B), the band intensity equivalence point corresponded to 1 × 104 to 2 × 104 copies of pMNTS2. Therefore, the number of NTS copies in 1 pg of P. piscicida DNA would correspond to approximately 20 to 40 copies.
FIG. 9.
Validation of QC-PCR with pMNTS2 as a competitor. A constant amount of pTNTS2 (104 copies) (A) or P. piscicida DNA (500 pg) (B) was amplified with 10-fold dilutions of pMNTS2. ○, target amplicon (523 bp); □, pMNTS2 amplicon (418 bp); rfu, relative fluorescence units. The error bars indicate the standard deviation.
To assess the number of NTS copies in a single P. piscicida zoospore, cells were manually isolated; distributed as one, two, or three cells per tube (n = 7 for each cell number); lysed by a freeze-thaw-boil procedure in sterile water; and subjected to PCR amplification. The band intensities of PCR products of one to three P. piscicida cells were plotted against the cell numbers (Fig. 10A), suggesting linear correlation (r = 0.98). Total DNA, extracted from 770 P. piscicida cells by glass bead-phenol extraction, was tested by QC-PCR to determine the target copy number in a single P. piscicida zoospore (Fig. 10B). It was estimated that ∼100 copies of the NTS sequence are present per P. piscicida zoospore. Because this number may vary with the efficiency of the DNA extraction method selected, three procedures (glass bead-phenol extraction, BIO101 kit, and freeze-thaw-boil in TE) were compared by QC-PCR. The DNA yields of the first two methods were similar and were 100-fold higher than that of the last method (data not shown).
FIG. 10.
Correlation of one, two, and three cells of P. piscicida zoospore amplification and assessment of NTS region copy number in single P. piscicida zoospore by QC-PCR. (A) Cells were collected with a capillary tube and then added directly to the PCR mixture (20 μl). The averages of individual band intensities corresponding to each cell number are plotted, with error bars indicating standard deviation (n = 7). (B) Band intensities of PCR amplicons corresponding to 770 P. piscicida zoospores with 10-fold dilutions of pMNTS2 are plotted, with error bars indicating standard deviation (n = 3). ○, target amplicon (523 bp); □, pMNTS2 amplicon (418 bp).
DISCUSSION
In this study, we characterized the rRNA gene cluster of a P. piscicida isolate from Maryland. Sequence information for the Dinophyceae available in GenBank (as of 29 April 2002) comprised 1,516 nucleotide sequences, of which 186 were annotated as SSU; 114 were annotated as ITS1, 5.8S, and/or ITS2; and 65 were annotated as LSU (most of them partial sequences), including 50 sequences identified as Pfiesteria spp. (35 as P. piscicida for rDNA) (29), mitochondrial cytochrome b (4), mitogen-activated protein kinase (1), and cyclin 1 (1); 10 sequences identified as P. shumwayae; and 5 sequences identified as Pfiesteria sp. We expanded the sequence information on the P. piscicida rDNA almost to completion of the locus, including a 2.8-kb stretch of LSU and the first NTS sequence reported for the Dinozoa, 1.25 kb upstream from the SSU. Thus, this study provides new information that may be relevant to the identification Pfiesteria subspecies and strains, as well as for phylogenetic studies. Finally, we selected the NTS to develop and validate qualitative and quantitative PCR-based assays for the assessment of P. piscicida in water and sediment environmental samples.
The sequences of the P. piscicida SSU, ITS1, 5.8S region, ITS2, and partial LSU reported here are identical to 21 sequences which stretch from the 5′ end of the SSU to 350 bp of the 5′-end LSU annotated as P. piscicida in GenBank (accession no. AF330600 to AF330620). Small differences (one or two nucleotide positions) were observed with five additional P. piscicida sequences (accession no. AF352333 to AF352335), but the consensus of all P. piscicida rRNA sequences available in GenBank was 100% identical to the sequence reported here. The NTS, considered the most variable region within the rRNA locus (4, 12), revealed differences in only two nucleotide positions among nine P. piscicida isolates tested. This low level of NTS variability observed was surprising in light of reports of the variability of the NTS from other alveolate species (11). In the oyster parasite P. marinus, two distinct NTS sequences, designated types I and II, that differed at six defined nucleotide positions were found in a 307-bp amplicon (52). It is noteworthy that the rather limited NTS variability among different P. piscicida isolates was also observed within a single isolate. This variability resembles that observed in some apicomplexan parasites that contain distinct sets of unlinked rRNA copies, which are differentially expressed in selected developmental stages (60, 66). Whether the differences found in P. piscicida rDNA may also reflect different stage-specifically transcribed rRNA gene units warrants further investigation and may reveal novel aspects of genome dynamics in dinoflagellates. Furthermore, a comparative study of sequences of NTS regions from a larger number of isolates from far-ranging geographic locations will reveal the true extent of the NTS variability within this species.
Pfiesteria blooms have been difficult to monitor. This has been mostly due to the lack of knowledge about the environmental factors that trigger proliferation of this species and sensitive and specific methods for year-round detection of zoospores and cysts in the environment. This has hampered our ability to understand the organism's possible role(s) in fish kill events and the associated human health problems. P. piscicida monitoring should include water samples that may carry flagellated motile forms (zoospores) and sediment samples that may contain the nonmotile resting forms (cysts). Therefore, the assay of choice for P. piscicida should be one that enables the detection of the dinoflagellate in both water and sediment environmental samples.
In recent years, detection assays for P. piscicida using molecular probes, such as standard PCR (53, 54, 67), the heteroduplex mobility assay (45), and real-time PCR (6), have been developed. Molecular identification of P. piscicida is useful to complement the morphological identification methods based on plate tabulation (61, 63) and fish mortality bioassays (9, 10). Molecular detection assays for P. piscicida have been developed based on nucleotide sequences of the SSU of the rRNA locus (6, 45, 53) and, most recently, of mitochondrial cytochrome b (67). Because the SSU is highly conserved among different species (38, 39), the possibility exists that closely related species, or yet-undescribed species that may exhibit similar target regions, may yield false-positive results, particularly in field studies (53). Alternatively, the mitochondrial cytochrome b gene has been reported as a suitable target for species identification and phylogenetic analysis, but limitations of assay sensitivity are expected due to the lower copy numbers relative to rRNA genes (41, 64). The purpose of this study was to develop and optimize a detection assay for P. piscicida that would overcome some of the limitations of the current tests by selecting a target sequence that would ensure species specificity and eliminate, as much as possible, the loss of sensitivity from PCR bias. Based on previous experience with the development of species- and strain-specific PCR assays for parasites of the genus Perkinsus (15, 35, 49, 50), the NTS of the rRNA locus was chosen as the target for the development of a species-specific assay for P. piscicida. The species specificity was demonstrated (no false-positive result was obtained by direct sequencing with any of the 37 Pfiesteria-like dinoflagellates and the 267 environmental samples tested), and higher sensitivity (one zoospore and/or 10 cysts in 1 g of sediment or 10 fg of P. piscicida DNA in 1 μg of heterologous DNA) of this detection PCR assay relative to current assays was shown (detection limit, 0.6 cell with real-time PCR based on SSU [6] or 0.2 cell with PCR targeted on mitochondrial cytochrome b [67]). Although the variability in the NTS was less than expected, possibly due to the small number of isolates examined, the fact that this region shows high interspecies variability suggests that the presence of other dinoflagellate species in the test sample should not diminish the species specificity of the assay. Our validation of assay specificity in the presence of large amounts of heterologous dinoflagellate DNA (>105-fold relative to P. piscicida DNA) supports this notion.
The results from the application of the PCR assay to clonal cultures and environmental water samples were reliable and highly reproducible. When sediments were tested, however, an inhibition prozone phenomenon was observed in those samples in which DNA was extracted by conventional methods (freeze-thaw followed by extraction in TE), with negative results turning positive when the test DNA sample was diluted. When DNA was extracted from sediments with the BIO101 kit for soil, in which DNA is isolated through binding to a matrix while potential coextractants are washed away, no prozone was observed. This suggested that the false-negative results in the undiluted DNA sample were most likely due to the presence of inhibitory substances extracted from the sediment that were carried over through the DNA extraction to the PCR mixture. The presence of substances in soil and sediment that may inhibit Taq DNA polymerase, such as humic acids, has been described as a common occurrence (40, 42). This potential problem may be avoided by selecting DNA extraction methods, such as the commercial kit tested in this study, which will prevent the coextraction of potential inhibitors. The DNA extraction was efficient and reproducible, enabling the detection by PCR of a single P. piscicida zoospore or 10 cysts in 1 g of sediment.
To facilitate the application of this PCR assay in laboratories where DNA extraction kits for soil are not available, an internal standard for the amplification reaction was constructed. This plasmid constitutes a quality control for the assay and can be used to identify false-negative results in environmental testing. The efficacy of this plasmid for the identification of inhibitory prozones in the PCR assay was experimentally validated. In addition, the same plasmid was used for the development of a QC-PCR assay. A requirement for this application is that the competitor and target DNAs should have similar amplification efficiencies. This requirement was met by the plasmid pTNTS2, for which the PCR product differs from the target amplicon in only 105 bp. In the QC-PCR assay format, known concentrations (copy numbers) of the competitor are added to the test samples, and their DNA content is estimated from the equivalent band intensities of the coamplified target and competitor PCR products on the gel. Based on the number of DNA target copies per cell (zoospores or cysts), the number of P. piscicida cells in the test samples can be easily calculated. In this study, uncut plasmid was used as a competitor; the spike-recovery validation of the QC-PCR with cut and uncut plasmid could also have informed of the supercoiled uncut plasmid competitor, which may lead to overestimation of the target DNA contents. The real-time PCR technology does not rely on the use of a competitor; nevertheless, it has been used for rapid environmental diagnosis.The presence of an inhibitor(s) in the test sample may yield inaccurate results, usually lower DNA content values than those expected from spike-recovery (19).
The QC-PCR format enabled the estimation of the copy numbers of the target DNA (the NTS of the rRNA gene cluster) per P. piscicida cell. The value of 20 to 40 copies of NTS per pg of P. piscicida DNA led to an estimate of 100 to 200 copies of NTS per P. piscicida cell (zoospore). This was calculated from the DNA content of a single P. piscicida cell (approximately 3 to 5 pg of DNA/zoospore) during the stationary phase, obtained from QC-PCR data. This estimate is consistent with the value obtained by DNA extraction and quantification by optical density at 260 nm (4 to 6 pg of DNA/cell), although it is in the lower range of DNA content (5.2 to 9.4 pg of DNA/cell) as measured by fluorescent staining of DNA (46). Dinoflagellates contain large amounts of DNA, ranging from 3.8 pg per cell in Crypthecodinium cohnii and 33 pg in Gyrodinium resplendens to 200 pg per cell in G. polyedra (1, 28). These values are greater than the DNA contents of any other unicellular eukaryotes characterized so far (0.046 to 3 pg/nucleus) (48). Furthermore, the DNA content can vary substantially not only from species to species but also between growth phases. In several autotrophic dinoflagellate species, cells in logarithmic phase were found to contain a twofold-larger amount of DNA than those in stationary phase (1). In most eukaryotes, rRNA genes are organized in tandemly repeated units (14), for example, 100 to 200 copies are present in yeast and 50 to 10,000 copies are present in mammals. Within the Alveolata, the number of rRNA copies ranges from 2 to >100. Two copies have been reported in Theileria sp. (30) and Cryptosporidium sp. (31), 3 copies in Babesia sp. (16), 4 to 8 copies in Plasmodium sp. (37, 56), and 110 copies in Toxoplasma sp. (22). Our estimate of rRNA gene copy numbers in P. piscicida (100 to 200 copies) is in the upper range of the known alveolates. With this information, the PCR-based assay targeted to the NTS developed and validated here can be used to assess the density of P. piscicida cells in environmental water and sediment samples. Finally, although the Taqman technology certainly constitutes the state of the art approach for quantification of DNA, the equipment and reagents are more costly than for standard PCR and it requires very well-trained operators. Therefore, the availability of a simple, inexpensive, and reliable QC-PCR assay that enables the assessment of P. piscicida cell densities in both water and sediment environmental samples should represent a valuable alternative to more costly quantitative assay formats.
Acknowledgments
The authors acknowledge K. A. Steidinger (Florida Department of Environmental Protection, St. Petersburg, Fla.), D. K. Stoecker (Center of Environmental Studies, University of Maryland, Cambridge), R. W. Litaker (University of North Carolina, Chapel Hill), and D. W. Coats (Smithsonian Institution, Edgewater, Md.) for providing dinoflagellate cultures.
This study was supported by grants NIEHS 5-P01-ES09563 and ECOHAB NA860P0192.
Keiko Saito and Tomás Drgon contributed equally to this study.
REFERENCES
- 1.Allen, J. R., M. Roberts, A. R. Loeblich III, and L. C. Klotz. 1975. Characterization of the DNA from the dinoflagellate Crypthecodinium cohnii and implications for nuclear organization. Cell 6:161-169. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., and B. A. Keafer. 1985. Dinoflagellate cyst dynamics in coastal and estuarine waters, p. 219-224. In D. M. Anderson, A. W. White, and D. G. Baden (ed.), Toxic dinoflagellates. Elsevier, New York, N.Y.
- 3.Ausubel, M. F., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Escherichia coli, plasmids, and bacteriophages, p. 1-38. In M. F. Ausubel, R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology, 4th ed. John Wiley & Sons, Inc., Hoboken, N.J.
- 4.Bena, G., M. F. Jubier, I. Olivieri, and B. Lejeune. 1998. Ribosomal external and internal transcribed spacers: combined use in the phylogenetic analysis of Medicago (Leguminosae). J. Mol. Evol. 46:299-306. [DOI] [PubMed] [Google Scholar]
- 5.Bever, C. T., Jr., L. Grattan, and J. G. Morris. 1998. Neurologic symptoms following Pfiesteria exposure: case report and literature review. Md. Med. J. 47:120-123. [PubMed] [Google Scholar]
- 6.Bowers, H. A., T. Tengs, H. B. Glasgow, Jr., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkholder, J. M., and H. B. Glasgow, Jr. 1997. Pfiesteria piscicida and other Pfiesteria-like dinoflagellates: behavior, impacts, and environmental controls. Limnol. Oceanogr. 42:1052-1075. [Google Scholar]
- 8.Burkholder, J. M., H. B. Glasgow, Jr., and C. W. Hobbs. 1995. Fish kills linked to a toxic ambush-predator dinoflagellate: distribution and environmental conditions. Mar. Ecol. Prog. Ser. 124:43-61. [Google Scholar]
- 9.Burkholder, J. M., H. G. Marshall, H. B. Glasgow, D. W. Seaborn, and N. J. Deamer-Melia. 2001. The standardized fish bioassay procedure for detecting and culturing actively toxic Pfiesteria, used by two reference laboratories for Atlantic and gulf coast states. Environ. Health Perspect. 109:745-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkholder, J. M., E. J. Noga, C. W. Hobbs, and H. B. Glasgow, Jr. 1992. New ′phantom' dinoflagellate is the causative agent of major estuarine fish kills. Nature 358:407-410. [DOI] [PubMed] [Google Scholar]
- 11.Chou, C.-H., Y.-C. Chiang, and T.-Y. Chiang. 1999. Within- and between-individual length heterogeneity of the rDNA-IGS in Miscanthus sinensis var. glaber (Poaceae): phylogenetic analyses. Genome 42:1088-1093. [DOI] [PubMed] [Google Scholar]
- 12.Ciliberto, G., G. Ragugei, F. Constanzo, L. Dente, and R. Cortese. 1983. Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase III. Cell 32:725-733. [DOI] [PubMed] [Google Scholar]
- 13.Coatas, E., R. Zardoya, J. Bautista, A. Garrido, C. Rojo, and V. López-Rodas. 1995. Morphospecies vs. genospecies in toxic marine dinoflagellates: an analysis of Gymnodinium catenatum/Gyrodinium impudicum and Alexandrium minutum/A. lusitanicum using antibodies, lectins, and gene sequences. J. Phycol. 31:801-807. [Google Scholar]
- 14.Cortadas, J., and M. C. Pavon. 1982. The organization of ribosomal genes in vertebrates. EMBO J. 1:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coss, C. A., J. A. Robledo, G. M. Ruiz, and G. R. Vasta. 2001. Description of Perkinsus andrewsi n. sp. isolated from the Baltic clam (Macoma balthica) by characterization of the ribosomal RNA locus, and development of a species-specific PCR-based diagnostic assay. J. Eukaryot. Microbiol. 48:52-61. [DOI] [PubMed] [Google Scholar]
- 16.Dalrymple, B. P. 1990. Cloning and characterization of the rRNA genes and flanking regions from Babesia bovis: use of the genes as strain discriminating probes. Mol. Biochem. Parasitol. 43:117-124. [DOI] [PubMed] [Google Scholar]
- 17.Davis, L. G., W. M. Kuehl, and J. F. Battey. 1994. Basic methods in molecular biology, 2nd ed. Appleton & Lange, Norwalk, Conn.
- 18.de la Herran, R., M. A. Garrido-Ramos, J. I. Navas, C. R. Rejon, and M. R. Rejon. 2000. Molecular characterization of the ribosomal RNA gene region of Perkinsus atlanticus: its use in phylogenetic analysis and as a target for a molecular diagnosis. Parasitology 120:345-353. [DOI] [PubMed] [Google Scholar]
- 19.Desjardin, L. E., Y. Chen, M. D. Perkins, L. Teixeira, M. D. Cave, and K. D. Eisenach. 1998. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J. Clin. Microbiol. 36:1964-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glasgow, H. B., Jr., J. M. Burkholder, D. E. Schmechel, P. A. Tester, and P. A. Rublee. 1995. Insidious effects of a toxic estuarine dinoflagellate on fish survival and human health. J. Toxicol. Environ. Health 46:501-522. [DOI] [PubMed] [Google Scholar]
- 21.Grattan, L. M., D. Oldach, T. M. Perl, M. H. Lowitt, D. L. Matuszak, C. Dickson, C. Parrott, R. C. Shoemaker, C. L. Kauffman, M. P. Wasserman, J. R. Hebel, P. Charache, and J. G. Morris, Jr. 1998. Learning and memory difficulties after environmental exposure to waterways containing toxin-producing Pfiesteria or Pfiesteria-like dinoflagellates. Lancet 352:532-539. [DOI] [PubMed] [Google Scholar]
- 22.Guay, J. M., A. Huot, S. Gagnon, A. Tremblay, and R. C. Levesque. 1992. Physical and genetic mapping of cloned ribosomal DNA from Toxoplasma gondii: primary and secondary structure of the 5S gene. Gene 114:165-171. [DOI] [PubMed] [Google Scholar]
- 23.Guillard, R. R. L. 1988. The Center for Culture of Marine Phytoplankton: history, structure, function and future. J. Protozool. 35:255-256. [Google Scholar]
- 24.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 25.Guillard, R. R. L., and J. H. Ryther. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8:229-239. [DOI] [PubMed] [Google Scholar]
- 26.Haley, S. T., J. F. Cavender, and T. E. Murray. 1999. Detection of Alexandrium tamarensis by rapid PCR analysis. BioTechniques 26:88-91. [DOI] [PubMed] [Google Scholar]
- 27.Haselow, D. T., E. Brown, J. K. Tracy, R. Magnien, L. M. Grattan, J. G. Morris, Jr., and D. W. Oldach. 2001. Gastrointestinal and respiratory tract symptoms following brief environmental exposure to aerosols during a Pfiesteria-related fish kill. J. Toxicol. Environ. Health A 63:553-564. [DOI] [PubMed] [Google Scholar]
- 28.Holm-Hansen, O. 1969. Algae: amounts of DNA and organic carbon in single cells. Science 163:87-88. [DOI] [PubMed] [Google Scholar]
- 29.Kane, A. S., D. Oldach, and R. Reimschuessel. 1998. Fish lesions in the Chesapeake Bay: Pfiesteria-like dinoflagellates and other etiologies. Md. Med. J. 47:106-112. [PubMed] [Google Scholar]
- 30.Kibe, M. K., O. K. ole-MoiYoi, V. Nene, B. Khan, B. A. Allsopp, N. E. Collins, S. P. Morzaria, E. I. Gobright, and R. P. Bishop. 1994. Evidence for two single copy units in Theileria parva ribosomal RNA genes. Mol. Biochem. Parasitol. 66:249-259. [DOI] [PubMed] [Google Scholar]
- 31.Le Blancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 32.Levin, E. D., A. H. Rezvani, N. C. Christopher, H. B. Glasgow, Jr., N. J. Deamer-Melia, J. M. Burkholder, V. C. Moser, and K. Jensen. 2000. Rapid neurobehavioral analysis of Pfiesteria piscicida effects in juvenile and adult rats. Neurotoxicol. Teratol. 22:533-540. [DOI] [PubMed] [Google Scholar]
- 33.Levin, E. D., B. B. Simon, D. E. Schmechel, H. B. Glasgow, Jr., N. J. Deamer-Melia, J. M. Burkholder, V. C. Moser, K. Jensen, and G. J. Harry. 1999. Pfiesteria toxin and learning performance. Neurotoxicol. Teratol. 21:215-221. [DOI] [PubMed] [Google Scholar]
- 34.Marin, I., A. Aguilera, B. Reguera, and J. P. Abad. 2001. Preparation of DNA suitable for PCR amplification from fresh or fixed single dinoflagellate cells. BioTechniques 30:88-90, 92-93. [DOI] [PubMed] [Google Scholar]
- 35.Marsh, A. G., J. D. Gauthier, and G. R. Vasta. 1995. A semiquantitative PCR assay for assessing Perkinsus marinus infections in the eastern oyster Crassostrea virginica. J. Parasitol. 81:577-583. [PubMed] [Google Scholar]
- 36.Matuszak, D. L., J. L. Taylor, C. Dickson, and G. C. Benjamin. 1998. Toxic-Pfiesteria—surveillance for human disease in Maryland. Md. Med. J. 47:144-147. [PubMed] [Google Scholar]
- 37.McCutchan, T. F., J. Li, C. A. McConkey, M. J. Rogers, and A. P. Waters. 1995. The cytoplasmatic ribosomal RNAs of Plasmodium spp. Parasitol. Today 11:134-138. [DOI] [PubMed] [Google Scholar]
- 38.Medina, M., A. G. Collins, J. D. Silberman, and M. L. Sogin. 2001. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc. Natl. Acad. Sci. USA 98:9707-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 40.Miller, D. N. 2001. Evaluation of gel filtration resins for the removal of PCR-inhibitory substances from soils and sediments. J. Microbiol. Methods 44:49-58. [DOI] [PubMed] [Google Scholar]
- 41.Miller, O. L., Jr., and B. R. Beatty. 1969. Portrait of a gene. J. Cell Physiol. 74:225. [DOI] [PubMed] [Google Scholar]
- 42.More, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nygren, M., M. Ronaghi, P. Nyrén, J. Albert, and J. Lundeberg. 2001. Quantification of HIV-1 using multiple quantitative polymerase chain reaction standards and bioluminometric detection. Anal. Biochem. 288:28-38. [DOI] [PubMed] [Google Scholar]
- 44.Oldach, D., E. Brown, and P. Rublee. 1998. Strategies for environmental monitoring of toxin producing phantom dinoflagellates in the Chesapeake. Md. Med. J. 47:113-119. [PubMed] [Google Scholar]
- 45.Oldach, D. W., C. F. Delwiche, K. S. Jakobsen, T. Tengs, E. G. Brown, J. W. Kempton, E. F. Schaefer, H. A. Bowers, H. B. Glasgow, Jr., J. M. Burkholder, K. A. Steidinger, and P. A. Rublee. 2000. Heteroduplex mobility assay-guided sequence discovery: elucidation of the small subunit (18S) rDNA sequences of Pfiesteria piscicida and related dinoflagellates from complex algal culture and environmental sample DNA pools. Proc. Natl. Acad. Sci. USA 97:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parrow, M. W., and J. M. Burkholder. 2002. Flow cytometric determination of zoospore DNA content and population DNA distribution in cultured Pfiesteria spp. (Pyrrhophyta). J. Exp. Mar. Biol. Ecol. 267:35-51. [Google Scholar]
- 47.Penna, A., and M. Magnani. 1999. Identification of Alexandrium (dinophtceae) species using PCR and rDNA-targeting probes. J. Phycol. 35:615-621. [Google Scholar]
- 48.Rizzo, P. J., and L. D. Nooden. 1974. Isolation and partial characterization of dinoflagellate chromatin. Biochim. Biophys. Acta 349:402-414. [DOI] [PubMed] [Google Scholar]
- 49.Robledo, J. A. F., C. A. Coss, and G. R. Vasta. 2000. Characterization of the ribosomal RNA locus of Perkinsus atlanticus and development of a polymerase chain reaction-based diagnostic assay. J. Parasitol. 86:972-978. [DOI] [PubMed] [Google Scholar]
- 50.Robledo, J. A. F., J. D. Gauthier, C. A. Coss, A. C. Wright, and G. R. Vasta. 1998. Species-specificity and sensitivity of a PCR-based assay for Perkinsus marinus in the eastern oyster Crassostrea virginica: a comparison with the fluid thioglycollate assay. J. Parasitol. 84:1237-1244. [PubMed] [Google Scholar]
- 51.Robledo, J. A. F., K. Saito, D. W. Coats, K. A. Steidinger, and G. R. Vasta. 2000. The non-transcribed spacer of the rRNA locus as a target for specific detection of the dinoflagellate Pfiesteria piscicida and protista parasite (Perkinsus spp.), p. 238-241. In Ninth International Conference on Harmful Algal Blooms. Intergovernmental Oceanographic Commission of UNESCO, Hobart, Australia.
- 52.Robledo, J. A. F., A. C. Wright, A. G. Marsh, and G. R. Vasta. 1999. Nucleotide sequence variability in the nontranscribed spacer of the rRNA locus in the oyster parasite Perkinsus marinus. J. Parasitol. 85:650-656. [PubMed] [Google Scholar]
- 53.Rublee, P. A., J. Kempton, E. Schaefer, J. M. Burkholder, H. B. Glasgow, Jr., and D. Oldach. 1999. PCR and FISH detection extends the range of Pfiesteria piscicida in estuarine waters. Va. J. Sci. 50:325-336. [Google Scholar]
- 54.Rublee, P. A., J. W. Kempton, E. F. Schaefer, C. Allen, J. Harris, D. W. Oldach, H. Bowers, T. Tengs, J. M. Burkholder, and H. B. Glasgow. 2001. Use of molecular probes to assess geographic distribution of Pfiesteria species. Environ. Health Perspect. 109:765-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampayo, M. A. M. 1985. Encystment and excystment of a Portuguese isolate of Amphidinium carterae in culture, p. 125-130. In D. M. Anderson, A. W. White, and D. G. Baden (ed.), Toxic dinoflagellates. Elsevier, New York, N.Y.
- 56.Shippen-Lentz, D. E., and A. C. Vezza. 1988. The three 5S rRNA genes from the human malaria parasite Plasmodium falciparum are linked. Mol. Biochem. Parasitol. 27:263-273. [DOI] [PubMed] [Google Scholar]
- 57.Shoemaker, R. C., and H. K. Hudnell. 2001. Possible estuary-associated syndrome: symptoms, vision, and treatment. Environ. Health Perspect. 109:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sholin, C. A., and D. M. Anderson. 1994. Identification of group and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). I. RELP analysis of SSU rRNA genes. J. Phycol. 30:744-754. [Google Scholar]
- 59.Siebert, P. D., and J. W. Larrick. 1993. PCR mimics: competitive DNA fragments for use as internal standard in quantitative PCR. BioTechniques 14:244-249. [PubMed] [Google Scholar]
- 60.Spaendonk, R. M. L., J. Ramesar, A. Wigcheren, W. Eling, A. L. Beetsma, G. Gemert, J. Hooghof, C. J. Janse, and A. P. Waters. 2001. Functional equivalence of structurally distinct ribosomes in the malaria parasite Plasmodium berghei. J. Biol. Chem. 276:22638-22647. [DOI] [PubMed] [Google Scholar]
- 61.Steidinger, K., J. Landsberg, R. W. Richardson, E. Truby, B. Blakesley, P. Scott, P. Tester, T. Tengs, P. Mason, S. Morton, D. Seaborn, W. Litaker, K. Reece, D. Oldach, L. Haas, and G. Vasta. 2001. Classification and identification of Pfiesteria and Pfiesteria-like species. Environ. Health Perspect. 109:661-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steidinger, K. A., J. M. Burkholder, H. B. Glasgow, Jr., C. W. Hobbs, J. K. Garrett, E. W. Truby, E. J. Noga, and S. A. Smith. 1996. Pfiesteria piscicida gen. et sp. nov. (Pfiesteriaceae fam. nov.), a new toxic dinoflagellate with a complex life cycle and behavior. J. Phycol. 32:157-164. [Google Scholar]
- 63.Steidinger, K. A., E. W. Truby, J. K. Garrett, and J. M. Burkholder. 1995. The morphology and cytology of a newly discovered toxic dinoflagellate. Lavoisier, Paris, France.
- 64.Stryer, L. 1995. Genes for ribosomal RNAs are tandemly repeated several hundred times, p. 992-993. In L. Stryer (ed.), Biochemistry, 4th ed. W. H. Freeman and Company, New York, N.Y.
- 65.Tyler, M. A., and J. F. Heinbokel. 1985. Cycles of red water and encystment of Gymnodinium pseudopalustre in Chesapeake Bay: effects of hydrography and grazing, p. 213-218. In D. M. Anderson, A. W. White, and D. G. Baden (ed.), Toxic dinoflagellates. Elsevier, New York, N.Y.
- 66.Waters, A. P., R. M. L. Spaendonk, J. Ramesar, R. A. W. Vervenne, R. W. Dirks, J. Thompson, and C. J. Janse. 1997. Species-specific regulation and switching of transcription between stage-specific ribosomal RNA genes in Plasmodium berghei. J. Biol. Chem. 272:3583-3589. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, H., and S. Lin. 2002. Detection and quantification of Pfiesteria piscicida by using the mitochondrial cytochrome b gene. Appl. Environ. Microbiol. 68:989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]