Abstract
Cyclopentanone 1,2-monooxygenase, a flavoprotein produced by Pseudomonas sp. strain NCIMB 9872 upon induction by cyclopentanol or cyclopentanone (M. Griffin and P. W. Trudgill, Biochem. J. 129:595-603, 1972), has been utilized as a biocatalyst in Baeyer-Villiger oxidations. To further explore this biocatalytic potential and to discover new genes, we have cloned and sequenced a 16-kb chromosomal locus of strain 9872 that is herein reclassified as belonging to the genus Comamonas. Sequence analysis revealed a cluster of genes and six potential open reading frames designated and grouped in at least four possible transcriptional units as (orf11-orf10-orf9)-(cpnE-cpnD-orf6-cpnC)-(cpnR-cpnB-cpnA)-(orf3-orf4 [partial 3′ end]). The cpnABCDE genes encode enzymes for the five-step conversion of cyclopentanol to glutaric acid catalyzed by cyclopentanol dehydrogenase, cyclopentanone 1,2-monooxygenase, a ring-opening 5-valerolactone hydrolase, 5-hydroxyvalerate dehydrogenase, and 5-oxovalerate dehydrogenase, respectively. Inactivation of cpnB by using a lacZ-Kmr cassette resulted in a strain that was not capable of growth on cyclopentanol or cyclopentanone as a sole carbon and energy source. The presence of σ54-dependent regulatory elements in front of the divergently transcribed cpnB and cpnC genes supports the notion that cpnR is a regulatory gene of the NtrC type. Knowledge of the nucleotide sequence of the cpn genes was used to construct isopropyl-β-thio-d-galactoside-inducible clones of Escherichia coli cells that overproduce the five enzymes of the cpn pathway. The substrate specificities of CpnA and CpnB were studied in particular to evaluate the potential of these enzymes and establish the latter recombinant strain as a bioreagent for Baeyer-Villiger oxidations. Although frequently nonenantioselective, cyclopentanone 1,2-monooxygenase was found to exhibit a broader substrate range than the related cyclohexanone 1,2-monooxygenase from Acinetobacter sp. strain NCIMB 9871. However, in a few cases opposite enantioselectivity was observed between the two biocatalysts.
Pseudomonas sp. strain NCIMB 9872, which is capable of growth on 0.1% cyclopentanol as sole carbon source, was isolated from a freshwater stream in Illinois by P. J. Chapman some three decades ago. Trudgill and coworkers (27) used this strain as a prototype organism to establish the biochemical pathway of cyclopentanol metabolism shown in Fig. 1. For reason of utility as a Baeyer-Villiger (BV) biocatalyst and its mechanistic aspects (2, 8, 13, 58, 71), the second biochemical step catalyzed by cyclopentanone 1,2-monooxygenase (CPMO) has been most extensively studied in this organism. The conversion of cyclopentanone to 5-valerolactone is NADPH and flavin adenine dinucleotide (FAD) dependent, and CPMO has been purified to near homogeneity. It is a tetramer of subunit molecular weight 50,000 to 54,000 (8, 66). Assay of the FAD/protein ratio gave values of 2 to 4 molecules of FAD bound to each tetrameric enzyme molecule (28, 66). An N-terminal 29-amino-acid sequence of CPMO has been determined (8).
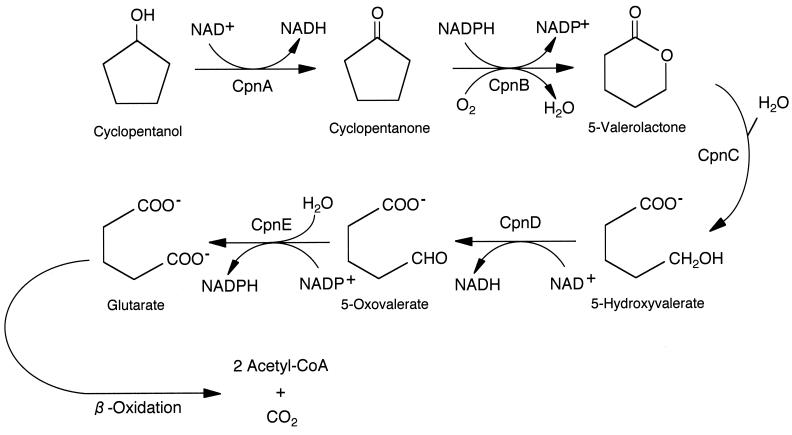
FIG. 1.
Degradation pathway for cyclopentanol by Comamonas sp. strain NCIMB 9872 (adapted from reference 27 with permission). CpnA, cyclopentanol dehydrogenase; CpnB, CPMO; CpnC, 5-valerolactone hydrolase; CpnD, 5-hydroxyvalerate dehydrogenase; CpnE, 5-oxovalerate dehydrogenase. An alternative name for 5-valerolactone is 5-pentanolide. Further oxidation of glutarate to acetyl-coenzyme A is believed to proceed via β-oxidation.
A chemical BV oxidation is the transformation of ketones into esters or of cyclic ketones into lactones by peracids, such as 3-chloroperbenzoic acid. This century-old reaction (for reviews, see references 54 and 63) continues to attract interest not only in broadening the spectrum of applications (ranging from the synthesis of steroids, antibiotics, and pheromones to the synthesis of monomers for polymerization, etc.) but also in developing oxidants that are more chemoselective and efficient and thus result in more product than waste (20).
The biological BV reactions catalyzed by BV monooxygenases (BVMOs) offer the prospect of eco-efficient chemical transformations in whole cells or improved expression systems via cloning. Moreover, BVMOs can provide products in optically active form which are difficult to obtain by other strategies (for reviews, see references 55, 61, 71, and 74). Notable stereospecific syntheses include the production of the natural product (R)-(+)-lipoic acid (2, 8) and the antidepressant Baclofen (47).
Before 1999, the only cloned BVMO-encoding gene (chnB) was that of the cyclohexanone 1,2-monooxygenase (CHMO) produced by Acinetobacter sp. strain NCIMB 9871 (18, 32, 61). This facilitated the construction of clones of CHMO in heterologous systems such as Saccharomyces cerevisiae and Escherichia coli (6, 15, 22, 38, 40, 48, 59). The new whole-cell engineered systems circumvented the potentially pathogenic Acinetobacter sp. in biotransformation, in addition to producing lactones with very high enantioselectivities in asymmetric BV oxidations of a large number of ketones (38, 48, 62). Another notable aspect of overexpression of a clone of CHMO in E. coli was the sequence determination by mass spectrometry (40) that confirmed the DNA-predicted sequence reported by Iwaki et al. (32). A correct CHMO sequence is important in view of its structure-function analysis, an exercise which has apparently begun in various laboratories (15, 40, 59).
The last three years have also witnessed new cloning activities of BVMO-encoding genes and characterization of the enzymes from several environmental isolates: a steroid monooxygenase from Rhodococcus (49); a strain SE19 almost identical to strain 9871 from Acinetobacter sp. (19); two related CHMO genes from one Brevibacterium sp. (11, 12); a novel 4-hydroxyacetophenone monooxygenase from two Pseudomonas spp. (37, 65); and CHMO activities from a black yeast (29) and the fungus Cunninghamella echinulata (3). To date, the complete pathway genes of only the cyclohexanol metabolism in Acinetobacter sp. strain SE19 and strain 9871 have been determined (19, 33).
Here, we report the cloning and nucleotide sequencing of the genes and the functional analysis of those enzymes involved in the complete cyclopentanol (cpn) degradation pathway in Pseudomonas (presently Comamonas) sp. strain NCIMB 9872.
(A portion of this work was reported at the ASM Conference on Biodegradation, Biotransformation and Biocatalysis (B3), San Juan, Puerto Rico, 2 to 6 Oct 2001.)
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas sp. strain NCIMB 9872 was purchased from the National Collections of Industrial and Marine Bacteria Ltd. (NCIMB; Aberdeen, Scotland) and grown at 30°C in Luria-Bertani (LB) broth (57) or mineral salt medium (MSM), pH 7.0, containing 0.2% cyclopentanone (Wako Pure Chemical Industries, Ltd., Osaka, Japan). The MSM recipe contains the following components per liter: 1.0 g of NH4NO3, 1.5 g of KH2PO4, 1.5 g of Na2HPO4, 0.2 g of MgSO4 · 7H2O, 0.01 g of CaCl2 · 2H2O, 0.005 g of FeSO4 · 7H2O, 0.002 g of MnSO4 · 4H2O, and 0.1 g of yeast extract. Agar was added to 1.5% for plates. E. coli strains were routinely cultured in LB media. When necessary, the media were supplemented with ampicillin (Ap; 100 μg/ml) or kanamycin (Km; 50 μg/ml for E. coli and 250 μg/ml for strain 9872).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Comamonas sp. strain NCIMB 9872 | Grows on cyclopentanol | 27 |
| Comamonas sp. strain 9872MB | cpnB::lacZ, Kmr | This study |
| C. testosteroni ATCC 11996 | Grows on testosterone, type strain | 46 |
| Plasmids | ||
| pKOK6.1 | lacZ-Kmr cassette | 41 |
| pSD80 | Expression vector Apr | 60 |
| pUC18 | Cloning vector, Apr | 77 |
| pUC19 | Cloning vector, Apr | 77 |
| pXcmkn12 | Direct cloning vector of PCR products, Apr Kmr | 14 |
| pMM4 | chnB from Acinetobacter sp strain NCIMB 9871 in pET22b(+) | 17 |
| pCMP10 | 116-bp PCR product in pXcmkn12 | This study |
| pCMP100 | 7.2-kb EcoRI fragment in pUC18 | This study |
| pCMP107 | 1.2-kb SmaI* fragment containing cpnD in pSD80a | This study |
| pCMP105 | 0.9-kb EcoRI*-PstI* fragment containing cpnC in pSD80 | This study |
| pCMP200 | 4.3-kb SphI fragment in pUC18 | This study |
| pCMP201 | 1.7-kb PstI-StuI fragment containing cpnB in pSD80 | This study |
| pCMP202 | 0.8-kb EcoRI*-PstI* fragment containing cpnA in pSD80 | This study |
| pCMP203 | 2.5-kb PstI* fragment containing cpnBA in pSD80 | This study |
| pCMP210 | cpnB::lacZ, Kmr from pCMP200 | This study |
| pCMP300 | 4.8-kb HindIII fragment in pUC18 | This study |
| pCMP400 | 5.7-kb Xhol fragment in pUC19 | This study |
| pCMP408 | 1.5-kb EcoRI* fragment containing cpnE in pSD80 | This study |
* indicates restriction sites introduced by PCR design.
Genetic methods and sequence analysis.
Total DNA from strain NCIMB 9872 was isolated by the method of Wilson (75). Plasmid isolation was performed by the method of Birnboim and Doly (9). Standard procedures such as Southern blot analysis, DNA subcloning, and DNA manipulations were performed according to the methods of Sambrook et al. (57). Hybridization was performed at 68°C. The gene probe was labeled by the digoxigenin-11-UTP system according to the manufacturer's instructions (Roche Molecular Biochemicals). DNA sequencing and analysis were carried out as previously described (4, 32).
16S rDNA sequencing.
The near full-length 16S ribosomal DNA (rDNA) gene of strain 9872 was amplified by PCR using the eubacteria 16S rDNA primers 27f and 1492r as described by Lane (44). PCRs were performed in a Perkin-Elmer model 2400 thermal cycler under conditions of 30 cycles consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min.
Cloning of the CPMO-encoding (cpnB) gene and associated open reading frames (ORFs).
Failing to obtain a positive signal in a Southern blot analysis of strain 9872 DNA when using the CHMO-encoding gene (chnB) from Acinetobacter sp. strain NCIMB 9871 as a probe (32), we resorted to purifying the CPMO protein from 100 g (wet weight) of cyclopentanone-grown cells as described previously (28), with the aim of determining an N-terminal amino acid sequence to design primers for PCR amplification. As a result, a 40-amino-acid-residue sequence, TTMTTMTTEQLGMNNSVNDKLDVLLIGAGFTGLYQLYHLR, was obtained by Edman degradation using an automated protein sequencer (model 477; Perkin-Elmer). Two primers with the following sequence were designed: 5′-ACIACIATGACIACNATGAC-3′ and 5′-ARRTGRTAIARYTGRTA-3′, where I = inosine; N = T, C, A, or G; R = A or G; and Y = C or T. The amplified 116-bp product was cloned directly in the PCR product cloning vector pXcmkn12 (14) and transformed in E. coli XL1-Blue, and the resulting plasmid was designated pCMP10 (Table 1; Fig. 2). Before using the amplified DNA as a hybridization probe, its nucleotide sequence was determined to confirm its identity. To clone the complete cpnB gene, the DNA insert from pCMP10 was amplified, labeled by the digoxigenin-11-UTP system, and used to probe a Southern hybridization of genomic DNA digested with the following restriction enzymes: BamHI, EcoRI, HindIII, KpnI, NheI, PstI, SalI, SphI, and XbaI. Three fragments (a 7.2-kb EcoRI fragment, 4.2-kb SphI fragment, and 4.8-kb HindIII fragment) were probed positive, and these were cloned in E. coli XL1-Blue cells by using pUC18 as a vector (77). These plasmid derivatives were designated pCMP100, pCMP200, and pCMP300, and both strands of the respective inserts were sequenced (Fig. 2). As the gene repertoire for the entire cpn pathway was found incomplete, a 5.7-kb XhoI fragment was cloned as a result of hybridization using probe 2 (a 0.6-kb EcoRI-PstI fragment) (Fig. 2). This plasmid was named pCMP400, and the sequence of its insert was determined.
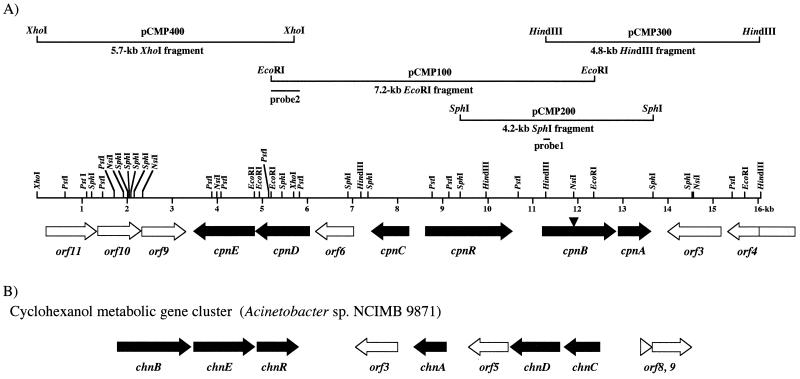
FIG. 2.
(A) Physical map of the cpn gene cluster in Comamonas sp. strain NCIMB 9872 and subclones in the 16,058-bp region. The orientation of the arrows indicates the direction of gene transcription. The black arrows are genes involved in cyclopentanol degradation. cpnR is a putative regulatory gene. Other ORFs are marked by open arrows. An arrowhead in the cpnB gene indicates the point of insertion of a lacZ-Kmr cassette at the NsiI site resulting in a mutant strain designated 9872MB. Probes 1 and 2 are PCR or restriction fragments used in hybridization experiments. (B) Gene organization of the cyclohexanol degradation (chn) pathway in Acinetobacter sp. strain 9871 (33). The chnABCDE genes (black arrows) encode cyclohexanol dehydrogenase, CHMO, caprolactone hydrolase, 6-hydroxyhexanoate dehydrogenase, and 6-oxohexanoate dehydrogenase, respectively; chnR is an AraC/XylS-type regulator (32, 33); orf5 is an equivalent of orf6 in the cpn pathway; orf3 encodes a potential pilin gene inverting protein; orf8 and -9 are putative transposases A and B, respectively (33). The chn gene cluster as shown is different from that present in strain SE19 (19) with the absence of “chnZ” in between chnR and orf3 (chnY designation in 19), and there are no equivalents of orf8 and -9 in the latter strain.
Inactivation of cpnB in Comamonas.
The cpnB disruption mutant was constructed by inserting a lacZ-Kmr cassette from pKOK6.1 (41). In pKOK6.1, the lacZ gene is promoterless and in addition to Kmr it is ampicillin resistant (Apr). The lacZ-Kmr cassette was excised as a PstI fragment and inserted into the NsiI site within the cpnB gene in pCMP200, yielding pCMP210. Electroporation of this plasmid into strain 9872 cells was carried out using a Gene Pulser (Bio-Rad). The parameters of electroporation were 2.5 kV, 25 μF, and 200 Ω. The cells were initially washed with 1 mM HEPES buffer and resuspended in 1 mM HEPES containing 10% glycerol. Kmr colonies were selected on LB plates containing Km (250 μg/ml).
Expression of cpn genes in E. coli.
The DNA fragments carrying cpnA, cpnB, cpnBA, cpnC, cpnD, and cpnE were amplified by using Pfu DNA polymerase (Stratagene) with the following pairs of PCR primers, with the desired restriction sites (SmaI, StuI, PstI, and EcoRI [underlined sequences]) to facilitate subsequent cloning: cpnB, 5′-AAAAGGCCTGAACTTCAATTATTTAGGAGAC-3′ and 5′-AAAACTGCAGGAGTTGCACAACAGAGTCTTAG-3′; cpnA, 5′-CGGAATTCATGGGACGTGTAAATGACAAAG-3′ and 5′-AAAACTGCAGGATGTTCCTTTCTCAGTTC-3′; cpnBA, 5′-AAAACTGCAGAACTTCAATTATTTAGGAGA-3′ and 5′-AAAACTGCAGGATGTTCCTTTCTCAGTTC-3′; cpnC, 5′-CGGAATTCATGAAACAACGCGAAGTGGCTA-3′ and 5′-AAAACTGCAGCCGAGTTGATGGGCGAATCC-3′; cpnD, 5′-TCCCCCGGGCCACTTCATCTGGAGTTCCAT-3′ and 5′-TCCCCCGGGTTCAGAAGGCACGGGCGTATA-3′; cpnE, 5′-CGGAATTCATGAACTCCTCTCTTCATTACA-3′ and 5′-CGGAATTCTCAGCCCGCAGACGTATAGGCC-3′.
In each case, the amplified DNA fragment was purified from an agarose gel, digested with the appropriate restriction enzyme(s), and cloned in the respective linearized pSD80 (60) vector as indicated in Table 1. E. coli BL21 containing each plasmid was cultivated in 20 ml of LB medium containing 100 μg of Ap/ml at 25°C. When the culture reached an A600 of 0.3 to 0.4, isopropyl-β-thio-d-galactoside (IPTG) was added to a final concentration of 0.5 mM in the medium. The cells were further cultured for 3 h. The resulting cells were harvested by centrifugation, washed in 50 mM phosphate buffer (pH 7.2), resuspended in the same buffer, and sonicated by two 30-s bursts with a Braun-Sonifier 250 apparatus. After centrifugation for 30 min at 18,000 × g at 4°C, the supernatant was used for the determination of enzyme activity.
Enzyme activities.
The various enzyme activities were assayed essentially as described by Trudgill (66) and Griffin and Trudgill (27). The CPMO activity was assayed at 25°C by measuring the decrease in absorbance at 340 nm due to the oxidation of NADPH to NADP+ in a solution containing 50 μmol of phosphate (pH 7.8), 1 μmol of cyclopentanone, 0.2 μmol of NADPH, and test extract. Cyclopentanol dehydrogenase (CpnA) and 5-hydroxyvalerate dehydrogenase (CpnD) activities were assayed at 25°C by measuring the increase in absorbance at 340 nm due to the reduction of NAD+ to NADH in a solution containing 50 μmol of glycine-NaOH (pH 9.0), 1 μmol of substrate (cyclopentanol or 5-hydroxyvalerate), 1 μmol of NAD+, and test extract. δ-Valerolactone hydrolase (CpnC) was assayed by measuring the rate of lactone consumption in a 200-μl reaction mixture that contained 2 μmol of δ-valerolactone, 50 μmol of phosphate buffer (pH 7.2), and test extract. Since the substrate 5-oxovalerate is not available commercially, NADP+-dependent 5-oxovalerate dehydrogenase (CpnE) activity was measured using the CpnD reaction mixture in the following manner. When the NAD+-dependent CpnD-driven reaction stopped, as evidenced by no further change in absorbance at 340 nm, NADP+ (0.2 μmol) and an aliquot of crude cell extract [e.g., E. coli(pCMP407)] were added. A concomitant increase in the A340 was used as a measure of CpnE activity.
One unit of activity is defined as the amount of enzyme required to convert 1 μmol of substrate in 1 min. Protein concentrations were determined by the method of Bradford (10).
Monooxygenase-catalyzed BV oxidations.
Among the ketones used in this study, compounds 1 to 5 (Fig. 3) are available commercially (Aldrich); the remaining ones were prepared as described previously (38, 48, 62, 72).
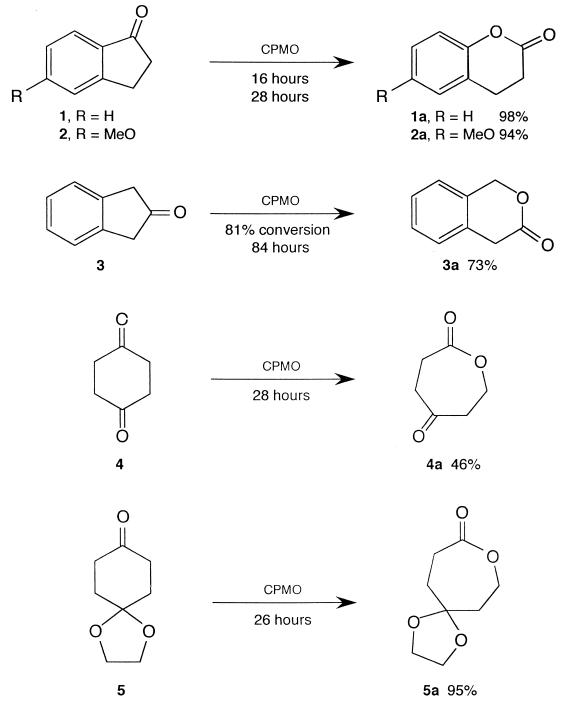
FIG. 3.
Biotransformations of selected ketones.
The recombinant E. coli(pCMP201) was maintained and propagated in the same way as the previously described E. coli BL21(DES)(pMM4) expressing CHMO from Acinetobacter sp. strain 9871 (17, 48). The pMM4 plasmid contains the chnB gene from strain 9871 under the control of a T7 promoter (17). An overnight culture of this strain was prepared and used at a 1:100 ratio to inoculate an LB-Ap medium supplemented with 2% glucose in a baffled Erlenmeyer flask. The culture (100 ml) was incubated at 30°C, 250 rpm, until the A600 was ca. 0.5 to 0.6. Ten microliters of IPTG stock solution (0.84 M) was added, followed by the substrate (10 mM). The culture was agitated at 30°C at 250 rpm and monitored by gas chromatography (GC), high-performance liquid chromatography, or thin-layer chromatography. The culture was saturated with NaCl and extracted with ethyl acetate. Combined extracts were washed once with brine and dried with anhydrous Na2SO4 or MgSO4. The solvent was removed on a rotary evaporator, and the residue was purified by flash chromatography.
Analytical techniques.
Optical rotations were measured on a Perkin-Elmer 241 polarimeter operating at room temperature. Infrared (IR) spectra were recorded on a Mattson Satellite FTIR spectrometer. 1H and 13C nuclear magnetic resonance (NMR) analyses were recorded in CDCl3 solution at room temperature, unless otherwise stated, on Varian XL-200 or Brüker AMX-400FT-NMR spectrometers. Capillary-column GC was performed on a Shimadzu GC-9A gas chromatograph employing a 5% OV-101 column (Supelco Inc.). Chiral GC was performed on a Hewlett-Packard 5890 instrument employing a 0.25 mm by 30 m (0.25-μm film) β-Dex 225 column (Supelco Inc.). Thin-layer chromatography was performed on precoated silica gel plates. Reaction products were purified by flash chromatography using 200- to 425-mesh silica gel. All solvents were distilled prior to use. Tetrahydrofuran was distilled from Na in the presence of benzophenone. Acetone was dried over CaSO4 and distilled from KMnO4. Methylene chloride was dried over anhydrous K2CO3, distilled, and stored over 3-Å molecular sieves.
Enantiomeric ratios and E-values were calculated as previously described (16). A calculation program is available at http://www.orgc.tugraz.at/orgc/.
Spectroscopic data.
Compounds 6, 7, 8, and 9 and their products have been described previously (48, 62) and therefore are not included below.
(i) Chroman-2-one (compound 1a) (52, 53).
Indan-1-one (compound 1) (35 mg, 0.26 mmol) was oxidized by CPMO in a 100-ml culture to give compound 1a (32 mg, 81% yield). 1H NMRδ: 6.82 to 7.29 (4H, m), 2.71 to 3.03 (4H, m) ppm; 13C NMRδ: 179.5, 154.1, 130.4, 127.9, 124.4, 120.8, 116.5, 29.2, 23.6 ppm. Some opened form (phenolic acid) was also isolated; 13C NMRδ: 168.9, 151.9, 128.3, 128.0, 126.9, 122.6, 116.9, 34.6, 24.8 ppm.
(ii) 6-Methoxy-chroman-2-one (compound 2a) (52, 53).
5-Methoxy-indan-1-one (compound 2) (20 mg, 0.12 mmol) was oxidized by CPMO in a 100-ml culture to afford compound 2a, mixed with the open hydroxy acid (21 mg, 95%). 1H NMRδ: 6.63 to 7.00 (3H, m), 3.79 (3H, s), 2.72 to ∼3.00 (4H, m) ppm; lactone 13C NMRδ: 179.2, 169.0, 147.9, 123.6, 117.7, 115.8, 113.2, 55.7, 29.2, 24.0 ppm; acid 13C NMRδ: 156.8, 153.7, 146.2, 128.1, 117.5, 113.1, 112.9, 55.7, 34.6, 24.9 ppm.
(iii) Oxepan-2,5-dione (compound 4a) (1).
1,4-Cyclohexanedione (compound 4) (100 mg, 0.89 mmol) was oxidized by CPMO in a 100-ml culture. The reaction was monitored by GC until almost 100% conversion was attained (24 h). Extraction with 25-ml portions of CHCl3 (five times) gave lactone (4a) (52 mg, 46% yield). The corresponding acid was not extracted. 13C NMRδ: 207.8, 129.2, 60.9, 47.1, 38.1, 27.2 ppm.
(iv) 1,4,8-Trioxa-spiro-(4,6)-undecan-9-one (compound 5a).
1,4-Dioxa-spiro-(4,5)-decan-8-one (compound 5) (0.30 mmol, 80 mg) was oxidized by CPMO in a 100-ml culture to give lactone (5a) (81 mg, 95%). IR (neat) νmax: 2,919 (m), 1,724 (s), 1,409 (m), 1,200 (s), 1,111 (s) cm−1; 1H NMRδ: 4.22 (2H, dm, J1 = 5.1 Hz, J2 = 2.2 Hz), 3.92 (4H, m), 2.63 (2H, td, J1 = 5.3 Hz, J2 = 1.3 Hz), 1.94 (2H, m), 1.83 (2H, m) ppm. 13C NMRδ: 175.3, 107.9, 64.8(2C), 64.3, 39.1, 32.7, 28.8 ppm.
(v) 5-Ethyl-5-hydroxy-oxepan-2-one (compound 9a) (48).
4-Ethyl-4-hydroxy-cyclohexanone (compound 9) (100 mg, 0.70 mmol) was oxidized by CPMO in a 100-ml culture to give lactone (compound 9a) (92 mg, 82%). IR (neat) νmax: 3,435 (s), 2,958 (m), 1,764 (s), 1,467 (m), 1,187 (s) cm−1; 1H NMR δ: 3.80 (2H, m), 2.61 (2H, m), 2.15 (2H, m), 1.97 (2H, m), 1.74 (2H, m), 0.96 (3H, t, J = 8.9 Hz) ppm; 13C NMR δ: 177.0, 88.6, 58.3, 40.2, 31.6, 30.7, 28.9, 7.9 ppm; MS = m/e 1.29 (M+ −29, 33%), 113 (71%), 101 (100%), 57 (72%), 55 (60%); [α]D25 +1.7 (c 1.0 CH2Cl2).
(vi) 5-Methyl-5-propyl-oxepan-2-one (compound 10a) (48).
4-Methyl-4-propyl-cyclohexanone (compound 10) (48) (40 mg, 0.26 mmol) was oxidized by CPMO in a 100-ml culture to give lactone (compound 10a) (40 mg, 89%). IR (neat) νmax: 2,958 (s), 2,932 (s), 2,872 (m), 1,733 (s), 1,467 (m), 1,187 (m) cm−1; 1H NMR δ: 4.15 (2H, m), 2.60 (1H, m), 2.26 (1H, m), 1.60 (4H, m), 1.25 (4H, m), 0.976 (3H, s), 0.892 (3H, m) ppm. 13C NMR δ: 176.4, 64.5, 40.3, 34.3, 33.9, 29.6, 29.2, 24.7, 16.5, 14.8 ppm, [α]D25 −10.8 (c 1.9 CH2Cl2).
Nucleotide sequence accession number.
The DNA sequences reported in the paper have been submitted to DDBJ, EMBL, and GenBank, and the accession numbers are AB022102 (4.5-kb SphI fragment), AB073151 (entire sequence), and AF172067 (16S rDNA).
RESULTS
Reclassification of strain 9872.
As proper genus or strain identification is a prerequisite in a potential risk assessment scheme, strain 9872 was subjected to 16S ribosomal sequence analysis. As a result, the near-complete 1,449-base-long 16S sequence of strain 9872 was found to be 100% identical to that of Comamonas (previously Pseudomonas) testosteroni (ATCC 11996), a prototype strain in testosterone metabolism (46, 64, 76). The ability of strain 9872 to use testosterone as a sole carbon source was confirmed by the formation of clearing zones around colonies of growth on an agar plate (46) as well as growth in liquid medium supplemented by steroid only (data not shown). On the other hand, unlike strain 9872, ATCC 11996 was not able to grow on either cyclopentanol, cyclohexanol, cyclopentanone, or cyclohexanone as a sole carbon source. Also, the total protein profile of the two strains grown on LB or minimal medium containing testosterone was found to be different when separated on a polyacrylamide gel (data not shown). These results showed that although strain 9872 is related to C. testosteroni, it differs in certain characteristics. It is proposed that strain 9872 be reclassified as a Comamonas sp.
Cloning of a 16-kb chromosomal locus.
As described in Materials and Methods, four overlapping clones of strain 9872 genomic DNA were obtained. Using all four plasmids as templates, the inserts were sequenced to provide a contiguous segment of 16,058 bp from which the presence of 11 complete and one incomplete ORFs was deduced (Fig. 2; Table 2). Six are on one strand, and the remaining ones are in the opposite direction. It is apparent that at least four transcriptional units are possible due to the direction and organization of the ORFs or genes.
TABLE 2.
Homology of the Comamonas sp. strain NCIMB 9872 gene products or ORFs
| Gene designation | Function of gene product | Position in sequencea | aa | Most similar gene products [species] (accession no.) [% amino acid identity (identical aa/overlapped aa)]b |
|---|---|---|---|---|
| orf11 | Possible reductase | 213-1,331 | 372 | N-Ethylmaleimide reductase [Vibrio cholerae N16961] (AE004426) [40.2 (146/363)], Morphinone reductase MorB [Pseudomonas putida M10] (U37350) [43.0 (156/362)] |
| orf10 | Unknown | 1,345-2,280 | 311 | Hypothetical protein [Arabidopsis thaliana] (AC011665) [36.7 (18/49)], Polyprotein [Maize dwarf mosaic virus] (AJ001691) [26.3 (25/95)] |
| orf9 | Possible monooxygenase | 2,320-3,309 | 329 | Hypothetical protein [Pseudomonas aeruginosa PAO1] (AE004549) [45.2 (149/330)], Hypothetical protein AGR_L_1143p[Agrobacterium tumefaciens C58] (AE008255) [41.6 (131/315)] |
| cpnE | 5-Oxovalerate dehydrogenase | 3,435-4,883 | 482 | Putative aldehyde dehydrogenase [Mesorhizobium loti MAFF303099] (AP003006) [61.5 (292/475)], Putative aldehyde dehydrogenase [Sinorhizobium meliloti 1021] (AL603646) [60.3 (288/478)] |
| cpnD | 5-Hydroxyvalerate dehydrogenase | 4,912-6,066 | 384 | Alcohol dehydrogenase homolog [Agrobacterium tumefaciens C58] (U59485) [47.8 (184/385)], Probable iron-containing alcohol dehydrogenase [Pseudomonas aeruginosa PAO1] (AE004931) [48.8 (191/391)] |
| orf6 | Unknown | 6,158-7,030 | 290 | Unknown Orf1 [Pseudomonas sp. strain TW3] (AF043544) [41.5 (117/282)], Unknown Orf3 [Pseudomonas sp. strain YH102] (AF187880) [32.4 (89/275)] |
| cpnC | 5-Valerolactone hydrolase | 7,376-8,276 | 299 | Unknown [Mesorhizobium loti MAFF303099] (AP003012) [41.2 (121/294)], AGR_pAT_685p [Agrobacterium tumefaciens C58] (AE007915) [28.7 (74/258)] |
| cpnR | NtrC-type regulator | 8,617-10,590 | 657 | Transcription activator of acetoin dehydrogenase operon AcoR [Bacillus subtilis AC327] (D78509) [31.0 (195/629)], Acetoin catabolism regulatory protein AcoR [Ralstonia eutropha H16] (M90471) [33.2 (213/642)] |
| cpnB | CPMO | 11,235-12,887 | 550 | Cyclohexanone monooxygenase [Brevibacterium sp. strain HCU] (AF257215) [49.5 (255/515)], Baeyer-Villiger monooxygenase homologue [Rhodococcus erythropolis DCL14] (AJ303350) [39.1 (203/519)] |
| cpnA | Cyclopentanol dehydrogenase | 12,920-13,672 | 250 | Short-chain alcohol dehydrogenase family protein FabG3 [Mycobacterium tuberculosis H37Rv] (Z74025) [46.4 (115/248)], Putative oxidoreductase [Mycobacterium tuberculosis CDC1551] (AE007057) [46.4 (115/248)] |
| orf3 | Possible integrase | 13,995-15,209 | 402 | Putative integrase [Salmonella enterica serovar Typhimurium LT2] (AF001386) [27.6 (108/392)], Integrase [Shigella dysenteriae type 1; 3818T] (AF153317) [26.4 (106/402)] |
| orf4 (incomplete) | Possible cytoplasmic axial filament protein | 15,353->16,058 | 234 | Cytoplasmic axial filament protein, authentic frameshift [Neisseria meningitidis MC58] (AE002529) [62.0 (142/229)], Cytoplasmic axial filament protein homolog NMA0672 [Neisseria meningitidis Z2491] (AL162753) [62.0 (142/229)] |
The numbers are nucleotide positions in the sequenced segment (DDBJ/EMBL/GenBank accession no. AB073151).
Nonredundant protein databases in the National Center for Biotechnology Information.
Localization of cpnB and characteristics of downstream sequences.
The 1,650-bp ORF in pCMP200, characterized by a perfect repeat of three consecutive sequences of ATGACCACC at its 5′ end (coding for M-T-T), was identified as the CPMO-encoding (cpnB) gene, since its N-terminal 40-amino-acid sequence was determined in this study. As well, a shorter sequence of 29 amino acids has been reported (8). The deduced amino acid sequence of the CPMO consists of 550 residues characterized by homology to known BVMOs and to motifs of related flavoproteins (25, 68). The highest score given by the BLAST search and the most related sequence by phylogenetic analysis was CHMO2 of Brevibacterium sp. strain HCU with an identity of 49.5%. Another notable homolog, showing 41% identity, is an unannotated sequence of Thermobifida (formerly Thermomonaspora) fusca referred to as the gene36 protein in the unfinished genome database (gnl/DOE2021 contig48). In contrast, the CPMO sequence shows 37.2% identity with the 543-residue CHMO of strain NCIMB 9871 (32). As previously noted, a characteristic FAD-binding fingerprint, with the sequence GxGxxG is found at amino acids 28 to 33 of CPMO (8, 74). However, the complete sequence of CPMO shows the presence of a repeated GxGxxG motif at amino acids 197 to 202, a situation similarly found in other BVMOs (25, 37). Also conserved in CPMO is a hexapeptide 393-ATGFDA-398, a sequence that is described to be common among FAD- and NAD(P)H-binding proteins (68), and a sequence at residues 171 to 182 of the sequence FKGQWYHTALWP, which fits a so-called BVMO-identifying sequence motif (25). Griffin and Trudgill (27) reported the presence of six titratable sulfhydryl groups in CPMO, which is in good agreement with the presence of four cysteines in the CPMO sequence.
Thirty-two base pairs downstream of cpnB and preceded by a consensus ribosome-binding sequence, GGAGA, is a 750-bp ORF designated CpnA that encodes a polypeptide of 247 residues. A homology search indicated that CpnA belongs to the short-chain dehydrogenase/reductase (SDR) family of proteins (36). The highest score of the BLAST search was the FabG3 of Mycobacterium tuberculosis H37Rv with an identity of 46.4% (Table 2). Phylogenetic analysis (data not shown) showed that CpnA clustered with the ChnA of Brevibacterium sp. strain HCU (12). This sequence is 39.1% identical to the cyclohexanol dehydrogenase (ChnA) of Acinetobacter sp. strain NCIMB 9871. Notable sequence features of CpnA are the conservation of a coenzyme-binding motif (GxxxGxG) at positions 13 to 19 and potential active-site residues S142, Y155, and K159 (36).
Downstream of cpnA, but transcribed in the reverse orientation, is an ORF of 402 codons whose translated product resembles an integrase of Salmonella spp. or Vibrio cholerae (Table 2). Also, separated by a sequence of 143 bp is ORF3, which is preceded by an incomplete ORF4 (Table 2).
cpnB upstream sequences.
Upstream of cpnB in the same direction and separated by a 244-bp intergenic sequence is a potential NtrC-type transcriptional activator (Table 2). This ORF, designated cpnR, consists of 653 residues with an appropriately positioned ribosome-binding site 5 bp from the putative initiator codon. CpnR is most closely related to AcoR, a transcription activator of the acetoin dehydrogenase operon of Bacillus subtilis AC327 (Table 2). Characteristic of this family of proteins is a weakly conserved N terminus (the effector binding sequence), a strongly conserved central region (the ATPase activity and interaction site with the σ54 RNA polymerase), and a C terminus that contains a helix-turn-helix motif required for DNA binding (42). In CpnR, the potential ATP-binding motif is present at amino acids 364 to 371 (GETGTGKE), and the helix-turn-helix motif is predicted between amino acids 623 and 642.
Remaining cpn pathway genes and unknown ORFs.
Divergently transcribed from the cpnRBA genes are four ORFs, three of which were designated cpnC, cpnD, and cpnE and which are responsible for the third (5-valerolactone hydrolase), fourth (5-hydroxyvalerate dehydrogenase), and fifth (5-oxovalerate dehydrogenase) biochemical steps of the cpn degradation pathway, respectively (Fig. 1). The predicted amino acid sequence of CpnC does not share a significant identity with hydrolases of the cyclohexanol pathways (19, 33) but shows homology with gluconolactonases that catalyze the hydrolysis of d-glucono-1,5-lactone (Table 2) as well as paraoxonases and arylesterases that catalyze the hydrolysis of paraoxon [o,o-diethyl-4(nitrophenyl) phosphate] to diethylphosphoric acid and p-nitrophenol (26, 30).
Following cpnC with an intergenic space of 345 bp is a 290-codon ORF designated ORF6, with significant homology to an unknown sequence (Orf1) found in the 4-nitrotoluene-degrading Pseudomonas sp. strain TW3 (34) and an unknown Orf3 of Pseudomonas sp. strain YH102 (Table 2). A homologous sequence is located between chnA and chnD in the cyclohexanol-degrading strain NCIMB 9871 (33).
The predicted coding region of CpnD (5-hydroxyvalerate dehydrogenase) is 87 bp downstream of Orf6. This 384-residue sequence possibly belongs to the iron-containing (type III) alcohol dehydrogenase family of enzymes (5); the closest homolog is the alcohol dehydrogenase of Agrobacterium tumefaciens C58, bearing 47.8% identity (Table 2). The presence of an iron-binding motif with the sequence GxxHxxAHxxGxxxxxPHG is characteristic of this family of proteins (5). In CpnD, this motif is present at amino acid positions 260 to 278, with the exception that the second His is replaced by Tyr. Interestingly, like most type III alcohol dehydrogenases (5), a conserved NAD-binding motif (GxGxxG) is not present in the sequence of CpnD.
Only 28 bp away is the predicted coding region of cpnE, which consists of 482 residues. This is a member of the superfamily of NAD(P)+-dependent aldehyde dehydrogenases that act on a broad variety of aldehyde and semialdehyde substrates by transforming them to carboxylic acids (31). The CpnE sequence shows 28.2% identity with the 6-oxohexanoate dehydrogenase (ChnE) of strain NCIMB 9871. But the highest score given by the BLAST search was a putative aldehyde dehydrogenase of Mesorhizobium loti MAFF303099 with an identity of 61.5%, followed by that of Sinorhizobium meliloti (Table 2). Based on what is generally known about aldehyde dehydrogenases, Cys284 of CpnE is predictably an active-site residue, Asp177 is a potential adenine ribose-binding residue, and the GxAxxG sequence at positions 228 to 233 is a putative NAD-binding motif (31, 45).
Upstream of cpnE and transcribed in the reverse orientation is a potential operon of at least three ORFs; homologous sequences of ORFs 9 and 11 in the database include a putative alkanal monooxygenase and several reductases, respectively (Table 2).
Inactivation of the cpnB gene.
We constructed Comamonas sp. strain 9872MB by chromosomal inactivation of the cpnB gene, using a lacZ-Kmr cassette from the pKOK6.1 vector (41). The lacZ-Kmr cassette was excised as a PstI fragment and inserted into the NsiI site within the cpnB gene in pCMP200, yielding pCMP210. The inactivation of cpnB (Fig. 2) was confirmed by PCR (data not shown). The resulting mutant 9872MB was found to be unable to grow on cyclopentanol, cyclopentanone, cyclohexanol, or cyclohexanone as a sole carbon and energy source.
Potential σ54-dependent regulatory elements for cpnB and cpnC.
Identification of a NtrC-type transcriptional activator CpnR implies that the −24/−12 promoters (also known as the RpoN- or σ54-dependent promoters [7, 42, 43]) would regulate cpn gene expression. Hence, the 5′ noncoding sequences of cpnB and cpnC were analyzed. These two regions (234 bp of cpnB and 261 bp of cpnC just in front of the respective start codons) were found not only to share 54.5% sequence identity but also to contain characteristic signatures of the σ54-dependent regulatory sequences (data not shown). These are the upstream activator sequence (characterized by a TGT-N9/11-ACA motif), presumably a binding site for CpnR, and the dinucleotide doublets GG and GA, representing the −24/−12 promoter elements. Note that replacement of the consensus −12 cytosine (C) by an adenine (A) in the second motif is allowable among the mapped or unmapped transcriptional start sites of the −24/−12 sequences (7). Also, although found in numerous RpoN promoters, the presence of an integration host factor sequence motif is not evident in the cpnBC promoter region.
Expression of cpnABCDE genes in E. coli and enzyme activity assays.
Knowledge of the cpn nucleotide sequence was used to construct clones of the respective genes in the IPTG-inducible plasmid pSD80 (60) in order to verify its protein production and enzyme activities. The constructed plasmids in E. coli BL21 cells contained cpnB, cpnA, chnBA in tandem, cpnC, cpnD, and cpnE. Figure 4A shows the overexpression of the CpnA and CpnB proteins (lanes 2 and 3; CpnAB is in lane 4) and the observed Mrs, estimated at 30 and 57 kDa, respectively. No enhanced protein band of these sizes was detectable in the control cells containing pSD80 vector only. The overproduction of a 34-kDa CpnC, a 40-kDa CpnD, and a 49-kDa CpnE protein band in the respective clones is evident in Fig. 4B. In all cases, the experimental Mr values were in good agreement with the predicted ones (CpnA, 26,626.1; CpnB, 62,109.8; CpnC, 32,241.07; CpnD, 40,490.3; CpnE, 51,063.95). Of the total cellular protein, the production ranged from 5% (CpnA) to 24.5% (CpnB), with about 8% in the case of CpnCDE, as estimated by densitometric scans of the Coomassie blue-stained proteins (data not shown). The level of coexpressed CpnB and CpnA was slightly lower than that expressed alone. The production of CpnE could reach 30% of the total cell proteins, but this resulted in an insoluble protein as inclusion bodies (results not shown).
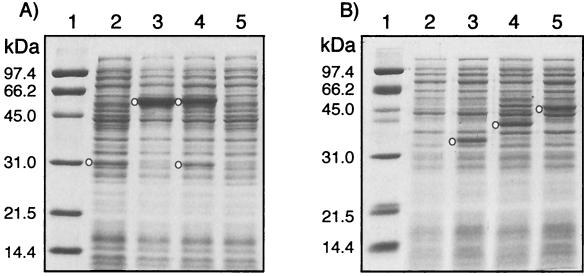
FIG. 4.
Coomassie blue-stained protein profiles of recombinant E. coli crude extracts separated on sodium dodecyl sulfate-10% polyacrylamide electrophoresis gel. Lane 1 in panels A and B shows molecular size markers as indicated alongside in kilodaltons; the open circles in lanes 2, 3, and 4 in panel A and lanes 3, 4, and 5 in panel B indicate the positions of the overexpressed CpnA, CpnB, CpnBA, CpnC, CpnD, and CpnE proteins in the respective clones pCMP202, pCMP201, pCMP203, pCMP105, pCMP107, and pCMP408 (see Table 1). Lane 5 in panel A and lane 2 in panel B are controls with E. coli cells containing the pSD80 vector only.
Crude protein extracts from the various E. coli BL21 cells were used to determine the enzyme activities as described in Materials and Methods. For comparison, the respective activities in the native strain 9872 were also determined (reported in parentheses below as specific activities [in units per milligram of protein]). As a result, the activities of NAD+-dependent cyclopentanol dehydrogenase, NADPH-dependent CPMO, 5-valerolactone hydrolase, NAD+-dependent 5-hydroxyvalerate dehydrogenase, and NADP+-dependent 5-oxovalerate dehydrogenase activities were found to be 0.58 (0.54), 0.28 (0.29), 34 (23), 2.5 (1.3), and 0.6 (0.12), respectively. Although overproduced, these activities in E. coli were only improved 1.5- to 2-fold, and in the case of CpnB and CpnA the specific activities were virtually unchanged. We have not assessed the kinetic parameters, such as lowered kcat values in the case of recombinant proteins compared to the Comamonas-produced proteins, as a possible reason for lack of a greatly improved specific activity after heterologous expression in E. coli (23). It is noteworthy that the recombinant CPMO, although overproduced in twice the amount in E. coli, appeared not as yellow as the native protein. In this regard, the ratio of FAD bound to the tetrameric subunit of CPMO needs to be evaluated.
The expression of CPMO in an alternative E. coli strain DH5α was found to be the same as that in BL21. In terms of cofactor dependence, we confirmed that NADP+ could not substitute for NAD+ in the CpnA and CpnD reactions; also, NADH could not replace NADPH in the CpnB activity (data not shown).
Substrate specificity of CpnA.
Besides cyclopentanol and cyclohexanol as known substrates for CpnA, we investigated eight other compounds. The activity toward cyclohexanol is 60% that of cyclopentanol. Interestingly, cell extracts of E. coli(pCMP202) were able to reduce NAD+ in the presence of 4-methylcyclohexanol better than the canonical cyclopentanol. On the other hand, 2-methylcyclohexanol and 1,4-cyclohexanediol gave only 20 to 30% of the cyclopentanol activity. Other alcohols such as 1-cyclohexane ethanol, trans-1,2-cyclohexanediol, isopropanol, and 5-hydroxyvalerate are weak or nonsubstrates under the same experimental conditions.
CPMO-catalyzed BV oxidations of miscellaneous ketones.
Induction of E. coli(pCMP201) cells by IPTG led to some 25% production of CPMO of the total cellular proteins (Fig. 4). This level is comparable to that of CHMO (20 to 30%) expressed in E. coli BL21(DE3)pMM4 as described in previous studies (17, 48).
The oxidations of various ketones were carried out in shake flasks, and the reactions were monitored by chiral-phase GC as described in Materials and Methods. Control experiments in which DH5α or BL21(DE3) cells were used instead of the plasmid-containing strains showed no lactone formation, confirming that CPMO or CHMO was indeed responsible for the observed BV reactions. The oxidation of cyclopentanone by E. coli DH5α(pCPM201) was rapid and complete (data not shown). Within 6 h, all of the starting material was converted to valerolactone. Cyclohexanone was equally well accepted, and the complete conversion to caprolactone was achieved in 7 h. Although no optimization was attempted, both lactones were isolated with better than 70% yields.
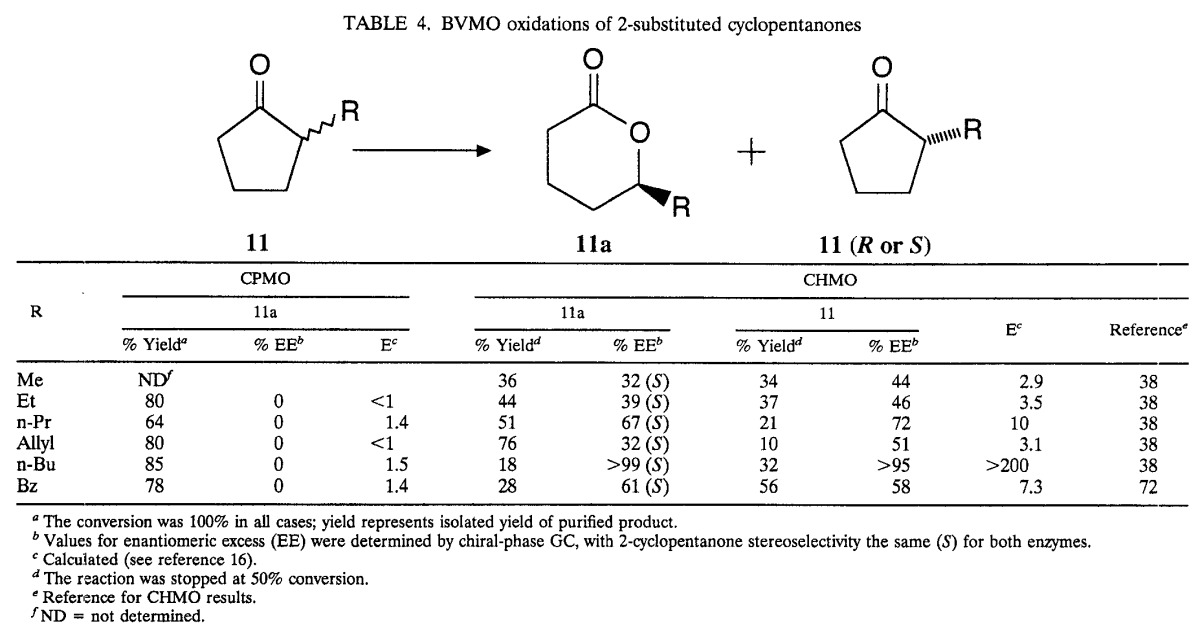
To establish CPMO substrate base and enantioselectivity, a constellation of cyclopentanones and cyclohexanones was investigated. These are shown in Fig. 3 and Tables 3 and 4. The yields refer to the isolated yields of chromatographically purified samples. The chemical and optical purities were established by comparing the chiral GC analyses of the lactonic products from biotransformations with those from chemical oxidations.
TABLE 3.
BVMO oxidations of 4-substituted cyclohexanones
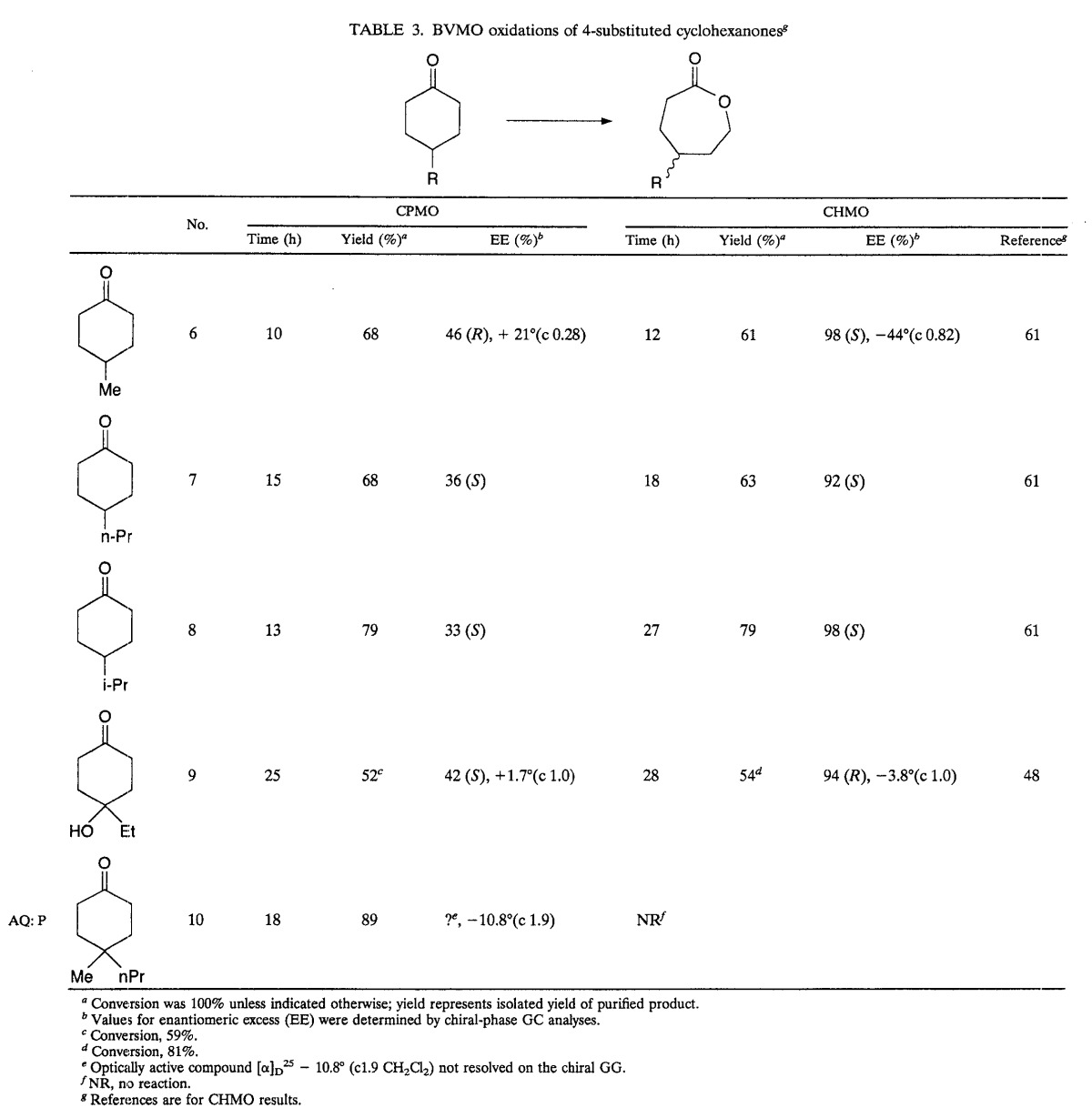
TABLE 4.
BVMO oxidations of 2-substituted cyclopentanones
The commercially available ketones 1 to 5 (Fig. 3) were found to be excellent substrates and were rapidly and completely converted to the corresponding lactones. The isolated yields were very high except for oxepan-2,5-dione (compound 4a), which was partially hydrolyzed. The oxidations of 1-indanones 1 and 2 were regioselective, and only the corresponding products 1a and 2a could be detected in their GC traces at the end of the transformation. Among these ketones, only 2-indanone (compound 3) was also oxidized by CHMO, and even that compound was a considerably poorer substrate for CHMO than it is for CPMO (62% conversion after 120 h versus 81% after 84 h).
Several prochiral 4-substituted cyclohexanones tested in BV oxidations with recombinant E. coli BL21(DE3)(pMM4) proved to be highly enantioselective, but only cyclohexanones with relatively small substituents were the acceptable substrates (48). Table 3 shows that CPMO rapidly oxidized the same substrates, but the enantioselectivity was generally much lower. The enantioselectivity of the two enzymes with 4-substituted substrates was not always the same. For example, 4-methylcyclohexanone (compound 6) was converted by CHMO to the (S)-lactone and by CPMO to the (R)-lactone. Similarly, 4-ethyl-4-hydroxycyclohexanone (compound 9) was converted by two enzymes to the antipodes, as shown by chiral GC and confirmed by the opposite signs for the rotation [α]. The absolute configuration for the product of CHMO oxidation was assigned by analogy with other related compounds as R (48) and, therefore, we tentatively identified the product of CPMO transformation as S. However, it should be stressed that these configurations have not been established unambiguously. 4-Methyl-4-n-propylcyclohexanone (compound 10) is a substrate for CPMO but not for CHMO. The lactonic product (compound 10a) is optically active ([α]D25, −10.8°, c 1.9 CH2Cl2), but as we were unable, so far, to resolve it on chiral chromatography or by NMR, the optical purity is not known.
CPMO-catalyzed transformations of 2-substituted cyclopentanones are consistently nonenantioselective, as shown in Table 4. The best enantiomeric ratio E (16), obtained for 2-n-butylcyclopentanone [11 (n-Bu)], was only 1.5, compared to the E value of >200 for the same reaction catalyzed by CHMO (38).
DISCUSSION
Despite its isolation some 30 years ago, NCIMB strain 9872 remains the sole member of the Comamonas genus that is known to metabolize cyclopentanol and related compounds. A polyphasic approach would be necessary to establish it as C. testosteroni or a new species, since there are examples of different species with identical or nearly identical 16S rDNA sequences (for a review, see reference 56).
This is the first delineation of a microbial gene locus that encompasses the cpn degradation pathway for the conversion of cyclopentanol to glutaric acid. It follows the cyclohexanol-to-adipic acid pathway (chn) of two Acinetobacter spp. (19, 33), strain SE19 that was recently isolated from an industrial wastewater bioreactor and the classical strain 9871 that came from a freshwater reservoir in Illinois (NCIMB). A comparison of the chn and cpn gene organization in strains 9871 and 9872 shows both common and different features (Fig. 2). First, the requisite genes are clustered within a ca 10-kb region; second, three genes, including a regulatory candidate (cpnRBA, chnBER), are encoded on one strand and the remaining three (cpnCDE, chnCDA) are on another. However, as indicated, the order and arrangement of the gene clusters are different, the two putative cpn operons are arranged outwards to each other (divergent), and those of the chn pathway are arranged inwards (convergent).
Another notable difference is the apparent mode of regulation. Expression of chnB in Acinetobacter sp. strain 9871 has been shown to be regulated by the transcriptional activator ChnR of the AraC/XylS type (19, 32). In strain 9872, it is predicted that both the putative cpnBA and cpnC(orf6)cpnDE operons are regulated by the σ54-dependent promoters, as evidenced by the identification of CpnR and the characteristic −24/−12 sequences. Future experiments are needed to dissect this positive regulatory circuit, the complexity of which is illustrated by the exhaustive analysis of the phenol-responding DmpR activator (73). At the moment, CpnR appears to be the first example of an NtrC-type regulator in cycloaliphatic metabolism. Many transcriptional regulators of aromatic catabolic pathways belong to the NtrC family of enhancer-binding proteins (21).
At the level of amino acid identity, an obvious relatedness of CpnB to ChnB2 of Brevibacterium sp. strain HCU is not unexpected, since the latter protein has been shown to have a preference for cyclopentanone as substrate (11). The relatedness of CpnB to an unannotated gene or ORF in Thermobifida fusca predicts that this cellulolytic actinomycete could possibly metabolize cyclopentanone.
Although CPMO has been long recognized as a potentially useful enzyme, only limited efforts were made to exploit it for the BV reactions of unnatural substrates, since it was assumed that its stereoselectivity is similar to that of CHMO (39). Extensively investigated and often found to be highly enantioselective, CHMO eclipsed all other monooxygenases (61). It is interesting that the CHMO of Acinetobacter sp. strain 9871 is part of a metabolic pathway that does not involve chiral discrimination, yet it is frequently highly enantioselective. In this respect, the behavior of CPMO vis à vis unnatural substrates merited investigation. In addition, the discovery that CHMO manifests lower regio- and enantioselectivities in biotransformation of 2- and 3-substituted cyclopentanones (38) prompted us to check out the substrate range and specificity of CPMO.
The results of this study confirmed that CPMO accepts a broad variety of substrates and that the fermentations are fast, clean, and efficient. One of the earlier studies of the CPMO-catalyzed BV transformations reported a successful oxidation of 5-hexyl-2-cyclopentenone to the corresponding lactone (8). Since conjugated ketones are apparently not suitable substrates for CHMO, the ability of CPMO to oxidize such compounds is of considerable interest. As shown in Fig. 3, indan-1-one (compound 1) and 5-methoxy-indan-1-one (compound 2) were excellent substrates and were efficiently converted to the single lactones. These compounds were not transformed by CHMO. A nonconjugated indan-2-one (compound 3) was oxidized by both strains, but the reaction with CPMO was faster and gave a higher yield. The CHMO-catalyzed reaction gave the same lactone at a lower yield (57%) after 96 h. A longer fermentation time did not improve the yield.
Chemical BV oxidation of 1,4-cyclohexandione (compound 4) is tedious under standard conditions (m-chloroperbenzoic acid and trifluoroacetic acid; room temperature; 7 days; only 20% conversion) and the yield is low. The recombinant CPMO gave, after 28 h, lactone 4a in a better yield (46% after purification by chromatography). Although the isolated CHMO enzyme was reported to perform this transformation, neither conversion nor the yield was reported (1), and the engineered CHMO strain did not show any product after the 48-h reaction (data not shown).
The BVMO-catalyzed reactions have become an important tool in organic synthesis. The CHMO-catalyzed transformations are of particularly great value since they provide the desired lactones in high optical purities (55, 61). The CPMO-catalyzed oxidations in this study did not yield highly optically pure products, but they may still be advantageous under certain circumstances. For example, in cases when optical purity is not an issue because the product is not chiral (compounds 1 to 5), or when chemical oxidation takes a very long time, as in the oxidation of 4 to 4a, the reactions with CPMO may be an excellent, environmentally preferred alternative (72). Granted that these preliminary findings are based on a small number of substrates, certain CPMO-catalyzed reactions may provide the antipodes of lactones produced by CHMO, as in the case of compounds 6 and 9. CPMO merits further development as a reagent for organic synthesis, and it is clear from this and a related study (72) that the substrate base and the enantioselectivities of the two biocatalysts must not be considered as identical.
Besides CPMO, the cloned cpn genes are a useful resource for biocatalyst development. In particular, δ-valerolactone hydrolase of strain 9872 has been shown to be enantioselective towards the R enantiomer of δ-decanolactone and δ-nonalactone, whereas the corresponding caprolactone hydrolase of strain 9871 showed little such preference (51). Clearly, resolution of racemic lactones is a property to be explored further. Tandemly cloned cpnBA opens the possibility of using a relatively less toxic alcohol than an aldehyde as an initial substrate. The cloned cpnD is of potential use towards the preparation of 5-oxovalerate, as there is no commercial source of this substrate. Finally, CpnE in combination with the Cpn pathway proteins can afford a “green” route for glutaric acid preparation. A known organic route is oxidation of cyclopentane with air at high temperature and pressure, giving cyclopentanone and cyclopentanol, which are then oxidized further with nitric acid (generating nitrous oxide) to give a mixture of glutaric acid and succinic acid (35). Unfortunately, this biological route remains theoretical at best, since both the value and economics of glutaric dicarboxylic acid do not seem to be a fertile ground (35). On the other hand, interestingly at the time of this writing, the status of glutaric acid has been given a boost as a possible lucrative new market in niche applications, such as its use as a pharmaceutical additive, solder flux, and food addulant (67).
It is not an uncommon feature among catabolic pathways, especially the better-studied pathways of aromatic metabolism, to find ORFs or genes of unknown functions or that appear to have no apparent contribution to the pathway of interest. In the cpn pathway, Orf6 stands out, having clear homology to Orf1, which is found in a nitrotoluene and 4-nitrobenzyl alcohol pathway of Pseudomonas sp. strain TW3 (34), and to Orf3 of strain YH102, which is capable of 4-nitrobenzoate degradation. Interestingly, a homolog of Orf6 is also found in the degradation pathway of cyclohexanol in strain 9871 and SE19 (ChnX). The significance, if any, of this ORF in the respective pathways is yet to be determined. In addition, the presence of an integrase-like sequence (Orf3) in the cpn degradative gene locus possibly signifies a gene integration or transfer event. It is noteworthy that phage-like integrase sequences have been identified in the chlorocatechol and chlorobiphenyl degradation pathways that enable integration into specific sites on the chromosome and excision and transfer to other hosts (50, 69). At present, we have no evidence that the chromosomal-encoded cpn pathway is on a mobile element. On the other hand, it is interesting that, in Acinetobacter sp. strain 9871, a putative insertion sequence is located divergently from the chnC gene that encodes a caprolactone hydrolase (33). However, this sequence is absent in strain SE19 (19).
Finally, we conclude that the cpn pathway has both documented and untapped possibilities that need to be further explored in the framework of sustainable development and/or the promotion of green chemistry. Although compounds like cyclopentanol or cyclohexanol and their ketones are not priority pollutants (24), there are numerous naturally occurring molecules in the environment that contain these core or cycloalkane structures (70). Steroids and fossil fuel hydrocarbons are just two examples, and their biotransformation or functionalization can yield value-added products.
Acknowledgments
We thank W. Lotz for supplying plasmid pKOK6.1.
Footnotes
This publication is issued as NRCC number 44669.
REFERENCES
- 1.Abril, O., C. C. Ryerson, C. T. Walsh, and G. M. Whitesides. 1989. Enzymic Baeyer Villiger type oxidations of ketones catalyzed by cyclohexanone oxygenase. Bioorg. Chem. 17:41-52. [Google Scholar]
- 2.Adger, B., M. T. Bes, G. Grogan, R. McCague, S. Pedragosa-Moreau, S. M. Roberts, R. Villa, P. W. Wan, and A. J. Willetts. 1997. The synthesis of (R)-(+)-lipoic acid using a monooxygenase-catalysed biotransformation as the key step. Bioorg. Med. Chem. 5:253-261. [DOI] [PubMed] [Google Scholar]
- 3.Alphand, V., and R. Furstoss. 2000. Microbiological transformations 44. Optimisation of a new Baeyer-Villigerase activity: application to the stereospecific oxidation of 3-phenylcyclobutanone. J. Mol. Catal. B 9:209-217. [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch, A. 1991. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 19(Suppl.):2241-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barclay, S. S., J. M. Woodley, M. D. Lilly, P. L. Spargo, and A. J. Pettman. 2001. Production of cyclohexanone monooxygenase from Acinetobacter calcoaceticus for large scale Baeyer-Villiger monooxygenase reactions. Biotechnol. Lett. 23:385-388. [Google Scholar]
- 7.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54 dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bes, M. T., R. Villa, S. M. Roberts, P. W. H. Wan, and A. Willetts. 1996. Oxidative biotransformations by microorganisms: production of chiral synthons by cyclopentanone monooxygenase from Pseudomonas sp. NCIMB 9872. J. Mol. Catal. B 1:127-134. [Google Scholar]
- 9.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of protein utilizing the principle of the protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Brzostowicz, P. C., K. L. Gibson, S. M. Thomas, M. S. Blasko, and P. E. Rouviere. 2000. Simultaneous identification of two cyclohexanone oxidation genes from an environmental Brevibacterium isolate using mRNA differential display. J. Bacteriol. 182:4241-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzostowicz, P. C., M. S. Blasko, and P. E. Rouviere. 2002. Identification of two gene clusters involved in cyclohexanone oxidation in Brevibacterium epidermidis strain HCU. Appl. Microbiol. Biotechnol. 58:781-789. [DOI] [PubMed] [Google Scholar]
- 13.Carnell, A. J., S. M. Roberts, V. Sik, and A. J. Willetts. 1990. Enzyme-catalysed Baeyer Villiger oxidations of some substituted bicyclo[3.2. 0]heptanones. J. Chem. Soc. Chem. Commun. 1990:1438-1439. [Google Scholar]
- 14.Cha, J., W. Bishai, and S. Chandrasegaran. 1993. New vectors for direct cloning of PCR products. Gene 136:369-370. [DOI] [PubMed] [Google Scholar]
- 15.Cheesman, M. J., M. B. Kneller, E. J. Kelly, S. J. Thompson, C. K. Yeung, D. L. Eaton, and A. E. Rettie. 2001. Purification and characterization of hexahistidine-tagged cyclohexanone monooxygenase expressed in Saccharomyces cerevisiae and Escherichia coli. Protein Expr. Purif. 21:81-86. [DOI] [PubMed] [Google Scholar]
- 16.Chen, C. S., Y. Fujimoto, G. Girdaukas, and C. J. Sih. 1982. Quantitative analyses of biochemical kinetic resolutions of enantiomers. J. Am. Chem. Soc. 104:7294-7299. [Google Scholar]
- 17.Chen, G., M. M. Kayser, M. D. Mihovilovic, M. E. Mrstik, C. A. Martinez, and J. D. Stewart. 1999. Asymmetric oxidations at sulfur catalyzed by engineered strains that overexpress cyclohexanone monooxygenase. New J. Chem. 23:827-832. [Google Scholar]
- 18.Chen, Y.-C. J., O. P. Peoples, and C. T. Walsh. 1988. Acinetobacter cyclohexanone monooxygenase: gene cloning and sequence determination. J. Bacteriol. 170:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng, Q., S. M. Thomas, K. Kostichka, J. R. Valentine, and V. Nagarajan. 2000. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J. Bacteriol. 182:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corma, A., L. T. Nemeth, M. Renz, and S. Valencia. 2001. Sn-zeolite beta as a heterogeneous chemoselective catalyst for Baeyer-Villiger oxidations. Nature 412:423-425. [DOI] [PubMed] [Google Scholar]
- 21.Diaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11:467-475. [DOI] [PubMed] [Google Scholar]
- 22.Doig, S. D., L. M. O'Sullivan, S. Patel, J. M. Ward, and J. M. Woodley. 2001. Large scale production of cyclohexanone monooxygenase from Escherichia coli TOP10 pQR239. Enzyme Microb. Technol. 28:265-274. [DOI] [PubMed] [Google Scholar]
- 23.Duetz, W. A., J. B. van Beilen, and B. Witholt. 2001. Using proteins in their natural environment: potential limitations of microbial whole-cell hydroxylations in applied biocatalysis. Curr. Opin. Biotechnol. 12:419-425. [DOI] [PubMed] [Google Scholar]
- 24.Fisher, W. B., J. F. VanPeppen, et al. 1993. Cyclohexanol and cyclohexanone, p. 410-416. In M. Bickford and J. I. Kroschwitz (ed.), Kirk-Othmer encyclopedia of chemical technology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 25.Fraaije, M. W., N. N. Kamerbeek, W. J. H. van Berkel, and D. B. Janssen. 2002. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 518:43-47. [DOI] [PubMed] [Google Scholar]
- 26.Furlong, C. E., L. G. Costa, C. Hassett, R. J. Richter, J. A. Sundstrom, D. A. Adler, C. M. Disteche, C. J. Omiecinski, C. Chapline, J. W. Crabb, and R. Humbert. 1993. Human and rabbit paraoxonases: purification, cloning, sequencing, mapping and role of polymorphism in organophosphate detoxification. Chem.-Biol. Interact. 87:35-48. [DOI] [PubMed] [Google Scholar]
- 27.Griffin, M., and P. W. Trudgill. 1972. The metabolism of cyclopentanol by Pseudomonas NCIB 9872. Biochem. J. 129:595-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffin, M., and P. W. Trudgill. 1976. Purification and properties of cyclopentanone oxygenase of Pseudomonas NCIB 9872. Eur. J. Biochem. 63:199-209. [DOI] [PubMed] [Google Scholar]
- 29.Hasegawa, Y., Y. Nakai, T. Tokuyama, and H. Iwaki. 2000. Purification and characterization of cyclohexanone 1,2-monooxygenase from Exophiala jeanselmei strain KUFI-6N. Biosci. Biotechnol. Biochem. 64:2696-2698. [DOI] [PubMed] [Google Scholar]
- 30.Hassett, C., R. J. Richter, R. Humbert, C. Chapline, J. W. Crabb, C. J. Omiecinski, and C. E. Furlong. 1991. Characterization of cDNA clones encoding rabbit and human serum paraoxonase: the mature protein retains its signal sequence. Biochemistry 30:10141-10149. [DOI] [PubMed] [Google Scholar]
- 31.Hempel, J., H. Nicholas, and R. Lindahl. 1993. Aldehyde dehydrogenases: widespread structural and functional diversity within a shared framework. Protein Sci. 2:1890-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwaki, H., Y. Hasegawa, M. Teraoka, T. Tokuyama, H. Bergeron, and P. C. K. Lau. 1999. Identification of a transcriptional activator (ChnR) and a 6-oxohexanoate dehydrogenase (ChnE) in the cyclohexanol catabolic pathway in Acinetobacter sp. strain NCIMB 9871 and localization of the genes that encode them. Appl. Environ. Microbiol. 65:5158-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwaki, H., Y. Hasegawa, M. Teraoka, T. Tokuyama, L. Bernard, and P. C. K. Lau. Cyclohexanol biodegradation genes: a pathway of opportunities. In R. A. Gross and H. N. Cheng (ed.), Biocatalysis: frontiers in polymer reactions. ACS Press, Dallas, Tex., in press.
- 34.James, K. D., M. A. Hughes, and P. A. Williams. 2000. Cloning and expression of ntnD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J. Bacteriol. 182:3136-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, R. W., C. M. Pollock, and R. R. Cantrell. 1993. Dicarboxylic acids, p. 614-628. In M. Bickford and J. I. Kroschwitz (ed.), Kirk-Othmer encyclopedia of chemical technology, 3rd ed. John Wiley & Sons Inc., New York, N.Y.
- 36.Jornvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzalez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 37.Kamerbeek, N. M., M. J. Moonen, J. G. Van Der Ven, W. J. Van Berkel, M. W. Fraaije, and D. B. Janssen. 2001. 4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB. A novel flavoprotein catalyzing Baeyer-Villiger oxidation of aromatic compounds. Eur. J. Biochem. 268:2547-2557. [DOI] [PubMed] [Google Scholar]
- 38.Kayser, M. M., G. Chen, and J. D. Stewart. 1998. Enantio- and regioselective Baeyer-Villiger oxidations of 2- and 3-substituted cyclopentanones using engineered bakers' yeast. J. Org. Chem. 63:7103-7106. [DOI] [PubMed] [Google Scholar]
- 39.Kelly, D. R., P. W. H. Wan, and J. Tang. 1998. Biotransformations I. Bio/Technology 8:536-587. [Google Scholar]
- 40.Kneller, M. B., M. J. Cheesman, and A. E. Rettie. 2001. ESI- and MALDI-MS analysis of cyclohexanone monooxygenase from Acinetobacter NCIB 9871. Biochem. Biophys. Res. Commun. 282:899-903. [DOI] [PubMed] [Google Scholar]
- 41.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84:467-471. [DOI] [PubMed] [Google Scholar]
- 42.Kustu, S., A. K. North, and D. S. Weiss. 1991. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem. Sci. 16:397-402. [DOI] [PubMed] [Google Scholar]
- 43.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 45.Liu, Z. J., Y. J. Sun, J. Rose, Y. J. Chung, C. D. Hsiao, W. R. Chang, I. Kuo, J. Perozich, R. Lindahl, J. Hempel, and B. C. Wang. 1997. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4:317-326. [DOI] [PubMed] [Google Scholar]
- 46.Marcus, P., and P. Talalay. 1956. Induction and purification of α- and β-hydroxysteroid dehydrogenase. J. Biol. Chem. 218:661-674. [PubMed] [Google Scholar]
- 47.Mazzini, C., J. Lebreton, V. Alphand, and R. Furstoss. 1997. A chemoenzymatic strategy for the synthesis of enantiopure (R)-(−)-baclofen. Tetrahedron Lett. 38:1195-1196. [Google Scholar]
- 48.Mihovilovic, M. D., G. Chen, S. Wang, B. Kyte, F. Rochon, M. M. Kayser, and J. D. Stewart. 2001. Asymmetric Baeyer-Villiger oxidations of 4-mono- and 4,4-disubstituted cyclohexanones by whole cells of engineered Escherichia coli. J. Org. Chem. 66:733-738. [DOI] [PubMed] [Google Scholar]
- 49.Morii, S., S. Sawamoto, Y. Yamauchi, M. Miyamoto, M. Iwami, and E. Itagaki. 1999. Steroid monooxygenase of Rhodococcus rhodochrous: sequencing of the genomic DNA, and hyperexpression, purification, and characterization of the recombinant enzyme. J. Biochem. 126:624-631. [DOI] [PubMed] [Google Scholar]
- 50.Nishi, A., K. Tominaga, and K. Furukawa. 2000. A 90-kilobase conjugative chromosomal element coding for biphenyl and salicylate catabolism in Pseudomonas putida KF715. J. Bacteriol. 182:1949-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onakunle, O. A., C. J. Knowles, and A. W. Bunch. 1997. The formation and substrate specificity of bacterial lactonases capable of enantioselective resolution of racemic lactones. Enzyme Microbiol. Technol. 21:245-251. [Google Scholar]
- 52.Pouchert, C. J. 1983. The Aldrich library of NMR, vol. 2, p. 308d.
- 53.Reiner, L. 1984. Dihydrocoumarin. Beilstein Handb. Organic Compounds. 17:315. [Google Scholar]
- 54.Renz, M., and B. Meunier. 1999. 100 years of Baeyer-Villiger oxidations. Eur. J. Org. Chem. 1999:737-750. [Google Scholar]
- 55.Roberts, S. M., and P. W. H. Wan. 1998. Enzyme-catalysed Baeyer-Villiger oxidations. J. Mol. Catal. B 4:111-136. [Google Scholar]
- 56.Rossello-Mora, R., and R. Amann. 2001. The species concept of prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Sandey, H., and A. Willetts. 1989. Biotransformation of cycloalkanones by microorganisms. Biotechnol. Lett. 11:615-620. [Google Scholar]
- 59.Sheng, D., D. P. Ballou, and V. Massey. 2001. Mechanistic studies of cyclohexanone monooxygenase: chemical properties of intermediates involved in catalysis. Biochemistry 40:11156-11167. [DOI] [PubMed] [Google Scholar]
- 60.Smith, S. P., K. R. Barber, S. D. Dunn, and G. S. Shaw. 1996. Structural influence of cation binding to recombinant human brain S100b: evidence for calcium-induced exposure of a hydrophobic surface. Biochemistry 35:8805-8814. [DOI] [PubMed] [Google Scholar]
- 61.Stewart, J. D. 1998. Cyclohexanone monooxygenase: a useful reagent for asymmetric Baeyer-Villiger reactions. Curr. Org. Chem. 2:195-216. [Google Scholar]
- 62.Stewart, J. D., K. W. Reed, J. Zhu, G. Chen, and M. M. Kayser. 1996. A “designer yeast” that catalyzes the kinetic resolutions of 2-alkyl-substituted cyclohexanones by enantioselective Baeyer-Villiger oxidations. J. Org. Chem. 61:7652-7653. [DOI] [PubMed] [Google Scholar]
- 63.Strukul, G. 1998. Transition metal catalysis in the Baeyer-Villiger oxidation of ketones. Angew. Chem. Int. Ed. 23:1198-1209. [DOI] [PubMed] [Google Scholar]
- 64.Tamaoka, J., D.-M. Ha, and K. Komagata. 1987. Reclassification of Pseudomonas acidovorans den Dooren de Jong 1926 and Pseudomonas testosteroni Marcus and Talalay 1956 as Comamonas acidovorans comb. nov. and Comamonas testosteroni comb. nov., with an emended description of the genus Comamonas. Int. J. Syst. Bacteriol. 37:52-59. [Google Scholar]
- 65.Tanner, A., and D. J. Hopper. 2000. Conversion of 4-hydroxyacetophenone into 4-phenyl acetate by a flavin adenine dinucleotide-containing Baeyer-Villiger-type monooxygenase. J. Bacteriol. 182:6565-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trudgill, P. W. 1990. Cyclopentanone 1,2-monooxygenase from Pseudomonas NCIMB 9872. Methods Enzymol. 188:77-81. [DOI] [PubMed] [Google Scholar]
- 67.Tullo, A. 2002. Glutaric acid debuts. Chem. Eng. News 80:13. [Google Scholar]
- 68.Vallon, O. 2000. New sequence motifs in flavoproteins: evidence for common ancestry and tools to predict structure. Proteins 38:95-114. [DOI] [PubMed] [Google Scholar]
- 69.van der Meer, J. R., R. Ravan, and V. Sentchilo. 2001. The clc element of Pseudomonas sp. strain B13 and other mobile degradative elements employing phage-like integrases. Arch. Microbiol. 175:79-85. [DOI] [PubMed] [Google Scholar]
- 70.Wackett, L. P., and C. D. Hershberger. 2001. Biocatalysis and biodegradation: microbial transformation of organic compounds. ASM Press, Washington, D.C.
- 71.Walsh, C. T., and Y.-C. J. Chen. 1988. Enzymic Baeyer-Villiger oxidations by flavin-dependent monooxygenases. Angew. Chem. Int. Ed. 27:333-343. [Google Scholar]
- 72.Wang, S., G. Chen, M. M. Kayser, H. Iwaki, P. C. K. Lau, and Y. Hasegawa. 2002. Baeyer-Villiger oxidations catalyzed by engineered microorganisms: enantioselective synthesis of δ-valerolactones with functionalized chains. Can. J. Chem. 80:613-621. [Google Scholar]
- 73.Wikstrom, P., E. O'Neill, L. C. Ng, and V. Shingler. 2001. The regulatory N-terminal region of the aromatic-responsive transcriptional activator DmpR constrains nucleotide-triggered multimerisation. J. Mol. Biol. 314:971-984. [DOI] [PubMed] [Google Scholar]
- 74.Willetts, A. 1997. Structural studies and synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 15:55-62. [DOI] [PubMed] [Google Scholar]
- 75.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 76.Yang, D., Y. Oyaizu, H. Oyaizu, G. J. Olsen, and C. R. Woese. 1985. Mitochondrial origins. Proc. Natl. Acad. Sci. USA 82:4443-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strain: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:10311-103119. [DOI] [PubMed] [Google Scholar]