Abstract
The nuclear envelope is one of the chief obstacles to the translocation of macromolecules that are larger than the diameter of nuclear pores. Heterochromatin protein 1 (HP1) bound to the lamin B receptor (LBR) is thought to contribute to reassembly of the nuclear envelope after cell division. Human polyomavirus agnoprotein (Agno) has been shown to bind to HP1α and to induce its dissociation from LBR, resulting in destabilization of the nuclear envelope. Fluorescence recovery after photobleaching showed that Agno increased the lateral mobility of LBR in the inner nuclear membrane. Biochemical and immunofluorescence analyses showed that Agno is targeted to the nuclear envelope and facilitates the nuclear egress of polyomavirus-like particles. These results indicate that dissociation of HP1α from LBR and consequent perturbation of the nuclear envelope induced by polyomavirus Agno promote the translocation of virions out of the nucleus.
Keywords: agnoprotein, heterochromatin protein 1, JC virus, lamin B receptor
Introduction
The nuclear envelope (NE) consists of an outer nuclear membrane, an inner nuclear membrane (INM), nuclear pore complexes and the peripheral nuclear lamina located adjacent to the INM. Various proteins of chromatin (histones, heterochromatin protein 1 (HP1), barrier-to-autointegration factor), the nuclear lamina (lamins A, B and C) and the INM (emerin, lamina-associated polypeptide 2β, lamin B receptor (LBR); Salina et al, 2001) associate with each other during reconstitution and stabilization of the NE. LBR is an integral protein of the INM and binds to HP1 proteins (Ye & Worman, 1996; Ye et al, 1997). The NE presents one of the chief obstacles to transport from the nucleus to the cytoplasm of macromolecules, including the virions of most DNA viruses (Pante & Kann, 2002). The β-herpesvirus gene products UL31 and UL34 induce depolymerization of the nuclear lamina to facilitate the nuclear egress of viral capsids (Muranyi et al, 2002). However, the mechanisms by which other DNA viruses exit the nucleus remain unclear, including the JC virus (JCV), which is the causative agent of progressive multifocal leukoencephalopathy and belongs to the family of polyomaviruses. The JCV agnoprotein (Agno) comprises 71 amino acids, is localized predominantly in the perinuclear region and the cytoplasm of infected cells (Okada et al, 2002) and is related to DNA-damage-induced cell-cycle regulation (Darbinyan et al, 2004). The amino-acid sequence of JCV Agno shares ∼60% homology with those of agnoproteins of other polyomaviruses; however, the sequence of the amino-terminal portion is ∼90% identical to those of other polyomaviruses, which is suggestive of conservation of function. We show that the N-terminal region of JCV Agno associates with HP1 in vivo, resulting in dissociation of HP1 from LBR. This effect alters the NE and thereby facilitates the release of progeny virions from the nucleus into the cytoplasm without nuclear disintegration.
Results
Human JCV Agno binds to HP1
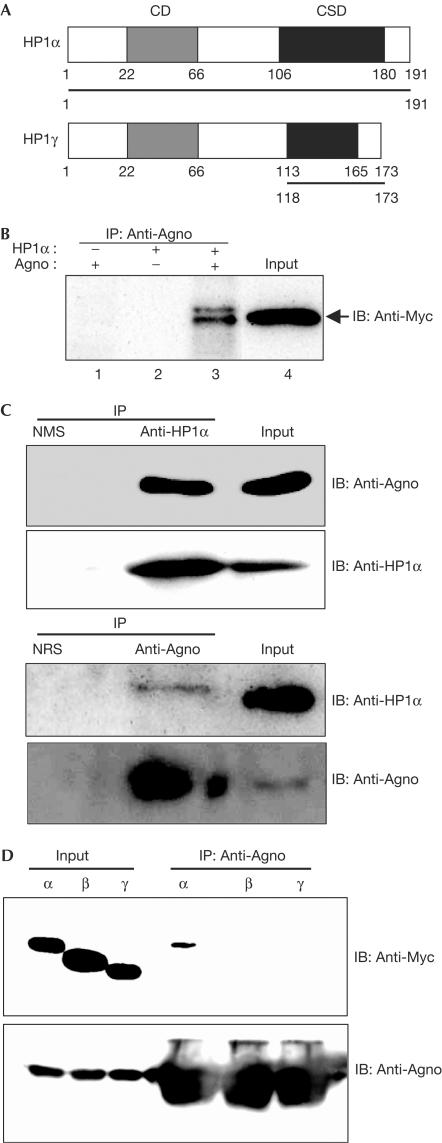
We initially performed a yeast two-hybrid screen with the conserved N-terminal 24 amino acids of JCV Agno as the bait. From a complementary DNA library of human embryonic kidney (HEK) 293 cells, which are permissive for JCV infection (Suzuki et al, 2001), six positive clones were isolated and were found to encode the entire sequence of HP1α and the chromoshadow domain (CSD) of HP1γ (Fig 1A). Interaction between Agno and either Myc-epitope-tagged human HP1α (Fig 1B) or endogenous HP1α (Fig 1C) was also shown in HEK293 cells. We also performed an immunoprecipitation assay to examine whether Agno binds to HP1β and HP1γ in vivo, and found that Agno did not interact with HP1β and HP1γ (Fig 1D), although the CSD of HP1γ was identified by a yeast two-hybrid screening.
Figure 1.
Interaction of JCV Agno with HP1 in vivo. (A) Schematic representation of human HP1α and HP1γ showing the regions (bars) encoded by cDNA fragments isolated by a yeast two-hybrid assay with the N-terminal region of Agno as a bait. CD and CSD, chromo and chromo-shadow domains, respectively. (B) 293AG cells (HEK293 cells in which the expression of JCV Agno is inducible by Dox) were transfected with a vector for Myc-tagged human HP1α and were incubated in the absence or presence of Dox for 24 h. Cell lysates were subjected to immunoprecipitation with antibodies to Agno (anti-Agno), and the resulting precipitates were subjected to immunoblotting (IB) with anti-Myc. (C) Lysates prepared from 293AG cells after treatment with Dox for 48 h were subjected to immunoprecipitation with anti-HP1α or anti-Agno, and the resulting precipitates and cell lysates (Input) were subjected to immunoblotting with the same antibodies, as indicated. NMS, normal mouse serum; NRS, normal rabbit serum. (D) 293T cells were transfected with a vector for Agno and Myc-tagged human HP1α, β or γ, and were subjected to immunoprecipitation with anti-Agno. The resulting precipitates were subjected to immunoblotting with anti-Myc and anti-Agno.
Agno interacts with HP1α at the NE
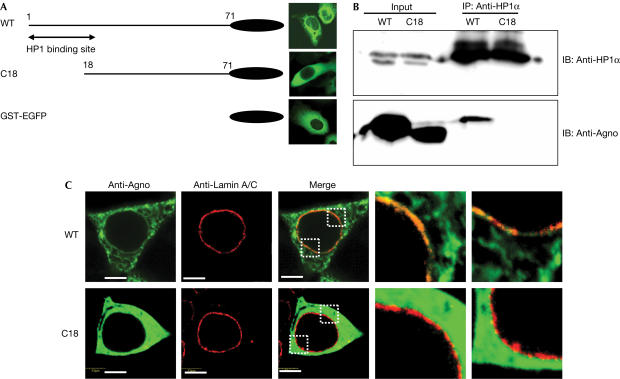
We have reported that the Agno is mainly expressed in the cytoplasm of infected cells (Okada et al, 2001), whereas HP1α is expressed in the nucleus. To examine where Agno and HP1α become associated, a mutant of Agno lacking most of the N-terminal HP1α-binding motif, designated C18, was constructed. Subcellular localizations of wild-type (WT) and a C18 mutant of Agno were predominantly observed in the cytoplasm (Fig 2A). Immunoprecipitation analysis showed that C18 did not interact with HP1α, whereas WT Agno bound to HP1α (Fig 2B). The C18 showed cytoplasmic distribution that was similar to WT; however, the C18 did not colocalize with lamin A/C at the NE (Fig 2C). These results indicated that the interaction between Agno and HP1α occurs at the NE.
Figure 2.
Impaired ability of an Agno mutant to colocalize or interact with HP1. (A) Schematic representation of GST–EGFP fusion constructs of wild-type (WT) and mutant (C18) forms of Agno and EGFP fluorescence images showing their subcellular localizations in transfected HEK293 cells. (B) Lysates of 293T cells transiently expressing the WT or C18 forms of Agno were subjected to immunoprecipitation (IP) with anti-HP1α, and the resulting precipitates were subjected to immunoblotting (IB) with anti-Agno or anti-HP1α. (C) Immunofluorescence analysis of the expression of GST–EGFP-fused WT or C18 forms of Agno and of endogenous Lamin A/C in HEK293 cells. Enlarged dotted rectangles of the merged images are represented. Scale bars, 5 μm.
Agno disrupts the interaction between HP1α and LBR
Some HP1 proteins may be partially localized in the perinuclear region and this may be because of LBR binding (Minc et al, 1999). Our yeast two-hybrid analysis indicated that Agno interacts with the CSD of HP1, which is also responsible for the association of these proteins with LBR (Ye et al, 1997). These observations led us to propose that Agno might interfere with the interaction between HP1 and LBR and thereby alter the structure of the NE.
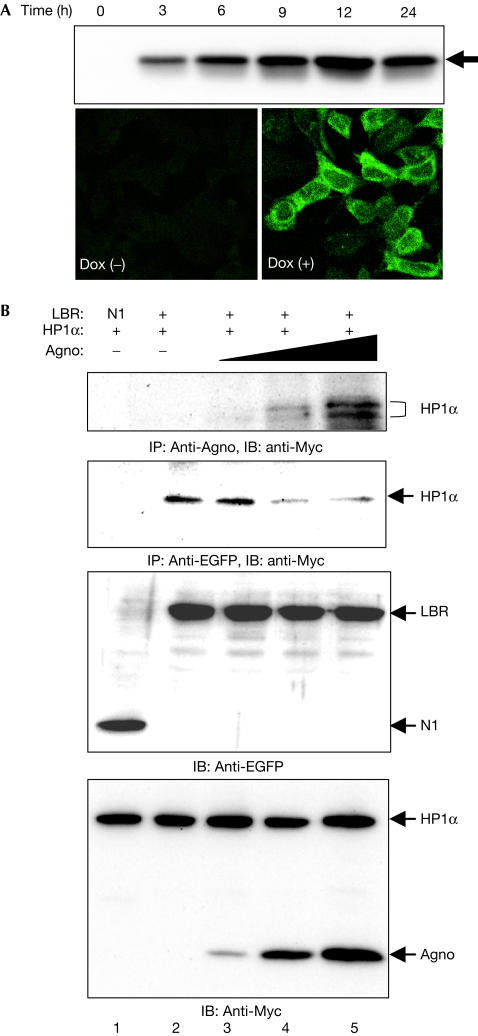
To examine this possibility, we established a cell line, designated 293AG, that is derived from HEK293 cells and in which the expression of JCV Agno subcloned into pcDNA4/TO/Myc–His is under the control of a tetracycline-responsive promoter. All 293AG cells expressed the recombinant Agno (∼14 kDa) in 3 h of exposure to doxycycline (Dox), and the level of expression increased gradually with time (Fig 3A). In the absence of Dox, Agno was not detected by immunoblot or immunofluorescence analysis.
Figure 3.
Agno-induced dissociation of the HP1α–LBR complex in vivo. (A) 293AG cells were incubated with Dox for the indicated times, after which cell lysates were subjected to immunoblotting with anti-Agno. Cells incubated with or without Dox for 24 h were also subjected to immunofluorescence analysis with anti-Agno. (B) 293AG cells were transfected with pEGFP-N1 or pLBR–EGFP together with pCMV–MycHP1α. The transfected cells were treated with Dox for 0, 3, 7 or 24 h. Cell lysates were then subjected to immunoprecipitation (IP) with anti-Agno or anti-EGFP, and the resulting precipitates were subjected to immunoblotting (IB) with anti-Myc. Cell lysates were also subjected directly to immunoblotting with anti-EGFP or anti-Myc.
We next transfected 293AG cells with a vector for Myc-epitope-tagged HP1α (pCMV–MycHP1α) and subjected lysates prepared from Dox-treated or untreated cells to immunoprecipitation with antibodies to Agno. Immunoblot analysis with antibodies to Myc of precipitates prepared from the Dox-treated cells, but not of those from the untreated cells, showed the presence of HP1α (Fig 3B). We then transfected 293AG cells with both pCMV–MycHP1α and pLBR–EGFP (enhanced green fluorescent protein), and induced Agno expression with Dox treatment. The amount of HP1α that co-precipitated with Agno in these cells was directly related to the level of Agno expression (Fig 3B). Conversely, the amount of HP1α that co-precipitated with LBR–EGFP was inversely related to the level of Agno expression.
Agno increases the lateral mobility of LBR in the NE
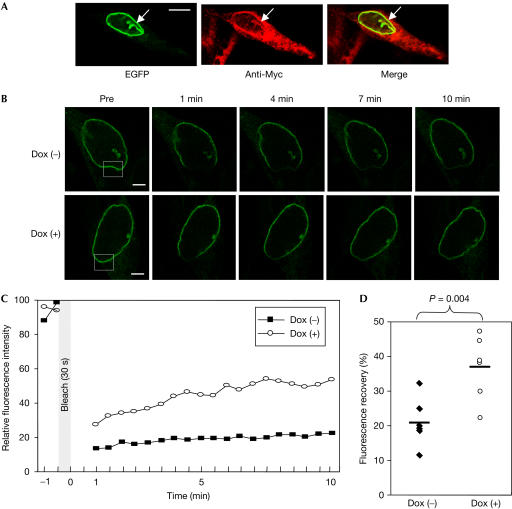
Immunofluorescence analysis also showed that Agno colocalized with LBR–EGFP at the NE, and this colocalization was pronounced in invaginated regions of the NE (Fig 4A). The dissociation of LBR from heterochromatin results in structural changes in the NE, including the formation of invaginations and protrusions (Mazlo et al, 2001); such changes are also characteristic of JCV-infected oligodendrocytes in the brains of patients with progressive multifocal leukoencephalopathy.
Figure 4.
Agno-induced increase in the lateral mobility of LBR. (A) Colocalization of LBR–EGFP detected by EGFP fluorescence and Agno detected with anti-Myc in 293AG cells. Arrows indicate an invagination of the NE in which LBR–EGFP and Agno are colocalized. Scale bar, 10 μm. (B) FRAP analysis of 293AG cells expressing LBR–EGFP and incubated in the absence or presence of Dox for 24 h. The fluorescence of LBR–EGFP in the boxed regions of the nuclear rim was irreversibly photobleached, and the recovery of fluorescence in these regions was monitored for 10 min. Representative images before (Pre) and at 1, 4, 7 and 10 min after bleaching are shown. Scale bars, 2 μm. (C) Quantification of the fluorescence recovery shown in (B). (D) Summary of fluorescence recovery ratios at 10 min after photobleaching. Each point represents an individual cell and the horizontal bars indicate the median values. The statistical significance of the difference between the two mean values was calculated by Student's two-tailed t-test.
Disruption of the interaction between HP1 and LBR by Agno might be expected to increase the lateral mobility of LBR in the NE. We examined this possibility by fluorescence recovery after photobleaching (FRAP) analysis in 293AG cells stably expressing LBR–EGFP. Photobleaching was induced at the nuclear rim, and recovery of fluorescence was monitored with a series of low-intensity scans. LBR–EGFP fluorescence in the bleached region of the nuclear rim recovered faster in cells expressing Agno than in those not exposed to Dox (Fig 4B,C); the mean recovery ratio at 10 min after photobleaching was significantly higher in the presence of Agno (Fig 4D). We also subjected the EGFP-fused full-length LBR (Haraguchi et al, 2000) to the FRAP analyses, and obtained results similar to those using the LBR–EGFP (data not shown). These observations indicated that Agno-induced dissociation of the LBR–HP1 complex resulted in an increase in the lateral mobility of LBR in the INM.
Agno facilitates nuclear egress of progeny virions
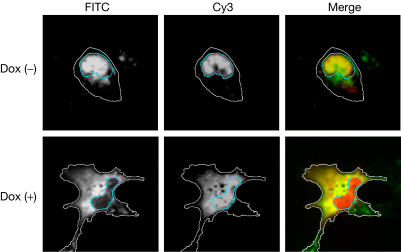
We proposed that the alteration of the NE induced by Agno might facilitate the nuclear release of progeny virions. To examine this possibility, we microinjected virus-like particles (VLPs) consisting of recombinant JCV VP1 into the nuclei of 293AG cells with or without Dox treatment. Such VLPs manifest a virion-like structure and physiological functions, including cellular attachment, intracytoplasmic transport and nuclear entry (Goldmann et al, 1999; Suzuki et al, 2001; Komagome et al, 2002; Qu et al, 2004), similar to those of JCV virions. We used Cy3 as a nuclear injection marker, which usually remained at the injection site. In the absence of Agno expression, most of the VLPs injected into the nucleus remained there 1 h later (Fig 5). In contrast, VLP fluorescence was not detected at this time in the nucleus of cells expressing Agno, suggesting that Agno promoted translocation of VLPs from the nucleus to the cytoplasm. Simultaneously, a certain amount of Cy3 also leaked into the cytoplasm in Dox (+) cells; however, it remained in the nucleus without Dox treatment. These observations indicate that the alteration of NE by Agno might facilitate nuclear egress of the progeny virions, and considering the behaviour of Cy3, this effect was not specific to virus particles.
Figure 5.
Agno facilitates the nuclear egress of VLPs. The nuclei of Dox-treated (lower panels) or untreated (upper panels) 293AG cells were microinjected with VLPs and Cy3 (injection site marker; red fluorescence), and, after 1 h, the cells were subjected to immunofluorescence analysis with anti-VP1 (green). Blue lines indicate the rim of the nuclei and white lines indicate the edge of the cytoplasm.
Discussion
The INM is linked to lamina and chromatin through interactions of its integral membrane proteins, including LBR. In the late stage of JCV infection, the nuclei of infected cells are filled with progeny virions and show structural alterations of the NE, such as invaginations and protrusions (Mazlo et al, 2001). We have shown that JCV Agno competes with LBR for binding to HP1, resulting in destabilization of the NE. Although viral protein-induced destabilization of the NE and a role for LBR have previously been described in herpesvirus infection (Scott & O'Hare, 2001; Muranyi et al, 2002), the role of LBR in this instance might be an indirect consequence of the depolymerization of lamin A/C that results from the virus-induced recruitment of protein kinase C to the INM. The association between LBR and its main ligand lamin B is also preserved during herpesvirus infection. LBR binds to a range of proteins (Ye & Worman, 1994, 1996), of which chromatin and chromatin-related proteins are thought to function as nucleoplasmic ligands that immobilize LBR in the INM (Ellenberg et al, 1997). LBR thus has a key role in the association of HP1 with the NE. Our data show that JCV Agno induces dissociation of HP1 from LBR by competitive binding to HP1, which caused an increase in the lateral mobility of LBR. This is the first evidence, to our knowledge, of a viral-protein-induced alteration of the NE through dissociation of HP1 and LBR.
In addition, expression of Agno also facilitated the nuclear export of VLPs without inducing nucleolysis, which is thought to mimic the nuclear egress of progeny virions in virus-infected cells. Consistent with this notion, agnogene-deficient simian virus 40 (SV40) shows a reduced capacity for cell-to-cell spread (Resnick & Shenk, 1986). Furthermore, we recently showed by RNA interference that JCV Agno is required for viral propagation (Orba et al, 2004). Together, our observations suggest that Agno facilitates viral propagation by promoting viral spreading. This function of JCV Agno is probably shared by agnoproteins of SV40 and human polyomavirus BK (BKV), given that the N-terminal region of JCV Agno is highly homologous to those of the SV40 and BKV proteins and is responsible for binding to HP1α.
Methods
Plasmid construction. A cDNA encoding the first 238 amino acids of LBR was amplified by PCR from an HEK293 cell cDNA library and subcloned into pEGFP-N1 (BD Biosciences Clontech, Franklin Lakes, NJ, USA), yielding pLBR–EGFP (Gerlich et al, 2001; Beaudouin et al, 2002). In addition, the pEGFP vector containing the full-length LBR, designated as hLBR in pEGFP-N3, was provided by Dr Haraguchi (Haraguchi et al, 2000). A full-length HP1α cDNA was also amplified from the HEK293 cell cDNA library and was subcloned into pCMV–Myc (Clontech). For expression of JCV Agno, the cDNA was subcloned into either pGST–EGFP.pcDNA4HisMax (GST: glutathione-S-transferase) or pcDNA4/TO/Myc–His (Invitrogen, Carlsbad, CA, USA).
FRAP analysis. We performed the FRAP analysis according to Ellenberg et al (1997) and Scott & O'Hare (2001), with several technical modifications, because the performance of our laser scanning microscope (LSM) system was different from the LSM systems that they used. As the laser intensity of our LSM system was much lower than their system, our bleaching time was longer than their experiments. FRAP analysis of 293AG cells stably expressing LBR–EGFP was performed with the Olympus FV300 confocal microscope fitted with a × 60 oil-immersion objective (numerical aperture, 1.25). The cells (2 × 105) were plated on 35-mm dishes coated with type I collagen and were incubated with or without Dox for 24 h before analysis. Images were recorded at 488 nm (at 3% power) and an intense pulse of laser light (100% power) was then applied for 30 s to bleach the fluorescent molecules in a defined region (2 × 2 μm). Recovery of fluorescence was monitored at 30 s intervals for 10 min by scanning with attenuated laser light (3% power). The FV300 software was used to generate a time series from the images and to plot the pixel intensity in the defined regions of interest for the duration of the experiment. At each time point, the intensity of an equivalent background area was subtracted and values were normalized for total loss of fluorescence. The relative recovery of fluorescence intensity was calculated as 100% × ((intensity at the recovery plateau)−(intensity at 1 min after bleaching))/((average intensity of the bleached region before bleaching)−(intensity at 1 min after bleaching)). We also subjected the pEGFP vector containing the full-length LBR (provided by Dr Haraguchi) to the FRAP analyses (Haraguchi et al, 2000).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400406s1.pdf).
Supplementary Material
Supplementary Data
Acknowledgments
We thank M. Satoh and M. Sasada for technical assistance; A. Kihara, M. Matsuda and N. Mochizuki for helpful suggestions; and T. Haraguchi for the hLBR in pEGFP-N3 plasmid. Y.O. is a research fellow of the Japan Society for the Promotion of Science. This study was supported in part by grants from the Ministry of Education, Science, Technology, Sports, and Culture of Japan, the Ministry of Health, Labor, and Welfare of Japan, the Japan Human Science Foundation.
References
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J (2002) Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108: 83–96 [DOI] [PubMed] [Google Scholar]
- Darbinyan A, Siddiqui KM, Slonina D, Darbinian N, Amini S, White MK, Khalili K (2004) Role of JC virus agnoprotein in DNA repair. J Virol 78: 8593–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincottschwartz J (1997) Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 138: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Gebhard M, Ellenberg J, Eils R (2001) Four-dimensional imaging and quantitative reconstruction to analyse complex spatiotemporal processes in live cells. Nat Cell Biol 3: 852–855 [DOI] [PubMed] [Google Scholar]
- Goldmann C et al. (1999) Molecular cloning and expression of major structural protein VP1 of the human polyomavirus JC virus: formation of virus-like particles useful for immunological and therapeutic studies. J Virol 73: 4465–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci 113: 779–794 [DOI] [PubMed] [Google Scholar]
- Komagome R, Sawa H, Suzuki T, Suzuki Y, Tanaka S, Atwood WJ, Nagashima K (2002) Oligosaccharides as receptors for JC virus. J Virol 76: 12992–13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazlo M, Ressetar HG, Stoner GL (2001) The neuropathology and pathogenesis of progressive multifocal leukoencephalopathy. Hum Polyomaviruses: Mol and Clin Perspect 257–335 [Google Scholar]
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108: 220–234 [DOI] [PubMed] [Google Scholar]
- Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH (2002) Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297: 854–857 [DOI] [PubMed] [Google Scholar]
- Okada Y, Endo S, Takahashi H, Sawa H, Umemura T, Nagashima K (2001) Distribution and function of JCV agnoprotein. J Neurovirol 7: 302–306 [DOI] [PubMed] [Google Scholar]
- Okada Y, Sawa H, Endo S, Orba Y, Umemura T, Nishihara H, Stan AC, Tanaka S, Takahashi H, Nagashima K (2002) Expression of JC virus agnoprotein in progressive multifocal leukoencephalopathy brain. Acta Neuropathol (Berl) 104: 130–136 [DOI] [PubMed] [Google Scholar]
- Orba Y, Sawa H, Iwata H, Tanaka S, Nagashima K (2004) Inhibition of virus production in JC virus-infected cells by postinfection RNA interference. J Virol 78: 7270–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M (2002) Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Sawa H, Suzuki T, Semba S, Henmi C, Okada Y, Tsuda M, Tanaka S, Atwood WJ, Nagashima K (2004) Nuclear entry mechanism of the human polyomavirus JC virus-like particle: role of importins and the nuclear pore complex. J Biol Chem 279: 27735–27742 [DOI] [PubMed] [Google Scholar]
- Resnick J, Shenk T (1986) Simian virus 40 agnoprotein facilitates normal nuclear location of the major capsid polypeptide and cell-to-cell spread of virus. J Virol 60: 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Enarson P, Raharjo WH, Burke B (2001) Nuclear envelope dynamics. Biochem Cell Biol 79: 533–542 [PubMed] [Google Scholar]
- Scott ES, O'Hare P (2001) Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J Virol 75: 8818–8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Sawa H, Komagome R, Orba Y, Yamada M, Okada Y, Ishida Y, Nishihara H, Tanaka S, Nagashima K (2001) Broad distribution of the JC virus receptor contrasts with a marked cellular restriction of virus replication. Virology 286: 100–112 [DOI] [PubMed] [Google Scholar]
- Ye Q, Worman HJ (1994) Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J Biol Chem 269: 11306–11311 [PubMed] [Google Scholar]
- Ye Q, Worman HJ (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem 271: 14653–14656 [DOI] [PubMed] [Google Scholar]
- Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ (1997) Domainspecific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem 272: 14983–14989 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data