Abstract
The frequency of recovery of atypical mycobacteria was estimated in two treatment plants providing drinking water to Paris, France, at some intermediate stages of treatment. The two plants use two different filtration processes, rapid and slow sand filtration. Our results suggest that slow sand filtration is more efficient for removing mycobacteria than rapid sand filtration. In addition, our results show that mycobacteria can colonize and grow on granular activated carbon and are able to enter distribution systems. We also investigated the frequency of recovery of mycobacteria in the water distribution system of Paris (outside buildings). The mycobacterial species isolated from the Paris drinking water distribution system are different from those isolated from the water leaving the treatment plants. Saprophytic mycobacteria (present in 41.3% of positive samples), potentially pathogenic mycobacteria (16.3%), and unidentifiable mycobacteria (54.8%) were isolated from 12 sites within the Paris water distribution system. Mycobacterium gordonae was preferentially recovered from treated surface water, whereas Mycobacterium nonchromogenicum was preferentially recovered from groundwater. No significant correlations were found among the presence of mycobacteria, the origin of water, and water temperature.
Environmental mycobacteria, which are also called atypical mycobacteria or nontuberculous mycobacteria (NTM), are common saprophytes in all natural ecosystems, including water, soil, food, dust, and aerosols (8, 9, 11, 16, 23, 37, 59). Some species are also pathogenic for humans or animals, causing pulmonary and cutaneous disease, lymphadenitis, and disseminated infections (11, 19, 36, 55, 57). These organisms are an increasing health risk, especially in the growing immunodeficient population. Before the introduction of protease inhibitors for antiretroviral therapy, disseminated infections due to NTM, especially Mycobacterium avium, were frequent in AIDS patients. NTM infection is currently one of the criteria used to diagnose AIDS in human immunodeficiency virus-infected patients (20).
NTM infections are transmitted by ingestion, inhalation, and inoculation from environmental sources rather than from person to person (11, 57). The environmental sources may include aerosols, water, soil, dust, food, and equipment. The mode of transmission can be identified only when the same Mycobacterium species is isolated from both a source and a patient, but mycobacteria are rarely isolated from a putative infection source. There is increasing evidence which suggests that tap water is the vehicle by which mycobacteria infect or colonize the human body (3, 4, 15, 34, 41, 47, 52-54, 56). In a number of cases, mycobacterial species (M. kansasii, M. fortuitum, M. chelonae, M. avium, and M. xenopi) have been recovered from tap water some time after they were recovered from patients (2, 3, 22, 26, 52, 58). Mycobacteria can contaminate inadequately disinfected equipment used for clinical investigations or surgery, resulting in nosocomial infections (2, 4, 53, 54, 56, 60). The presence of mycobacteria in tap water may lead to pseudoinfection diagnoses (i.e., false-positive cases related to isolation of tap water mycobacteria exogenous to clinical samples). Moreover, pseudoinfection can be traced to laboratory contamination, which can occur at any stage of the laboratory procedures used to isolate mycobacteria. Any fluid reservoir that is not routinely and effectively decontaminated and sterilized can act as a source of organisms. Pseudoinfection caused by contaminated bronchoscopes should be suspected when mycobacterial species are more frequently isolated from bronchoscopy specimens than from other bronchopulmonary specimens (14, 32, 35, 38).
Mycobacteria have been isolated from public water distribution systems and from samples from various other sources, including home distribution systems, hot and cold water taps, ice machines, heated nebulizers, and showerhead sprays (6, 9, 11, 15, 26, 31, 44, 53). They may be present at high densities in biofilms on the insides of pipes and taps (45). NTM can colonize, survive, persist, grow, and multiply in tap water (6). Mycobacteria are not killed by common disinfectants and can tolerate wide ranges of pHs and temperatures, which allows them to persist in drinking water systems for long periods of time (6, 11, 28, 40).
According to the guidelines of the World Health Organization, a European Commission directive (European Union Council Directive 98/83/EC) states that drinking water should not contain pathogenic microorganisms in a quantity or at a concentration able to adversely affect human health. However, the routine bacteriological examinations that are carried out with drinking water do not include a search for NTM. Moreover, few publications have reported isolation and identification of environmental mycobacteria from drinking water networks (9, 12, 17, 51).
The frequencies of recovery of mycobacteria from two treatment plants at some intermediate stages of treatment and from the water distribution system of Paris were investigated in this study. Between March 2000 and July 2001, water samples were collected from 12 sites located across Paris and from the treatment lines of two treatment plants in the Paris area. The aims of this study were (i) to estimate the frequency of recovery of NTM in the distribution system, (ii) to determine whether there were any relationships between the presence and distribution of mycobacterial species and the origin of the water (treated surface water or groundwater), (iii) to evaluate the efficiencies of different processes for removing mycobacteria, and (iv) to determine whether the source of the mycobacteria recovered from treated water in the distribution system could be traced to treatment plants.
(This work is part of the doctoral thesis of C. Le Dantec.)
MATERIALS AND METHODS
Collection of water samples.
Water samples were collected every month between July 2000 and July 2001 from 12 sites within the distribution system of the city of Paris (Fig. 1). All of the collection sites were located outside buildings. Paris is supplied by groundwater and treated surface water. Depending on the geographical location, three different types of areas can be defined: areas supplied specifically by groundwater, areas supplied specifically by treated water from a river, and areas supplied nonspecifically by groundwater, treated water, or a mixture of the two (depending on the management of the distribution system, which is highly cross-connected).
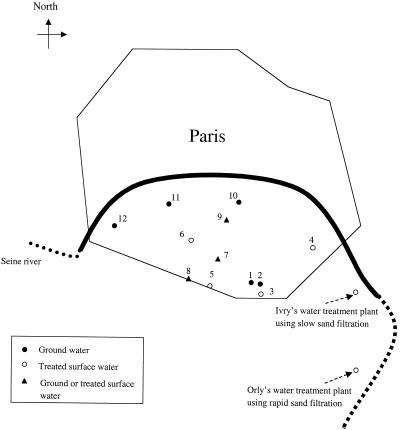
FIG. 1.
Locations of the 12 sampling sites within the Paris water distribution system. All the sites were located outside buildings. The origins of the water collected at the 12 selected sites are indicated. The locations of the two water treatment plants are indicated by arrows.
Groundwater samples were collected from sites 1, 2, 10, 11, and 12. The groundwater, which comes from natural sources located 150 km from Paris, is treated with chlorine before it is transported to Paris through an aqueduct. This type of water does not enter water treatment plants but is transferred directly into the distribution system. Treated surface water samples, from the water treatment plants described below, were collected from sites 3 to 6. Sites 7 to 9 were supplied by groundwater, treated water, or a mixture of the two as described above. Samples could not be collected in November 2000. In January 2001, culture results could not be interpreted because of technical problems with the incubation of NTM cultures.
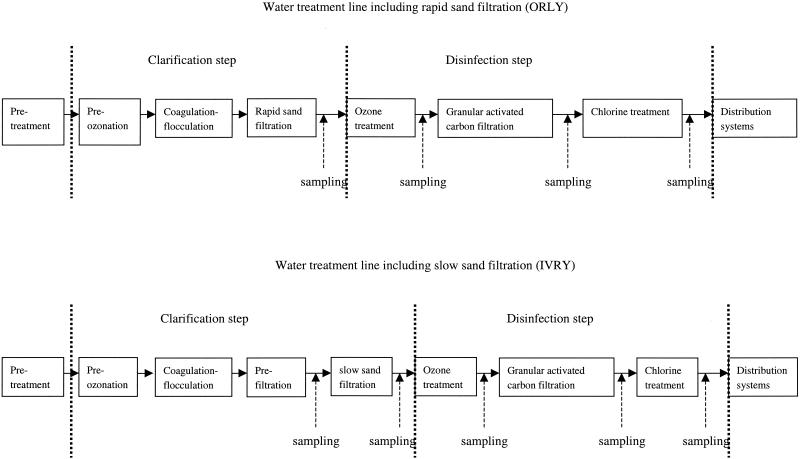
Water samples were collected every 3 months between March 2000 and July 2001 from several sites in two treatment plants. The treatment plants are located at Orly and Ivry on the Seine River and use different preliminary treatment processes. Samples were collected from the Orly plant after clarification (which consists of preozonation and coagulation-flocculation combined with rapid sand filtration), ozonation, granular activated carbon (GAC) filtration, and chlorination (Fig. 2). Samples were collected from the Ivry plant after prefiltration, slow sand filtration, ozonation, GAC filtration, and chlorination (Fig. 2). The major difference between the two water treatment plants is the type of sand filtration, which is rapid at Orly and slow at Ivry.
FIG. 2.
Schematic diagram of the two treatment plants in which different preliminary treatment processes (rapid and slow sand filtration processes) are used. The dotted lines delimit the main clarification and disinfection steps. The locations of the sampling sites in the two treatment lines are indicated by dashed arrows.
All samples either from treatment plants or from the distribution system were collected in sterile, 1-liter, wide-mouth plastic bottles containing 2 g of sodium thiosulfate to neutralize chlorine at the ambient temperature. The bottles were dipped into the water and filled completely before they were transported to the Hygiene Laboratory of Paris. Samples were always processed on the day of collection. The temperature of each sample was determined with a mercury thermometer with a 1°C scale.
Processing of water samples.
One liter of water was passed through a membrane filter (pore size, 0.45 μm; Millipore). If the sample was too turbid, a smaller sample (100 ml) was filtered. Turbid samples were obtained at the early stages at the treatment plants (prefiltration at the Ivry plant and rapid sand filtration at the Orly plant [see Tables 3 and 4]). The membrane was put into a sterile tube containing 10 ml of the water sample and was sonicated for 5 min at 47 Hz. The membrane was decontaminated by adding 15 ml of 3% sodium dodecyl sulfate and 1% NaOH and was shaken for 30 min at room temperature. The pH was adjusted to 7 with 40% phosphoric acid. The membrane was removed, and the neutralized solution was centrifuged for 30 min at 3,000 × g. After centrifugation, most of the supernatant was removed. The sediment was then resuspended in about 2 ml of the remaining supernatant and plated on Lowenstein-Jensen medium. For every sample, 10 tubes were inoculated with 0.2 ml of the resuspended sediment. The tubes were incubated at 30, 37, and 42°C and examined every 2 days for the first 10 days and once a week thereafter for 3 months. The number of colonies per liter of original sample and the colony morphology were noted. After Ziehl-Neelsen staining, acid-fast bacilli were subcultured on Lowenstein-Jensen medium. Mycobacteria were identified by molecular methods as described below.
TABLE 3.
Concentrations of mycobacteria isolated from water from the Orly treatment plant at some intermediate stages of treatment, in which rapid sand filtration is used
| Date (mo/day/yr) | Process | Organism(s) | Concn (CFU · ml−1) |
|---|---|---|---|
| 6/20/2000 | Rapid sand filtration | M. peregrinum | >20,000 |
| M. gordonae | >6,000 | ||
| Ozone treatment | NDa | ||
| GAC filtration | Unidentified mycobacteriab | 20 | |
| Treated water (chlorination) | Unidentified mycobacteria | 20 | |
| 9/19/2000 | Rapid sand filtration | Unidentified mycobacteria | 20 |
| M. gordonae | 3,000 | ||
| M. gordonae | 400 | ||
| Ozone treatment | ND | ||
| GAC filtration | Unidentified mycobacteriab | 20 | |
| Unidentified mycobacteriab | 10 | ||
| Treated water (chlorination) | ND | ||
| 12/19/2000 | Rapid sand filtration | Unidentified mycobacteria | 3,000 |
| Ozone treatment | ND | ||
| GAC filtration | Unidentified mycobacteriab | 10 | |
| Treated water (chlorination) | Unidentified mycobacteriab | 10 | |
| 3/20/2001 | Rapid sand filtration | M. gordonae | 500 |
| Ozone treatment | ND | ||
| GAC filtration | Unidentified mycobacteriab | 10 | |
| Treated water (chlorination) | ND | ||
| 6/19/2001 | Rapid sand filtration | M. gordonae | 100 |
| Ozone treatment | M. gordonae | 10 | |
| GAC filtration | Unidentified mycobacteriab | 20 | |
| Treated water (chlorination) | ND |
ND, not detected.
Identical mycobacteria.
TABLE 4.
Concentrations of mycobacteria isolated from the water from the Ivry treatment plant at some intermediate stages of treatment, in which slow sand filtration is used
| Date (mo/day/yr) | Process | Organism(s) | Concn (CFU · liter−1) |
|---|---|---|---|
| 3/28/2000 | Prefiltration | NDa | |
| Slow sand filtration | ND | ||
| Ozone treatment | ND | ||
| GAC filtration | M. peregrinum | >3,000 | |
| Treated water (chlorination) | ND | ||
| 7/18/2000 | Prefiltration | Contaminated | |
| Slow sand filtration | Unidentified mycobacteria | 10 | |
| Ozone treatment | M. nonchromogenicum | 10 | |
| GAC filtration | M. fortuitum | >>3,000 | |
| Treated water (chlorination) | ND | ||
| 10/17/2000 | Prefiltration | M. nonchromogenicum | 100 |
| Slow sand filtration | M. peregrinum | 800 | |
| M. flavescens | 10 | ||
| M. chelonae | 10 | ||
| Ozone treatment | ND | ||
| GAC filtration | M. fortuitum-M. peregrinum | 2,000 | |
| Treated water (chlorination) | ND | ||
| 1/23/2001 | Prefiltration | M. duvalii | 20 |
| Unidentified mycobacteria | 300 | ||
| Unidentified mycobacteria | 2,000 | ||
| M. flavescens | 20 | ||
| Slow sand filtration | M. peregrinum | >400 | |
| Ozone treatment | ND | ||
| GAC filtration | M. peregrinum | >3,000 | |
| Treated water (chlorination) | M. nonchromogenicum | 60 | |
| 4/17/2001 | Prefiltration | Contaminated | |
| Slow sand filtration | Unidentified mycobacteria | 10 | |
| Ozone treatment | ND | ||
| GAC filtration | M. peregrinum | 1,500 | |
| Treated water (chlorination) | ND | ||
| 7/17/2001 | Prefiltration | Contaminated | |
| Slow sand filtration | Contaminated | ||
| Ozone treatment | ND | ||
| GAC filtration | M. fortuitum | 2,000 | |
| Unidentified mycobacteria | 10 | ||
| Treated water (chlorination) | ND |
ND, not detected.
The detection limit of this decontamination method was determined by using cultures of M. gordonae. One liter of tap water, sterilized by filtration, was inoculated with a culture of M. gordonae to obtain final concentrations of 10, 102, 103, and 104 CFU · liter−1. The exact number of cells in each water culture was checked by plating on Middlebrook 7H11 plates. Water samples were treated as described above. Cell counting showed that the decontamination process reduced the number of mycobacteria to 1% of the original number.
Molecular identification techniques. (i) Sample preparation.
Nucleic acids were extracted by lysing bacterial cells. Briefly, colonies were transferred to a 2-ml microcentrifuge tube and incubated in 10 mM Tris-1 mM EDTA for 20 min at 80°C to inactivate the mycobacteria. The sample was centrifuged at 10,000 × g, and 5 μl of the thermolysate was used in a PCR.
(ii) Analysis of the 16S rDNA gene.
A 1,500-bp fragment corresponding to the 16S ribosomal DNA (rDNA) gene was amplified by PCR. PCR was performed with a Perkin-Elmer 480 thermal cycler by using a 50-μl reaction mixture containing 5 μl of crude DNA extract, 1× Taq buffer (Perkin-Elmer), 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.25 mM, 1.25 U of Taq polymerase (Perkin-Elmer), 0.5 μM primer 264, and 0.5 μM primer 285 (24). The PCR conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, 60°C for 1 min, and 70°C for 1 min and a final 10-min extension at 72°C. Amplicons were detected by electrophoresis in 1.5% agarose gels, followed by ethidium bromide staining.
PCR products were sequenced with sequencing primers 244 and 259 (24) by using an automatic sequencer, the dideoxy chain termination method (43), a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Perkin-Elmer Corp., Foster City, Calif.), a GenAmp 9600 PCR system (Perkin-Elmer), and a 373 stretch DNA analysis system (Applied Biosystems). Nucleotide sequences were compared to known sequences in the GenBank database by using the BlastN algorithm (1).
(iii) Analysis of the hsp65 gene.
The hsp65 gene was investigated by performing PCR restriction analysis as described by Telenti et al. (49). The PCR conditions were the same as those described above. The PCR products were sequenced with sequencing primer Tb11 (5′-ACCAACGATGGTGTGTCCAT-3′) (49).
(iv) Test for specific identification of M. gordonae.
M. gordonae was identified by amplifying the specific internal transcribed spacer region as described by Park et al. (39). The primers used to amplify this region were GORF (5′-CGACAACAAGCTAAGCCAGA-3′) and GORR (5′-CAACGCATACATTTTGATGC-3′).
RESULTS
Frequency of recovery of mycobacteria in the water distribution system.
A total of 72% of the water samples from the water distribution system (104 of 144 samples) were positive for mycobacteria; 68% of the groundwater samples (41 of 60 samples), 69% of the treated surface water samples (33 of 48 samples), and 83% of the samples having alternate origins (either groundwater or treated surface water) (30 of 36 samples) were positive for mycobacteria.
Concentrations between 1 and 50 CFU · liter−1 were found in 78% of the samples, concentrations between 51 and 500 CFU · liter−1 were found in 21% of the samples, and concentrations of more than 500 CFU · liter−1 were found in only one sample (Table 1). A concentration higher than 1,000 CFU · liter−1 was never found. Considering the low rate of recovery of mycobacteria (1%) after our decontamination procedure, we estimated that no water sample contained more than 105 CFU · liter−1. Of the 78% of the samples having mycobacterial concentrations between 1 and 50 CFU · liter−1, 34% contained saprophytic mycobacteria, 15% contained potentially pathogenic mycobacteria, and 51% contained unidentified mycobacteria (Table 1). Similarly, of the 21% of the samples having mycobacterial concentrations between 51 and 500 CFU · liter−1, 32% contained saprophytic mycobacteria, 14% contained potentially pathogenic mycobacteria, and 54% contained unidentified mycobacteria (Table 1).
TABLE 1.
Numbers and percentages of saprophytic, potentially pathogenic, and unidentified mycobacteria
| Organisms | 1-50 CFU · liter−1
|
51-50 CFU · liter−1
|
>500 CFU · liter−1
|
|||
|---|---|---|---|---|---|---|
| No. of positive samplesa | % | No. of positive samples | % | No. of positive samples | % | |
| Saprophytic mycobacteria | 35 | 34 | 9 | 32 | 0 | 0 |
| Potentially pathogenic mycobacteria | 15 | 15 | 4 | 14 | 0 | 0 |
| Unidentified mycobacteria | 52 | 51 | 15 | 54 | 1 | 100 |
| All mycobacteria | 102 | 78 | 28 | 21 | 1 | 1 |
One sample could contain different mycobacterial species.
Saprophytic mycobacteria accounted for 41% of all positive samples (Table 2). These saprophytic mycobacteria included both slow- and fast-growing mycobacteria, such as M. gordonae (29% of all positive samples), M. nonchromogenicum (11%), M. aurum (1%), and M. gadium (1%) (Table 2). Potentially pathogenic mycobacteria accounted for 16% of all positive samples. Most of these were fast-growing mycobacteria, including M. fortuitum (5% of the positive samples), M. peregrinum (6%), and M. chelonae (7%). The slow-growing organism M. intracellulare was detected in only one sample (Table 2). The other strains (55%) could not be identified as their 16S rDNA and hsp65 gene sequences did not consistently match sequences in the databases. Most of these organisms grew at 30°C. When all positive samples were considered, M. gordonae and M. nonchromogenicum were the most frequently identified species isolated from the water distribution system (Table 2).
TABLE 2.
Numbers and percentages of samples positive for mycobacterial species isolated from groundwater or treated surface water or from mixed water (ground or treated surface water samples) collected from the Paris water distribution system
| Species or group | Groundwater (41 positive samples)
|
Treated surface water (33 positive samples)
|
Mixture (ground or treated surface water) (30 positive samples)
|
All samples (104 positive samples)a
|
||||
|---|---|---|---|---|---|---|---|---|
| No. of positive samples | % | No. of positive samples | % | No. of positive samples | % | No. of positive samples | % | |
| Saprophytic mycobacteria | ||||||||
| M. gordonae | 4 | 9.8 | 16 | 48.5 | 10 | 33 | 30 | 28.8 |
| M. nonchromogenicum | 8 | 19.5 | 0 | 0.0 | 4 | 13 | 12 | 11.5 |
| M. aurum | 0 | 0.0 | 1 | 3.0 | 0 | 0 | 1 | 1.0 |
| M. gadium | 0 | 0.0 | 1 | 3.0 | 0 | 0 | 1 | 1.0 |
| Potentially pathogenic mycobacteria | ||||||||
| M. fortuitum | 3 | 7.3 | 1 | 3.0 | 1 | 3 | 5 | 4.8 |
| M. peregrinum | 3 | 7.3 | 0 | 0.0 | 3 | 10 | 6 | 5.8 |
| M. chelonae | 2 | 4.9 | 3 | 9.1 | 3 | 10 | 8 | 7.7 |
| M. intracellulare | 1 | 2.4 | 0 | 0.0 | 0 | 0 | 1 | 1.0 |
| Unidentified mycobacteria | 34 | 82.9 | 17 | 51.5 | 20 | 67 | 57 | 54.8 |
When all samples were examined, the percentages of positive samples for the groups were as follows: saprophytic mycobacteria, 41.3%; potentially pathogenic mycobacteria, 16.3%; and unidentified mycobacteria, 54.8%.
Correlation between mycobacterial species and origins of the water samples in the distribution system.
The water samples collected from selected sites had different origins. At five sampling sites the water was groundwater, at four sites the water was treated surface water from treatment plants, and at three sites the water was either treated surface water or groundwater depending on the management of the distribution system, as described above (Fig. 1).
To determine the origins of the mycobacterial species in the water distribution system, the correlation between the presence of mycobacterial species and the origin of the water samples was determined (Table 2). The percentages of positive samples were similar for groundwater and treated surface water (see above), but the percentages of each species isolated in the two types of water were different (Table 2). M. gordonae was found more frequently in treated surface water samples (48% of positive samples) than in groundwater samples (10% of positive samples). M. nonchromogenicum was found in groundwater samples (19% of positive samples) and in samples containing both types of water. However, M. nonchromogenicum was not found in treated surface water. M. fortuitum and M. chelonae were found in both types of water. M. peregrinum and M. intracellulare were not found in treated surface water. Treated surface water samples were more likely to contain M. gordonae, whereas groundwater samples were more likely to contain M. nonchromogenicum.
Efficiencies of the two water treatment plants in which different filtration processes are used to remove mycobacteria.
Several mycobacterial species, including saprophytic and potentially pathogenic species, were isolated from the two water treatment plants. The saprophytic mycobacteria identified were M. gordonae, M. nonchromogenicum, M. flavescens, and M. duvalii, whereas the potentially pathogenic mycobacteria were M. peregrinum, M. fortuitum, and M. chelonae. The concentrations of mycobacteria in the two water treatment lines decreased dramatically (Tables 3 and 4). The concentration of mycobacteria was between 20 and 20,000 CFU · liter−1 after the clarification step at the Orly plant, but it was between 10 and 20 CFU · liter−1 at the end of the water treatment process. Similarly, after the prefiltration step at the Ivry plant the concentration of mycobacteria was between 20 and 2,000 CFU · liter−1, whereas at the end of the treatment process a maximum concentration of 60 CFU · liter−1 was detected. In addition, the frequencies of recovery of mycobacteria and the spectra of mycobacterial species (especially at the Ivry plant) at the end of the two treatment processes were lower than the frequencies of recovery of mycobacteria and the spectra of mycobacterial species after the first treatment step. These results suggest that water treatment plants efficiently remove mycobacteria.
At the Orly water treatment plant, where rapid sand filtration was used, all samples collected after rapid sand filtration of water from the Seine River were positive for mycobacteria (Table 3). Only two mycobacterial species were identified, M. gordonae and M. peregrinum; the other species were not identified. The concentrations of mycobacteria were found to be between 20 and 20,000 CFU · liter−1. At the Ivry water treatment plant, where slow sand filtration was used, four of six samples were positive for mycobacteria after the slow sand filtration step (Table 4). The concentrations of mycobacteria ranged from 10 to 800 CFU · liter−1, values which are lower than the values obtained after rapid sand filtration at the Orly plant.
At each treatment plant, only one sample collected after ozonation contained mycobacteria. These samples each had a low mycobacterial concentration (10 CFU · liter−1). The postozonation samples contained M. nonchromogenicum at the Ivry plant (Table 4) and M. gordonae at the Orly plant (Table 3). When ozonated water was filtered through GAC, all samples were positive for mycobacteria; an unidentified mycobacterial species was isolated at the Orly plant, and potentially pathogenic mycobacteria were isolated at the Ivry plant (Tables 3 and 4). The carbon filter itself was probably contaminated by these mycobacteria. In addition, after water was filtered through GAC, the concentrations of mycobacteria were between 10 and >3,000 CFU · liter−1 at the Ivry plant, whereas concentrations between 10 and 20 CFU · liter−1 were detected at the Orly plant. Several mycobacterial species were eliminated by the final chlorination step at the Ivry plant (Table 4). However, one sample from the Orly plant contained the same unidentified mycobacterial taxon at the same concentration in water collected after GAC filtration and in water collected after the final chlorination step (Table 3). This suggests that this unidentified mycobacterial taxon is highly resistant to chlorine.
At the Ivry plant, M. nonchromogenicum was isolated from a sample after chlorination, whereas this species could not be detected after previous treatment steps (Table 4). This unexpected isolation from a single sample was likely due to contamination during sampling or laboratory processing.
No mycobacteria were found in most water samples at the ends of the water treatment processes, indicating that both water treatment processes efficiently removed mycobacteria and that mycobacteria found in the Paris water distribution system did not come from the water treatment plants.
Effect of temperature on the presence of mycobacteria in water.
The temperature of the groundwater was between 10 and 16°C, whereas the temperature of the treated surface water was between 8 and 22°C, depending on the season.
No correlation between the percentage of samples positive for mycobacteria and temperature was found (data not shown). The percentage of positive treated surface water samples (50%) was the same whether the temperature of the samples was 8 or 22°C. Similarly, 60% of groundwater samples were positive for mycobacteria when the temperature of samples was 10 or 15°C.
DISCUSSION
Our study indicated that the Paris water distribution system is colonized by several species of mycobacteria, such as M. gordonae, M. nonchromogenicum, M. aurum, M. gadium, M. fortuitum, M. peregrinum, and M. chelonae. Interestingly, many unidentifiable mycobacteria, possibly belonging to new taxa, were found in the water distribution system and in the water treatment plants. The use of new molecular identification methods greatly improved our taxonomic information and allowed us to distinguish between organisms that had previously been included in roughly defined complexes. Moreover, identification of mycobacteria has become more accurate and complex since the introduction of highly discriminatory techniques because sequencing of conserved genes can reveal new characteristic sequences suggestive of new taxa (50).
Although our isolation method was reproducible, the decontamination procedure eliminated large quantities of mycobacteria, and the yield was low (1%, as determined for M. gordonae). No selective medium is available for mycobacteria, and the associated flora, consisting of bacteria and fungi, has to be eliminated by decontamination procedures involving alkali or acid treatment. Mycobacteria are more resistant to these treatments than bacteria or fungi, but they are not fully resistant. The number of mycobacteria in our samples may have been lower than the limit of sensitivity of the isolation method. Direct use of molecular identification methods with water samples may improve the sensitivity of detection and contribute to a better description of the whole mycobacterial flora. We are currently designing such a procedure.
The lack of detectable mycobacteria at the ends of the water treatment lines and the presence of mycobacteria in the treated water distribution system could be explained by the presence of biofilms in pipes. The capacity of mycobacteria to form biofilms has been demonstrated recently (18, 33, 42). In Germany, 90% of biofilm samples from pipes of various distribution systems contained mycobacteria, suggesting that mycobacterial biofilms are present in almost all collection and piping systems (45).
The efficiency of removal of mycobacteria was determined at two water treatment plants in which different filtration processes are used; slow sand filtration is used at the Ivry plant, and rapid sand filtration is used at the Orly plant. The slow sand filtration process reproduces the natural filtration that occurs in soil. Water flows through different filters consisting of terracotta balls and grains of sand of various diameters. A biomass, composed of phytoplankton, zooplankton, and bacteria living both symbiotically and as mutual predators, develops in the filters and acts as a biological filter. Bacterial activity in the filters removes biodegradable organic compounds, some of which are precursors of chlorinated organic substances (e.g., trihalomethanes) (7, 10, 13). The combination of coagulation-flocculation with filtration is effective against microorganisms that are difficult to eliminate, such as Giardia cysts and Cryptosporidium oocysts (30). No Cryptosporidium oocysts were detected in the treated water from two water treatment plants in which slow sand filters were used, even though this parasite was frequently isolated from the raw water entering the plants (5). Similarly, our results show that both rapid sand filtration and slow sand filtration remove mycobacteria. However, the concentrations of mycobacteria were higher after rapid sand filtration. The coliform concentrations were similar in all raw water samples, approximately 5 × 105 CFU · liter−1. Based on these data, and because the two plants are a few kilometers apart along the Seine River, we assumed that the raw waters entering the two treatment plants were similar and that the differences in recovery of mycobacteria at the ends of the plants were due to the water treatment processes. Our results (Tables 3 and 4) suggest that the slow sand filtration used at the Ivry plant is more efficient for removing mycobacteria than the rapid sand filtration used at the Orly plant.
Ozonation effectively reduced the mycobacterial load. However, low numbers of M. gordonae and M. nonchromogenicum were still detected after ozonation. This is consistent with the results of a previous study of the survival of bacteria after ozonation, which showed that Mycobacterium spp. were the predominant surviving bacteria along with some other gram-positive bacteria (29). Taylor et al. recently showed that M. avium is 100-fold more resistant to ozone than Escherichia coli and that water-grown cells are 10-fold more resistant than medium-grown cells (48). The mycobacteria that survive ozonation may be more hydrophobic and able to form aggregates, which decrease the inactivation efficiency of ozone.
After filtration through GAC, the water samples contained high levels of potentially pathogenic mycobacteria and few unidentified mycobacteria. As previously reported, our results suggest that GAC is colonized by bacteria (27). Similarly, Lee and Deininger showed that the percentage of opportunistic pathogens, including Mycobacterium species and species belonging to other genera, was dramatically reduced after ozonation, whereas it was increased after GAC filtration (29). However, no potentially pathogenic mycobacteria were found after chlorination in our study, whereas unidentified mycobacteria belonging to a single taxon were still found. Recently, we showed that the resistance of mycobacteria to chlorine varies widely according to species (28). It is likely that the single, possibly new taxon of mycobacteria recovered from the water treated at the Orly plant is unusually resistant to chlorine.
In our study, no correlation was found between water temperature and the types of mycobacterial species found in water. Our hypothesis is that these organisms are permanent residents in the systems. However, Kubalek and Komenda showed that when the culture medium was incubated at 25°C, more mycobacteria were isolated from drinking water samples collected in the spring than from samples collected in the fall (25). However, like us, these authors obtained similar recovery frequencies for spring and fall samples when incubation was carried out at 37°C (25).
Our study shows that the mycobacterial load is generally effectively reduced by water treatment plants. However, high levels of many species were recovered from the water distribution system. Joret et al. hypothesized that bacteria can recover their abilities to grow after depletion of residual disinfectant (21). As mycobacteria are highly resistant to chlorine, this could explain why they were frequently recovered from distributed water. However, large studies are necessary to determine whether identical mycobacterial strains are present in raw water and in the distribution network.
Drinking water is a source of disseminated M. avium infection in hospitalized AIDS patients (52). Similarly, waterborne household infections caused by M. xenopi have been reported (46). In both instances, showers and drinking water have been suggested to be the sources of infection. Inclusion of mycobacteria on the list of bacteriological indicators of water quality could help improve documentation of the level of contamination. It is possible that exposure to tap water mycobacteria could be reduced by using the recommendations for preventing Legionella infection.
Acknowledgments
This work received support from the Lyonnaise des Eaux and the Société Anonyme de Gestion des Eaux de Paris.
We thank Laurence Giudicelli for valuable technical support.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astagneau, P., N. Desplaces, V. Vincent, V. Chicheportiche, A. Botherel, S. Maugat, K. Lebascle, P. Leonard, J. Desenclos, J. Grosset, J. Ziza, and G. Brucker. 2001. Mycobacterium xenopi spinal infections after discovertebral surgery: investigation and screening of a large outbreak. Lancet 358:747-751. [DOI] [PubMed] [Google Scholar]
- 3.Bolan, G., A. L. Reingold, L. A. Carson, V. A. Silcox, C. L. Woodley, P. S. Hayes, A. W. Hightower, L. McFarland, J. W. D. Brown, and N. J. Petersen. 1985. Infections with Mycobacterium chelonei in patients receiving dialysis and using processed hemodialyzers. J. Infect. Dis. 152:1013-1019. [DOI] [PubMed] [Google Scholar]
- 4.Campagnaro, R. L., H. Teichtahl, and B. Dwyer. 1994. A pseudoepidemic of Mycobacterium chelonae: contamination of a bronchoscope and autocleaner. Aust. N. Z. J. Med. 24:693-695. [DOI] [PubMed] [Google Scholar]
- 5.Chauret, C., N. Armstrong, J. Fisher, R. Sharma, S. Springthorpe, and S. Sattar. 1995. Correlating Cryptosporidium and Giardia with microbial indicators. J. Am. Water Works Assoc. 87:76-84. [Google Scholar]
- 6.Collins, C. H., J. M. Grange, and M. D. Yates. 1984. Mycobacteria in water. J. Appl. Bacteriol. 57:193-211. [DOI] [PubMed] [Google Scholar]
- 7.Collins, M. R., T. T. Eighmy, J. Fenstermacher, and S. K. Spanos. 1992. Removing natural organic matter by conventional slow sand filtration. J. Am. Water Works Assoc. 84:80-90. [Google Scholar]
- 8.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dailloux, M., C. Laurain, M. Weber, and P. Hartemann. 1999. Water and nontuberculous mycobacteria. Water Res. 33:2219-2228. [Google Scholar]
- 10.Eighmy, T. T., M. R. Collins, S. K. Spanos, and J. Fenstermacher. 1992. Microbial populations, activities and carbon metabolism in slow sand filters. Water Res. 26:1319-1328. [Google Scholar]
- 11.Falkinham, J. O., 3rd. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischeder, R., R. Schulze-Robbecke, and A. Weber. 1991. Occurrence of mycobacteria in drinking water samples. Zentralbl. Hyg. Umweltmed. 192:154-158. [PubMed] [Google Scholar]
- 13.Fox, K. R., R. J. Miltner, G. S. Logsdon, D. L. Dicks, and L. F. Drolet. 1984. Pilot-plant studies of slow-rate filtration. J. Am. Water Works Assoc. 76:62-68. [Google Scholar]
- 14.Fraser, V. J., M. Jones, P. R. Murray, G. Medoff, Y. Zhang, and R. J. Wallace, Jr. 1992. Contamination of flexible fiber optic bronchoscopes with Mycobacterium chelonae linked to an automated bronchoscope disinfection machine. Am. Rev. Respir. Dis. 145:853-855. [DOI] [PubMed] [Google Scholar]
- 15.Goslee, S., and E. Wolinsky. 1976. Water as a source of potentially pathogenic mycobacteria. Am. Rev. Respir. Dis. 113:287-292. [DOI] [PubMed] [Google Scholar]
- 16.Grange, J. M., M. D. Yates, and E. Boughton. 1990. The avian tubercle bacillus and its relatives. J. Appl. Bacteriol. 68:411-431. [DOI] [PubMed] [Google Scholar]
- 17.Haas, C. N., M. A. Meyer, and M. S. Palmer. 1983. The ecology of acid fast organisms in water supply, treatment and distribution systems. J. Am. Water Works Assoc. 75:139-144. [Google Scholar]
- 18.Hall-Stoodley, L., and H. Lappin-Scott. 1998. Biofilm formation by the rapidly growing mycobacterial species Mycobacterium fortuitum. FEMS Microbiol. Lett. 168:77-84. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, K. H. 1981. Atypical mycobacterial infections in children. Rev. Infect. Dis. 3:1075-1080. [PubMed] [Google Scholar]
- 20.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joret, J. C., V. Mennecart, C. Robert, B. Compagnon, and P. Cervantes. 1997. Inactivation of indigenous bacteria in water by ozone and chlorine. Water Sci. Technol. 35:81-86. [Google Scholar]
- 22.Kaustova, J., Z. Olsovsky, M. Kubin, O. Zatloukal, M. Pelikan, and V. Hradil. 1981. Endemic occurrence of Mycobacterium kansasii in water-supply systems. J. Hyg. Epidemiol. Microbiol. Immunol. 25:24-30. [PubMed] [Google Scholar]
- 23.Kazda, J. F. 1983. The principles of the ecology of mycobacteria, p. 323-341. In C. Ratledge and J. Stanford (ed.), The biology of mycobacteria, vol. 2. Academic Press, London, United Kingdom.
- 24.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubalek, I., and S. Komenda. 1995. Seasonal variations in the occurrence of environmental mycobacteria in potable water. APMIS 103:327-330. [DOI] [PubMed] [Google Scholar]
- 26.Laussucq, S., A. L. Baltch, R. P. Smith, R. W. Smithwick, B. J. Davis, E. K. Desjardin, V. A. Silcox, A. B. Spellacy, R. T. Zeimis, H. M. Gruft, et al. 1988. Nosocomial Mycobacterium fortuitum colonization from a contaminated ice machine. Am. Rev. Respir. Dis. 138:891-894. [DOI] [PubMed] [Google Scholar]
- 27.LeChevallier, M. W., T. S. Hassenauer, A. K. Camper, and G. A. McFeters. 1984. Disinfection of bacteria attached to granular activated carbon. Appl. Environ. Microbiol. 48:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Dantec, C., J. P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl. Environ. Microbiol. 68:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. Y., and R. A. Deininger. 2000. Survival of bacteria after ozonation. Ozone Sci. Eng. 22:65-75. [Google Scholar]
- 30.Logsdon, G. S., J. M. Symons, R. L. Hoye, and M. M. Arozarena. 1981. Alternative filtration methods for removal of Giardia cysts and cyst models. J. Am. Water Works Assoc. 73:111-118. [Google Scholar]
- 31.Ludovici, P. P., R. A. Phillips, and W. S. Jeter. 1977. Comparative inactivation of bacteria and viruses in tertiary-treated wastewater by chlorination, p. 359-390. In J. D. Jonhson (ed.), Disinfection: water and wastewater. Ann Arbor Science Publishers, Ann Arbor, Mich.
- 32.Maloney, S., S. Welbel, B. Daves, K. Adams, S. Becker, L. Bland, M. Arduino, R. Wallace, Jr., Y. Zhang, G. Buck, et al. 1994. Mycobacterium abscessus pseudoinfection traced to an automated endoscope washer: utility of epidemiologic and laboratory investigation. J. Infect. Dis. 169:1166-1169. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, A., S. Torello, and R. Kolter. 1999. Sliding motility in mycobacteria. J. Bacteriol. 181:7331-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan, C. M., P. A. Hashisaki, and D. F. Dundas. 1991. An outbreak of soft-tissue infections due to Mycobacterium fortuitum associated with electromyography. J. Infect. Dis. 163:1150-1153. [DOI] [PubMed] [Google Scholar]
- 35.Nye, K., D. K. Chadha, P. Hodgkin, C. Bradley, J. Hancox, and R. Wise. 1990. Mycobacterium chelonei isolation from broncho-alveolar lavage fluid and its practical implications. J. Hosp. Infect. 16:257-261. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien, R. J., L. J. Geiter, and D. E. Snider, Jr. 1987. The epidemiology of nontuberculous mycobacterial diseases in the United States. Results from a national survey. Am. Rev. Respir. Dis. 135:1007-1014. [DOI] [PubMed] [Google Scholar]
- 37.Pankhurst, C. L., N. W. Johnson, and R. G. Woods. 1998. Microbial contamination of dental unit waterlines: the scientific argument. Int. Dent. J. 48:359-368. [DOI] [PubMed] [Google Scholar]
- 38.Pappas, S. A., D. M. Schaaff, M. B. DiCostanzo, F. W. King, Jr., and J. T. Sharp. 1983. Contamination of flexible fiber optic bronchoscopes. Am. Rev. Respir. Dis. 127:391-392. [DOI] [PubMed] [Google Scholar]
- 39.Park, H., H. Jang, C. Kim, B. Chung, C. L. Chang, S. K. Park, and S. Song. 2000. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers. J. Clin. Microbiol. 38:4080-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelletier, P. A., G. C. du Moulin, and K. D. Stottmeier. 1988. Mycobacteria in public water supplies: comparative resistance to chlorine. Microbiol. Sci. 5:147-148. [PubMed] [Google Scholar]
- 41.Peters, M., C. Muller, S. Rusch-Gerdes, C. Seidel, U. Gobel, H. D. Pohle, and B. Ruf. 1995. Isolation of atypical mycobacteria from tap water in hospitals and homes: is this a possible source of disseminated MAC infection in AIDS patients? J. Infect. 31:39-44. [DOI] [PubMed] [Google Scholar]
- 42.Recht, J., A. Martinez, S. Torello, and R. Kolter. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182:4348-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulze-Robbecke, R., C. Feldmann, R. Fischeder, B. Janning, M. Exner, and G. Wahl. 1995. Dental units: an environmental study of sources of potentially pathogenic mycobacteria. Tuber. Lung Dis. 76:318-323. [DOI] [PubMed] [Google Scholar]
- 45.Schulze-Robbecke, R., B. Janning, and R. Fischeder. 1992. Occurrence of mycobacteria in biofilm samples. Tuber. Lung Dis. 73:141-144. [DOI] [PubMed] [Google Scholar]
- 46.Slosarek, M., M. Kubin, and M. Jaresova. 1993. Water-borne household infections due to Mycobacterium xenopi. Cent. Eur. J. Public Health 1:78-80. [PubMed] [Google Scholar]
- 47.Stine, T. M., A. A. Harris, S. Levin, N. Rivera, and R. L. Kaplan. 1987. A pseudoepidemic due to atypical mycobacteria in a hospital water supply. JAMA 258:809-811. [PubMed] [Google Scholar]
- 48.Taylor, R. H., J. O. Falkinham, 3rd, C. D. Norton, and M. W. LeChevallier. 2000. Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of Mycobacterium avium. Appl. Environ. Microbiol. 66:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tortoli, E., A. Bartoloni, E. C. Bottger, S. Emler, C. Garzelli, E. Magliano, A. Mantella, N. Rastogi, L. Rindi, C. Scarparo, and P. Urbano. 2001. Burden of unidentifiable mycobacteria in a reference laboratory. J. Clin. Microbiol. 39:4058-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsintzou, A., A. Vantarakis, O. Pagonopoulou, A. Athanassiadou, and M. Papapetropoulou. 2000. Environmental mycobacteria in drinking water before and after replacement of the water distribution network. Water Air Soil Pollut. 120:273-282. [Google Scholar]
- 52.Von Reyn, C. F., J. N. Maslow, T. W. Barber, J. O. Falkinham, 3rd, and R. D. Arbeit. 1994. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137-1141. [DOI] [PubMed] [Google Scholar]
- 53.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 54.Wallace, R. J., Jr., J. M. Musser, S. I. Hull, V. A. Silcox, L. C. Steele, G. D. Forrester, A. Labidi, and R. K. Selander. 1989. Diversity and sources of rapidly growing mycobacteria associated with infections following cardiac surgery. J. Infect. Dis. 159:708-716. [DOI] [PubMed] [Google Scholar]
- 55.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, R. C. Good, J. A. Tschen, and M. S. Stone. 1983. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5:657-679. [DOI] [PubMed] [Google Scholar]
- 56.Wenger, J. D., J. S. Spika, R. W. Smithwick, V. Pryor, D. W. Dodson, G. A. Carden, and K. C. Klontz. 1990. Outbreak of Mycobacterium chelonae infection associated with use of jet injectors. JAMA 264:373-376. [PubMed] [Google Scholar]
- 57.Wolinsky, E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107-159. [DOI] [PubMed] [Google Scholar]
- 58.Wright, E. P., C. H. Collins, and M. D. Yates. 1985. Mycobacterium xenopi and Mycobacterium kansasii in a hospital water supply. J. Hosp. Infect. 6:175-178. [PubMed] [Google Scholar]
- 59.Yajko, D. M., D. P. Chin, P. C. Gonzalez, P. S. Nassos, P. C. Hopewell, A. L. Reingold, C. R. Horsburgh, Jr., M. A. Yakrus, S. M. Ostroff, and W. K. Hadley. 1995. Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:176-182. [PubMed] [Google Scholar]
- 60.Zhang, Y., M. Rajagopalan, B. A. Brown, and R. J. Wallace, Jr. 1997. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J. Clin. Microbiol. 35:3132-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]