Abstract
Alpha-galactosides are abundant sugars in legumes such as soy. Because of the lack of α-galactosidase (α-Gal) in the digestive tract, humans are unable to digest these sugars, which consequently induce flatulence. To develop the consumption of the otherwise highly nutritional soy products, the use of exogenous α-Gal is promising. In this framework, we characterized the melA gene for α-Gal in Lactobacillus plantarum. The melA gene encodes a cytoplasmic 84-kDa protein whose enzymatically active form occurs as oligomers. The melA gene was cloned and expressed in Escherichia coli, yielding an active α-Gal. We show that melA is transcribed from its own promoter, yielding a monocistronic mRNA, and that it is regulated at the transcriptional level, i.e., it is induced by melibiose but is not totally repressed by glucose. Posttranscriptional regulation by the carbon source could also occur. Upstream of melA, a putative galactoside transporter, designated RafP, was identified that shows high homology to LacS, the unique transporter for both α- and β-galactosides in Streptococcus thermophilus. rafP is also expressed as a monocistronic mRNA. Downstream of melA, the lacL and lacM genes were identified that encode a heterodimeric β-galactosidase. A putative galM gene identified in the same cluster suggests the presence of a galactose operon. These results indicate that the genes involved in galactoside catabolism are clustered in L. plantarum ATCC 8014. This first genetic characterization of melA and of its putative associated transporter, rafP, in a lactobacillus opens doors to various applications both in the manufacture of soy-derived products and in probiotic and nutraceutical issues.
Human consumption of soy-derived products has been hampered by the presence of α-galactosides, mostly raffinose and stachyose, in soybeans. Alpha-galactosides are carbohydrate reserves in many plant tissues and particularly in seeds. They include one or several galactose units, linked together or to the glucose moiety of sucrose through α-1,6 linkages. Since humans and monogastric animals are deficient in pancreatic α-galactosidase (α-Gal), α-galactosides are not digested in the duodenum. Therefore, they pass into the large intestine where they are degraded by gas-producing intestinal bacteria, such as Clostridium spp. and Bacteroides spp., yielding considerable amounts of CH4, CO2, and H2. The abnormal accumulation of flatulent rectal gas thus provokes gastrointestinal distress, such as abdominal pain, nausea, diarrhea, and increased peristalsis (46).
To overcome these drawbacks of soy products and to boost the consumption of these otherwise highly nutritional food products, many attempts have been made to eliminate the α-galactosides from soybeans. They consist of tedious physical methods, including bean soaking, bean germination, water extraction, and ultrafiltration (28, 30, 32). Alternatively, the use of α-Gal as a biotechnological approach for removal of α-galactosides from leguminous seeds has been proposed (13, 18, 51). The enzyme α-Gal (α-d-galactoside-galactohydrolase [EC 3.2.1.22]) catalyzes the hydrolysis of α-1,6-galactoside links present in α-galactosides, such as raffinose. In this framework, the use of microbial α-Gal offers a promising solution for the degradation of α-galactosides, especially during soymilk fermentation by lactic acid bacteria (LAB).
Several microorganisms are known to produce α-Gal (3, 13). In LAB, Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus brevis, and Lactobacillus buchneri used in vegetable fermentations can hydrolyze α-galactosides to digestible carbohydrates. However, little is known about LAB α-Gals. Recently, the α-Gal-encoding genes from two lactic acid bacteria, Carnobacterium piscicola and Lactococcus raffinolactis, were cloned and characterized (10; I. Boucher, M. Parrot, C. Vadeboncoeur, and S. Moineau, Abstr. Int. Anim. Agric. Food Sci. Conf. Am. Dairy Sci. Assoc., Indianapolis, Ind., abstr. 24, p. 6, 2001). In lactobacilli, α-Gals have been characterized at the biochemical and physiological levels (1, 21, 22, 38, 40, 52, 57). No data are available regarding the molecular characterization of α-Gal in lactobacilli. In order to fill this gap, with the aim of developing probiotic microorganisms with high α-Gal activity, we undertook the genetic characterization of the α-Gal system in Lactobacillus. In this study we describe the cloning, sequencing, and characterization of an α-Gal gene (melA) from L. plantarum ATCC 8014 and its expression in Escherichia coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Lactobacilli were grown in MRS broth (15) supplemented with glucose, galactose, melibiose, or raffinose at a 2% (wt/vol) final concentration at 30 or 37°C without shaking. The α-Gal− strain E. coli RA11r (ΔlacZY melA recA) (24) is a K12 derivative and was used in expression experiments. E. coli DH5α [supE44Δ(lac)U169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used for all other genetic manipulations. E. coli strains were routinely grown in Luria-Bertani medium (49) at 37°C, in M9 minimal medium (49) supplemented with 1% melibiose (M9mel), or in M9 minimal medium enriched with 0.2% Casamino Acids-10 mM glycerol (50)-1% melibiose (EM9mel) at 30°C under aerobic conditions. When appropriate, ampicillin was added at a concentration of 100 μg/ml.
DNA manipulations.
Chromosomal DNA was extracted from lactobacilli as previously described (45). E. coli CaCl2-competent cells were used, and heat shock transformation and recombinant DNA techniques were performed according to the method of Sambrook et al. (49). Restriction endonucleases and DNA polymerases were purchased from New England Biolabs, (New England Biolabs, Inc., Beverly, Mass.) or Promega (Promega Co., Madison, Wis.) and used as recommended by the suppliers. For routine PCR amplification, Taq polymerase (Qbiogene, Illkirch, France) was used. For cloning and sequencing purposes, the high-fidelity Vent polymerase (New England Biolabs) was used. Amplified fragments were either cloned in pBluescript II SK+ (pBS) (Stratagene, La Jolla, Calif.) with E. coli DH5α as a host or directly sequenced using an automated DNA Sequencer 373 (Applied Biosystems Inc., Foster City, Calif.).
Probing, cloning, and sequencing of the gene for α-Gal and its flanking regions.
Based on the nucleotide sequence of the AGA-1 (α-Gal R) gene of Pediococcus pentosaceus (GenBank sequence accession no. Z32771) and an L. plantarum C3.8 partial sequence (courtesy of Evaristo Suarez, Oviedo, Spain), the degenerate primer P2 and nondegenerate Pal7R (Table 1) were used to amplify the genes for α-Gal in lactobacilli. Amplicons were cloned in pBS and sequenced. The upstream genomic sequence of the amplified fragment was obtained by Uneven PCR (8). For this purpose, nested primers were combined with a set of arbitrarily selected 10-mer primers from the B series of Bioprobe (Bioprobe Systems, Montreuil-sous-Bois, France). The resulting DNA fragments were sequenced, and new nested primers were designed to continue walking. The following oligonucleotides pairs were used in first and second PCR rounds, respectively: PXRb and P2Rb, Pal11R and Pal12R, Pal17 and Pal18, Pal19 and Pal20, Pal21 and Pal22, and Raf1 and Raf2 (Table 1). After assembly of the sequences obtained in six independent uneven PCRs, the entire region was amplified using Raf5 and Pal25 primers and the high-fidelity Vent DNA polymerase. The amplicons obtained were subsequently sequenced. The downstream sequence of the original P2-Pal7R amplicon was obtained by inverse PCR: chromosomal L. plantarum ATCC 8014 DNA was digested to completion with EcoRI and self-ligated to yield the template used with the divergent primers Pu02 and Pu04. Once the sequences were assembled, three complete open reading frames (ORFs) were detected. The primers Pal11b and Pal14Re with sites for restriction enzymes (BamHI and EcoRI, respectively) were then used to amplify a fragment spanning the entire ORF 2, which was then cloned in pBS to yield the plasmid p1α. In order to characterize the sequence lying downstream of the gene for α-Gal, the primers Lac1B and Lac5B were designed on the basis of the highly homologous lacL and lacM genes from Leuconostoc lactis (GenBank sequence accession no. M92281) (14), and the resulting amplicon was sequenced.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a,b |
|---|---|
| P2 | TTTGGTTMGARCCAGAAAWKATTTCA |
| Pal7R | CTAGTTTGAACGGGAGTCGTG |
| PXRb | GCGGATCCGGCTGGCTNAAATCAAGC |
| P2Rb | GCGGATCCGTATTCGTTCCGCGATAA |
| Pal11R | AACGACCATAACGGTATCATCC |
| Pal12R | TTGTACGGTGGTTCCTGAATC |
| Pal17 | TCATAGCAGCAGGTACACCG |
| Pal18 | CGGTTTCCAAGTGAGTTGACC |
| Pal19 | ACGACTAAACTTCTTTGCTAATGG |
| Pal20 | TAACAACTGCTAAGAAGAACA |
| Pal21 | AAATTGCAAAAATAATTAAATATAGAATGGGG |
| Pal22 | CACCAATGTTAGAGCCAACCCGGGCG |
| Raf1 | GAAGGCATCGTGCCCAAACGCCCC |
| Raf2 | CCACTACTCTTATCGAACAAGTGGGAGG |
| Raf5 | GGGATACTTGAATGATAAGCGCC |
| Pal25 | GTGGGCCAACACGTTGCCTGTCTCC |
| Pu02 | GGCTTCATCCTCATCTTCGAC |
| Pu04 | GGATGTTTGTTAGTCCGGATAAG |
| Lac1B | CAGCGGCGGTATAGGGAAGGC |
| Lac5B | CCAGGCCTATTCAGCGACGCCGCGG |
| 4A | CGGACTTTGGTTCGAGCCAG |
| 5AR | CGACCGTCCTAACAATTCATCCGG |
| Pal11b | GCGGATCCGGATGATACCGTTATGGTCGTT |
| Pal14Re | GCGAATTCTTATCAATCTACGGTGCCG |
M, R, W and K represent A/C, A/G, A/T, and G/T, respectively.
Sites for restriction enzymes (BamHI and EcoRI) are underlined.
DNA and protein sequences analyses.
Nucleotide and amino acid sequence homology searches were performed using the BLAST facility at the National Center of Biotechnology Information (2). Promoters were searched using the Neural Network Promoter Prediction algorithm (www.fruitfly.org/seq_tools/promoter.html) (48). DNA secondary structures were analyzed using the GeneBee server (http://www.genebee.msu.su/genebee.html) (7). Protein alignments were made using the BOXSHADE 3.21 software (http://www.ch.embnet.org/software/BOX_form.html), and predictions of protein signals or structures were obtained using the ExPASy Molecular Biology Server (http://expasy.ch).
Preparation of cell extracts and α-Gal activity measurement.
For lactobacilli, 7-ml cultures at an optical density at 600 nm (OD600) of 1.5 were harvested by centrifugation at 10,000 × g for 3 min at 4°C, washed twice with Mc Ilvaine buffer(Na2HPO4-citric acid, pH 5.8 [37]) and resuspended in 0.5 ml of the same buffer (final OD600, 20). Cells were disrupted with 500 mg of glass beads (0.10 to 0.11 mm; Sigma) upon shaking at maximum speed on a vortex mixer during four cycles of 2 min each with 5-min pauses on ice. Cellular debris was removed by centrifugation at 10,000 × g for 10 min at 4°C, and α-Gal activity in supernatants was determined by the method of Church et al. (9). One unit of α-Gal is defined as the amount of enzyme that releases 1.0 μmol of pNP per min. The protein concentration was determined by the method of Bradford using bovine serum albumin as a standard (6). The α-Gal activity in E. coli EM9mel cultures was determined using 10-ml samples harvested at regular time intervals, washed with Mc Ilvaine buffer, and further processed as described for lactobacilli.
Protein analyses.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out according to the method of Laemmli (33) using a Mini Protean II system (Bio-Rad, Hercules, Calif.). Proteins were stained using Coomassie brilliant blue R-250. Bovine serum albumin (New England Biolabs) was used as an internal standard for α-Gal quantitation in gels. Ultrafiltration studies were performed on L. plantarum ATCC 8014 cell extracts using Ultrafree-MC filters units as recommended by the manufacturer (Millipore, Guyancourt, France). Both α-Gal activity and the amount of protein were determined in the resulting retentates and ultrafiltrates, as indicated above.
RNA isolation, Northern hybridization, and 5′ RACE analysis.
Total RNA was extracted from L. plantarum as previously described for Lactococcus lactis (47). A 535-bp internal fragment of melA amplified by PCR with primers 4A and 5AR was used as a probe. For Northern blot analysis, 25-μg RNA samples were denatured, separated by electrophoresis along with an RNA ladder (Invitrogen, Cergy-Pontoise, France) in 1.25% agarose gels, and transferred to a Hybond-N membrane (Amersham Biosciences, Saclay, France). RNA was stained with methylene blue (56). DNA probe labeling and signal detection were carried out using the ECL kit as recommended by the manufacturer (Amersham Biosciences). To determine the transcription start region, rapid amplification of 5′-cDNA ends (5′ RACE) experiments were performed according to the instruction provided by the manufacturer (Gibco-BRL, Invitrogen). The 5′ RACE system is a procedure for amplification of nucleic acid sequences from an mRNA template between a defined internal site and unknown sequences at the 5′ end of the mRNA. This system includes synthesis of first-strand cDNA, homopolymeric tailing of the cDNA, and PCR amplification using a gene-specific primer and an adapter primer complementary to the homopolymeric tail. Briefly, total RNA was isolated, DNase treated, and reverse transcribed using a reverse primer downstream of the putative transcription initiation point (Pal18 and Pal7R primers for rafP and melA, respectively). After RNase treatment, a poly(G) tail was added to the 5′ end of cDNA by terminal deoxynucleotidyl transferase in the presence of dGTP, and PCR was performed using a specific nested reverse primer (Pal17 and 5AR for rafP and melA, respectively) combined with a poly(C) primer provided by the kit.
Nucleotide sequence accession numbers.
The DNA sequences described in this study are available in the GenBank database under accession numbers AY048859 (partial galM and rafP), AF189765 (melA), and AY048860 (partial lacL and lacM).
RESULTS
Selection of α-Gal producer strains.
Several Lactobacillus strains were screened for their capacity to use melibiose and raffinose by using the API 50CH test (bioMérieux, Marcy l'Etoile, France) (data not shown). A panel of positive strains including L. plantarum, L. fermentum, L. brevis, L. buchneri, and Lactobacillus reuteri was retained for assay of α-Gal activity. L. fermentum CRL251 and CRL722 showed the highest α-Gal activities. These two strains and the widely studied model strain L. plantarum ATCC 8014 were selected for further studies.
Organization and sequence analysis of the galactosidase gene cluster.
The three α-Gal+ strains, L. fermentum CRL722 and CRL251 and L. plantarum ATCC 8014, were probed for the presence of the gene for α-Gal using PCR with degenerate oligonucleotides. In L. plantarum only, an amplicon of 800 bp was obtained using P2-Pal7R primers, and we decided to characterize the gene for α-Gal in this bacterium. Sequence analysis of the PCR product showed high homology with the genes for α-Gal from various microorganisms. Using uneven and inverse PCR techniques, we walked the genome of L. plantarum on both sides of this fragment. Analysis of the sequence revealed the gene organization illustrated in Fig. 1. The central gene encoding α-Gal was designated melA, according to the nomenclature in E. coli (melA stands for melibiase). Upstream of melA, a complete ORF (rafP) corresponding to a putative raffinose transporter and a partial ORF (galM) similar to the microbial galactose-1-epimerase (mutarotase) gene were identified. Inspection of the sequence downstream of melA revealed the presence of two putative genes on the opposite strand that encode the two subunits of the heterodimeric β-galactosidase (lacL and lacM).
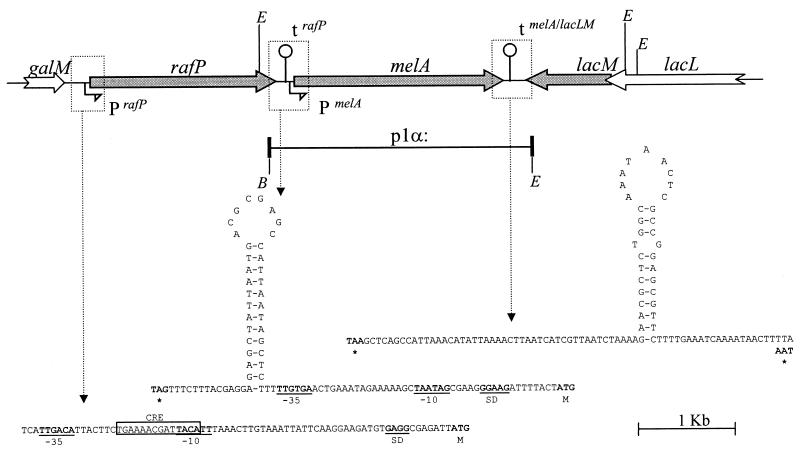
FIG. 1.
Galactoside metabolism gene cluster of L. plantarum ATCC 8014. The gene order and nucleotide sequence of intercistronic regions are shown. White arrows correspond to incomplete ORFs. The region cloned in p1α is represented as a bold segment. Relevant restriction enzymes sites are indicated: B, BamHI; E, EcoRI. Promoters (P) and terminators (t) are indicated. Secondary structures are given as corresponding DNA sequences. Stop (∗) and start codons (M) are in bold letters. Promoter sequences (−35, −10) and Shine Dalgarno sequence (SD) are underlined. The CRE site is boxed.
(i) The melA structural gene for α-Gal.
The 2,214-bp melA gene encodes a 738-amino-acid polypeptide with a deduced molecular weight of 84 kDa and an isoelectric point of 5.12. Based on its amino acid sequence, MelA belongs to family 36 of the glycosyl hydrolases according to the Henrissat classification (25, 26). Prediction of protein localization was performed using the Psort program from the ExPASy server. Neither a signal peptide nor a transmembrane segment was identified, suggesting that MelA is located in the cytoplasm. MelA and other α-Gals from family 36 of glycosyl hydrolases show strong conservation (>75%) in their central domain (Fig. 2).
FIG. 2.
Multiple alignment analysis of putative active sites of α-Gal sequences from microorganisms producing family 36 glycosyl hydrolases. Sequences from Lactobacillus plantarum ATCC 8014 (melA L.plan), Pediococcus pentosaceus (aga1 P.pent), Bacillus stearothermophilus (agaN B.stea), Thermoanaerobacter ethanolicus (melA T.etha), Carnobacterium piscicola (agaA C.pisc), Escherichia coli (rafA E.coli), and Bifidobacterium adolescentis (aga B.adol) are shown. The gene denomination precedes the species acronym at the beginning of each line. The last amino acid number is at the right end of each sequence.
Inspection of the sequence at the 5′ end of melA revealed a potential promoter sequence and a putative Shine-Dalgarno sequence (GGAAG) 8 nucleotides upstream of the ATG start codon of melA. Downstream of melA, inverted repeats were detected, indicative of a stem-loop structure with a free-energy change (ΔG0) value of −17 Kcal/mol. This structure could act as a transcriptional terminator for both the melA and the lacLM genes since it is preceded by a stretch of A bases and followed by a stretch of T bases (see below).
(ii) The rafP gene for a raffinose transporter.
The rafP gene specifies a 652-residue protein with a calculated molecular weight of 73 kDa. No putative signal peptide was detected in the RafP sequence, while 9 to 12 transmembrane segments (according to the prediction program used) are predicted, suggesting that it is an integral membrane protein. RafP exhibits a high level of homology with the raffinose transporter of P. pentosaceus (71% similarity) (GenBank sequence accession no L32093/Z32771). Also, RafP shows high homology (>70% similarity) with and a structural organization similar to that of LacS, the bifunctional lactose and α-galactoside transporter from Streptococcus thermophilus (44). Similar proteins have been found for Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus salivarius, and Leuconostoc lactis (34, 53, 54).
Inspection of the sequence upstream of the rafP gene revealed a putative promoter sequence and a consensus catabolite responsive element (CRE) located between the −10 and −35 boxes (Fig. 1). This organization suggests that this gene is subject to catabolic repression (27). A potential Shine-Dalgarno sequence (GAGG) is located 7 bp upstream of the ATG start codon. Downstream of rafP and just upstream of the −35 box of the potential melA promoter, the sequence shows a putative mRNA transcriptional terminator consisting of a stem-loop structure with a ΔG0 of −12.6 Kcal/mol followed by a stretch of T.
(iii) The lacL and lacM genes for β-galactosidase.
Downstream of melA and located on the opposite strand, the overlapping β-Gal genes lacM and lacL were identified. They showed 98% similarity with the LacL and LacM subunits of β-Gal from Leuconostoc lactis (14) (GenBank accession no. Q02603), 77% similarity with lacLM genes from Lactobacillus acidophilus (GenBank accession no. BAA20536/O07684), and 74% similarity with those from Lactobacillus sakei (41) (GenBank accession no 2208404A).
The L. plantarum ATCC 8014 melA gene can be highly expressed in E. coli.
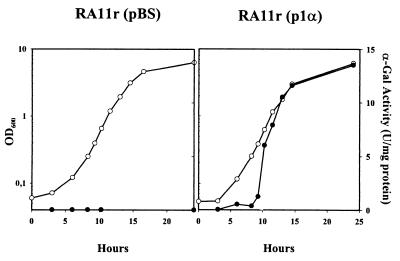
A 2,690-bp PCR fragment containing the melA gene from L. plantarum ATCC 8014 including the putative rafP terminator, melA promoter, and melA terminator was cloned in pBS in the same orientation as the lacZ promoter, yielding the plasmid p1α (Fig. 1). To determine whether the melA gene from L. plantarum ATCC 8014 was active in E. coli, p1α was introduced in the α-Gal− E. coli RA11r strain and grown in M9mel. In contrast with the native strain, E. coli RA11r(p1α) showed normal growth in M9mel, suggesting that melA is functional in E. coli (data not shown). In a second step, cultures of E. coli RA11r(pBS) and E. coli RA11r(p1α) were grown in the enriched EM9mel medium, allowing the growth of both strains (Fig. 3). Monitored α-Gal activity showed no α-Gal production in E. coli RA11r(pBS) and an α-Gal production kinetics in E. coli RA11r(p1α) that paralleled the growth curve (Fig. 3). Interestingly, α-Gal production reached amounts about 15 times higher than those found with the native producer L. plantarum ATCC 8014 (Fig. 3; compare with Fig. 6). This increase could be partly due to a gene dosage effect, since p1α that derives from pBS is a high-copy-number plasmid. The high level of expression strongly suggests that the putative promoter sequence identified upstream of melA is active in E. coli. The observed high expression levels of melA were obtained in cultures without the inducer isopropyl-β-d-thiogalactopyranoside. In addition, p1α contains the putative rafP terminator located upstream of the melA promoter sequence, and we presume that this terminator would arrest any transcript synthesized from the lacZ promoter. Altogether, these observations strongly suggest that the L. plantarum ATCC 8014 melA gene is functional and that it is transcribed from its own promoter.
FIG. 3.
Growth (white) in EM9mel at 30°C and α-Gal activity (black) of α-Gal− E. coli RA11r(pBS) and RA11r(p1α).
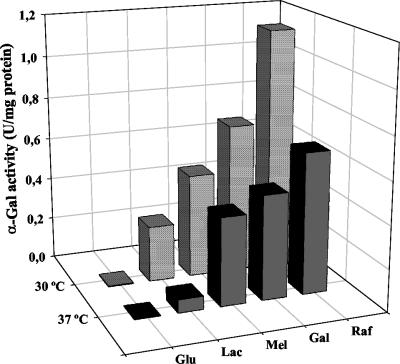
FIG. 6.
α-Gal activity in cell extracts of L. plantarum ATCC 8014 grown at 30 or 37°C in MRS plus glucose (Glu), lactose (Lac), melibiose (Mel), galactose (Gal), and raffinose (Raf) until reaching an OD600 of 1.5.
Transcriptional analyses of the melA and rafP genes of L. plantarum ATCC 8014.
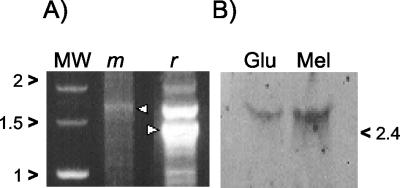
mRNA from L. plantarum ATCC 8014 was analyzed by both 5′ RACE and Northern blotting. The 5′RACE experiment was performed using reverse primers Pal17 and 5AR, located 1.4 and 1.8 kb downstream of the putative promoter sequences of rafP and melA, respectively. Major amplicons of 1.4 kb (rafP) and 1.8 kb (melA), respectively, were obtained (Fig. 4A). These correspond in size to monocystronic mRNA. In the case of rafP, additional amplicons of greater and lesser size were detected. The larger amplicon could be due to unspecific annealing of the rafP primer, or it could reflect the presence of polycistronic mRNA. The smaller amplicons could be due to the presence of degradation products.
FIG. 4.
(A) RNA analysis by 5′RACE experiment. PCR fragments were amplified from cDNA using internal primers for the rafP gene (r) or the melA gene (m). Arrowheads indicate specific products. MW, Marker Smart Ladder (Eurogentec). (B) Northern blot analysis of L. plantarum ATCC 8014 grown in glucose (Glu) or melibiose (Mel) using an internal melA fragment as a probe. Migration of molecular weight standards is indicated on the right edge of the figure.
In order to assess whether the melA gene is regulated in L. plantarum ATCC 8014, mRNA was hybridized with a melA probe generated by labeling a 535-bp PCR fragment. An approximately unique 2.5-kb band corresponding to the melA transcript was detected irrespective of whether glucose or melibiose was used (Fig. 4B). This result confirms that melA is transcribed from its own promoter. Interestingly, melA transcription was induced at a level of approximately fourfold when L. plantarum ATCC 8014 was grown with melibiose compared to results with glucose.
Protein analyses.
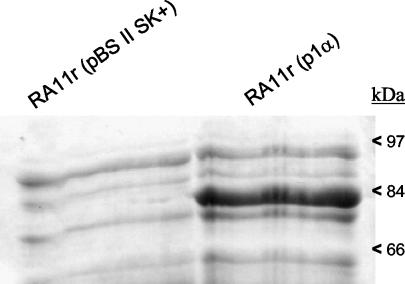
Comparison of the SDS-PAGE profiles from E. coli RA11r(p1α) with that of E. coli RA11r(pBS) (negative control) showed similar patterns except for one protein band migrating around 80 kDa that was absent in the negative control (Fig. 5). Most probably, this band represents α-Gal whose molecular weight deduced from the amino acid sequence is 84 kDa. Quantitation of α-Gal estimated by comparison with known amounts of bovine serum albumin indicates that the expression level is around 10 mg/liter of culture at an OD600 of 1 (data not shown).
FIG. 5.
SDS-PAGE analysis of total protein extracts from E. coli RA11r(p1α) expressing the L. plantarum melA gene. E. coli RA11r(pBS) was used as a negative control. The positions of molecular size standards are indicated on the right edge of the figure.
The molecular weight of native α-Gal was assessed from cell extracts of L. plantarum ATCC 8014. Using ultrafiltration units with a 100-kDA cutoff, all α-Gal activity was recovered in the retentate, suggesting a molecular weight of more than 100 kDa for α-Gal (data not shown). Using units with a 300-kDa cutoff, only half the activity was retained by the membrane, suggesting that α-Gal occurs as oligomers. This hypothesis was supported by zymogram analysis that showed that MelA monomers are inactive (data not shown).
Galactosides induce α-Gal activity in L. plantarum ATCC 8014.
The α-Gal activity was assessed for L. plantarum ATCC 8014 grown with various sugars at 30 and 37°C. With all sugars, α-Gal activity in cultures of L. plantarum ATCC 8014 grown at 30°C was higher than in those at 37°C (Fig. 6). Apart from glucose, all other sugars used induced α-Gal activity. Raffinose exhibited the strongest induction, followed by galactose, melibiose, and lactose. The relative induction levels for the different galactosides were similar at 30 and 37°C.
DISCUSSION
LAB have long been used as starters in food fermentation, where they exert technologically important functions such as acidification, aroma formation, and modification of rheological properties of raw agricultural products. As such, LAB have been widely studied in the fields of physiology and genetics. The impressive amount of knowledge acquired on these bacteria now opens the way to new applications, one of which is the production by LAB of compounds beneficial to health. A priori, LAB are excellent candidates to deliver such compounds in the gastrointestinal tract of humans and animals, since these bacteria are commonly ingested along with food products, and some, including lactobacilli, can survive in the gut, where they could exert a given biological activity.
The production of α-Gal by LAB is a biological activity whose delivery in the gastrointestinal tract is of potential interest since neither humans nor monogastric animals are able to synthesize digestive α-Gal. Since lactobacilli had been reported to exhibit α-Gal activity and given that some lactobacilli are part of the native intestinal microflora, we focused this work on α-Gals of lactobacilli. Upon probing the α-Gal-encoding gene using degenerate primers, the α-Gal structural gene of L. plantarum ATCC 8014 could be amplified, and it was designated melA. The melA gene is present as one unique copy in L. plantarum ATCC 8014 as indicated by Southern blot analysis using an internal melA fragment as a probe (data not shown). However, we cannot rule out the existence of additional α-Gals with different sequences in L. plantarum ATCC 8014.
DNA sequence analysis of the melA locus in L. plantarum ATCC 8014 identified five putative genes, galM-rafP-melA-lacM-lacL. This result shows that the genes required for the utilization of both α- and β-galactosides are in close vicinity. C. piscicola, a LAB previously classified as Lactobacillus carnosus, is the only other LAB for which clustered α- and β-Gals have been characterized so far. In contrast with L. plantarum ATCC 8014, all genes are on the same strand (10, 11). In several thermophilic bacteria, such as Thermus strain T2 (31), Bacillus stearothermophilus KVE39 (19), Thermoanaerobacter ethanolicus (GenBank sequence accession no. Y08557; unpublished data), and Thermotoga maritima (35), clustered α- and β-Gals have also been identified.
The structural melA gene for α-Gal of L. plantarum ATCC 8014 encodes an 84-kDa protein belonging to family 36 glycosyl hydrolases. This family is one of the three glycosyl hydrolase families defined for α-Gals, and it gathers α-Gals from eucaryotic and prokaryotic sources. BLAST and ClustalW analyses show high homology of α-Gals in their core domain (Fig. 2). Probably this region makes part of the active site of the enzymes, but this has not yet been demonstrated. In contrast with plant and yeast α-Gals that belong to family 27 and that can occur as monomers, the bacterial α-Gals so far characterized occur as oligomers (35). Analysis of the melA gene product using PSORT software from ExPASy did not show the presence of sorting signals (characteristic signal peptide or transmembrane segment), suggesting that MelA occurs as a soluble enzyme in the cytoplasm of L. plantarum ATCC 8014. This is consistent with MelA extraction experiments that showed that MelA was present in cell lysates from L. plantarum. This cytoplasmic localization of MelA implies that the degradation of α-galactosides in L. plantarum relies on the presence of a galactoside transporter.
A putative raffinose transporter, RafP, was identified for L. plantarum ATCC 8014 that could transport both α- and β-galactosides in L. plantarum as suggested by its homology with the unique galactoside transporter LacS of S. thermophilus. LacS has affinity not only for β-galactosides and galactose but also for melibiose and to a lesser extent for raffinose (17, 43, 44). LacS functions both as a lactose-galactose antiporter when sugars are in excess at either side of the membrane and as a proton symport system. It has an amino-terminal domain with homology to the melibiose carrier of various microorganisms and a carboxy-terminal domain with homology to domain I of enzyme EIIA of various phosphoenolypyruvate phosphotransferase systems (PEP-PTS) from gram-positive and -negative organisms (44). It will be of great interest to determine which type of α-galactosides can be transported by RafP and also if this is an active transporter or a PTS transporter. In particular, the transport of stachyose and raffinose, the two major α-galactosides of soy, should be assessed.
Expression and transcriptional studies demonstrate that both melA and rafP are expressed in L. plantarum ATCC 8014. The melA gene is transcribed from its promoter, yielding a monocistronic RNA. Therefore, the melA gene is not part of an operon, in contrast to melA genes in other microorganisms, e.g., E. coli (4), Klebsiella pneumoniae (23), Enterobacter cloacae (42), Citrobacter freundii (50), P. pentosaceus (GenBank sequence accession no L32093, unpublished data; inferred from the sequence data), T. maritima (35), and Thermoanaerobacter ethanolicus (GenBank accession no. Y08557, unpublished data; inferred from the sequence data). The melA gene is regulated at the transcriptional level, as shown by the fourfold increase of melA transcript in the presence of melibiose compared with that in the presence of glucose. However, melA is not totally repressed in the presence of glucose, and this feature together with the absence of a CRE site in the melA promoter region suggests that melA regulation is mediated by an induction mechanism rather than by catabolite repression (27).
The assessment of MelA activity in cultures of L. plantarum showed important differences according to the carbohydrate used. Interestingly, MelA activity in melibiose-containing cultures was more than 150-fold higher than that in glucose-containing cultures. This ratio is much higher than the ratio of 4 observed at the transcriptional level. These results suggest that posttranscriptional regulation could also occur. It could result from allosteric control mechanisms that have been shown to modulate the activity of glycolytic enzymes in LAB (20).
The 5′ RACE experiment showed that rafP is also expressed as a monocistronic mRNA, indicating the functionality of its promoter. However, due to the limitation of the 5′ RACE methodology at the PCR amplification step, this should be confirmed by Northern blot analysis, and we cannot rule out at the moment that rafP could also be part of a larger operon.
In view of future applications in the probiotic field, we decided to investigate the influence of different carbohydrates on the α-Gal activity at both 30°C (the optimal growth temperature for L. plantarum) and 37°C (the human body temperature). MelA activity assayed in cultures grown with various carbohydrates showed that lactose, melibiose, galactose, and raffinose increase MelA activity in L. plantarum ATCC 8014 (lactose and raffinose showed the lowest and highest increase, respectively). This suggests an activation at the enzyme level or an induction at the gene level. The same order in MelA activity was observed in cultures assayed at 30°C and at 37°C, although the level of activity was lower at 37°C than at 30°C. Although we cannot rule out that α-Gal is thermosensitive at 37°C, this result together with the observation of Tamura and Matsushita (52), who showed a decrease of both lactose and melibiose uptake at 37°C from that at 30°C for L. plantarum ATCC 8014, suggests that two types of regulation occur: (i) one at the transporter level, and this regulation would be thermosensitive; and (ii) another at the α-Gal level in response to the carbon source that is similar at 30 and 37°C.
In the course of this study, galM and lacLM were identified. The presence of galM that specifies a mutarotase suggests that rafP is preceded by a galactose operon, since galM is the last 3′ gene of the gal operon in various lactobacilli (Lactobacillus helveticus galKTM [GenBank accession no. X57248] [39], L. sakei galETM [GenBank accession no. AF401040; unpublished data], and in Lactobacillus casei 64H galKETRM [GenBank accession no. AF005933] [5]) as well as in S. thermophilus (GenBank accession no. U61402) (55) and in S. salivarius (GenBank accession no. AF389474) (53) (galR-galKTE-galM). The lacLM genes identified in this study specify a heterodimeric β-galactosidase, the main enzyme involved in lactose utilization (29). Although lacLM genes have been detected both in plasmids (36) and on the chromosome (16) in L. plantarum, the sequence described here corresponds to a chromosomal region, since no amplification was obtained using plasmid DNA as a template (data not shown). Our observation confirms that of Fernandez et al. (16), and this establishes that L. plantarum ATCC 8014 lacLM genes are present on the chromosome. Altogether, these data show that the genes involved in galactosides utilization are clustered in the same locus in L. plantarum ATCC 8014.
This work is the first step in the characterization of a melA gene involved in α-galactoside catabolism in lactobacilli. Further studies should now focus on the biochemical characterization of MelA. It is also of interest to investigate RafP functionality, its regulation aspect, and its substrate specificity. Control of MelA activity in lactobacilli should open doors to interesting applications. One would be to develop starters for soymilk fermentation, since there is actually a huge demand for this kind of product devoid of undesirable side effects. Also, the development of probiotic bacteria is foreseen to deliver α-Gal in the digestive tract of humans and animals. The objective is to deliver α-Gal in the duodenum so that α-galactosides can be degraded at this level. First, this would address the problem of flatulence associated with the presence of α-galactosides in the lower intestine. Second, this could increase the metabolizable energy of-soy derived products since the microflora of the lower gut would then utilize other carbohydrate substrates, such as fibers (12).
Acknowledgments
This work was supported by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), the Consejo de Ciencia y Técnica de la Universidad Nacional de Tucumán (CIUNT), Agencia Nacional de Promoción Científica y Tecnológica (UE 025), and contract QLK1-CT-2000-01376 from the EEC (http://www.nutracells.com). A.S. was the recipient of grants from Lallemand and from INRA.
We are grateful to Tadashi Shimamoto for providing us with E. coli RA11r, to Catherine Foucaud for help in Northern analyses, to Vincent Juillard for fruitfull discussions throughout this work, and to Maarten van de Guchte for reviewing the manuscript.
REFERENCES
- 1.Ahrne, S., and G. Molin. 1991. Spontaneous mutations changing the raffinose metabolism of Lactobacillus plantarum. Antonie Leeuwenhoek 60:87-93. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Annunziato, M. E., R. R. Mahoney, and R. E. Mudgett. 1986. Production of α-Galactosidase from Aspergillus oryzae grown in solid state culture. J. Food. Sci. 51:1370-1371. [Google Scholar]
- 4.Aslanidis, C., K. Schmid, and R. Schmitt. 1989. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J. Bacteriol. 171:6753-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettenbrock, K., and C. A. Alpert. 1998. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl. Environ. Microbiol. 64:2013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky, L. I., V. V. Ivanov, Y. L. Kalaidzidis, A. M. Leontovhich, V. K. Nikolaev, S. I. Feranchuk, and V. A. Drachev. 1995. Gene-Bee-NET: internet-based server for analyzing biopolymers structure. Biochemistry 60:923-928. [PubMed] [Google Scholar]
- 8.Chen, X., and R. Wu. 1997. Direct amplification of unknown genes and fragments by uneven polymerase chain reaction. Gene 185:195-199. [DOI] [PubMed] [Google Scholar]
- 9.Church, F. C., S. P. Meyers, and V. R. Srinivasan. 1980. Isolation and characterization of alpha-galactosidase from Pichia guilliermondii, p. 339-348. In L. A. Underkofler and M. L. Wulf (ed.), Developments in industrial microbiology, vol. 21. Lubrecht & Cramer, Arlington, Va.
- 10.Coombs, J., and J. E. Brenchley. 2001. Characterization of two new glycosyl hydrolases from the lactic acid bacterium Carnobacterium piscicola strain BA. Appl. Environ. Microbiol. 67:5094-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombs, J. M., and J. E. Brenchley. 1999. Biochemical and phylogenetic analyses of a cold-active beta-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl. Environ. Microbiol. 65:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coon, C. N., K. L. Leske, O. Akavanichan, and T. K. Cheng. 1990. Effect of oligosaccharide-free soybean meal on true metabolizable energy and fiber digestion in adult roosters. Poult. Sci. 69:787-793. [DOI] [PubMed] [Google Scholar]
- 13.Cruz, R., J. C. Bastistela, and G. Wosiacki. 1981. Microbial α-Galactosidase for soybean processing. J. Food Sci. 46:1196-1200. [Google Scholar]
- 14.David, S., H. Stevens, M. van Riel, G. Simons, and W. M. de Vos. 1992. Leuconostoc lactis beta-galactosidase is encoded by two overlapping genes. J. Bacteriol. 174:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Man, J. C., M. Rogosa, and M. E. Shape. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 16.Fernandez, M., A. Margolles, J. E. Suarez, and B. Mayo. 1999. Duplication of the beta-galactosidase gene in some Lactobacillus plantarum strains. Int. J. Food Microbiol. 48:113-123. [DOI] [PubMed] [Google Scholar]
- 17.Foucaud, C., and B. Poolman. 1992. Lactose transport system of Streptococcus thermophilus. Functional reconstitution of the protein and characterization of the kinetic mechanism of transport. J. Biol. Chem. 267:22087-22094. [PubMed] [Google Scholar]
- 18.Ganiats, T. G., W. A. Norcross, A. L. Halverson, P. A. Burford, and L. A. Palinkas. 1994. Does Beano prevent gas? A double-blind crossover study of oral alpha-galactosidase to treat dietary oligosaccharide intolerance. J. Fam. Pract. 39:441-445. [PubMed] [Google Scholar]
- 19.Ganter, C., A. Boeck, P. Buckel, and R. Mattes. 1988. Production of thermostable, recombinant alpha-galactosidase suitable for raffinose elimination from sugar beet syrup. J. Biotechnol. 8:301-310. [Google Scholar]
- 20.Garrigues, C., P. Loubiere, N. D. Lindley, and M. Cocaign-Bousquet. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garro, M. S., G. F. de Valdez, G. Oliver, and G. S. de Giori. 1996. Influence of carbohydrates on the alpha-galactosidase activity of Lactobacillus fermentum. Curr. Microbiol. 33:302-305. [DOI] [PubMed] [Google Scholar]
- 22.Garro, M. S., G. Savoy de Giori, G. Font de Valdez, and G. Oliver. 1993. Characterization of alpha-galactosidase from Lactobacillus fermentum. J. Appl. Bacteriol. 75:485-488. [Google Scholar]
- 23.Hama, H., and T. H. Wilson. 1992. Primary structure and characteristics of the melibiose carrier of Klebsiella pneumoniae. J. Biol. Chem. 267:18371-18376. [PubMed] [Google Scholar]
- 24.Hanatani, M., H. Yazyu, S. Shiota-Niiya, Y. Moriyama, H. Kanazawa, M. Futai, and T. Tsuchiya. 1984. Physical and genetic characterization of the melibiose operon and identification of the gene products in Escherichia coli. J. Biol. Chem. 259:1807-1812. [PubMed] [Google Scholar]
- 25.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 28.Ibrahim, S. S., R. A. Habiba, A. A. Shatta, and H. E. Embaby. 2002. Effect of soaking, germination, cooking and fermentation on antinutritional factors in cowpeas. Nahrung 46:92-95. [DOI] [PubMed] [Google Scholar]
- 29.Jeffrey, S. R., and W. J. Dobrogosz. 1990. Transport of β-galactosides in Lactobacillus plantarum NC2. Appl. Environ. Microbiol. 56:2484-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, W. J., C. J. B. Smith, and T. O. M. Nakayama. 1973. The removal of oligosaccharides from soybeans. Lebensm. Wiss. Technol. 6:201. [Google Scholar]
- 31.Koyama, Y., S. Okamoto, and K. Furukawa. 1990. Cloning of alpha- and beta-galactosidase genes from an extreme thermophile, Thermus strain T2, and their expression in Thermus thermophilus HB27. Appl. Environ. Microbiol. 56:2251-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku, S., L. S. Wei, M. P. Steinberg, A. I. Nelson, and T. Hymowitz. 1976. Extraction of oligosaccharides during cooking of whole soybeans. J. Food Sci. 41:361-364. [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Leong-Morgenthaler, P., Z. M. C. Zwahlen, and H. Hottinger. 1991. Lactose metabolism in Lactobacillus bulgaricus: analysis of the primary structure and expression of the genes involved. J. Bacteriol. 173:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebl, W., B. Wagner, and J. Schelhase. 1997. Properties of an α-galactosidase, and structure of its gene galA, within an α- and β-galactoside utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima. Syst. Appl. Microbiol. 21:1-11. [DOI] [PubMed] [Google Scholar]
- 36.Mayo, B., B. Gonzalez, P. Arca, and J. E. Suarez. 1994. Cloning and expression of the plasmid encoded beta-D-galactosidase gene from a Lactobacillus plantarum strain of dairy origin. FEMS Microbiol. Lett. 122:145-152. [DOI] [PubMed] [Google Scholar]
- 37.McIlvaine, T. C. 1921. A buffer solution for colorimetric comparison. J. Biol. Chem. 49:183-186. [Google Scholar]
- 38.Mital, B. K., R. S. Shallenberger, and K. H. Steinkraus. 1973. α-Galactosidase of lactobacilli. Appl. Microbiol. 26:783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mollet, B., and N. Pilloud. 1991. Galactose utilization in Lactobacillus helveticus: isolation and characterization of the galactokinase (galK) and galactose-1-phosphate uridyl transferase (galT) genes. J. Bacteriol. 173:4464-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nigatu, A., S. Ahrné, and G. Molin. 2000. Temperature-dependent variation in API 50 CH fermentation profiles of Lactobacillus species. Curr. Microbiol. 41:21-26. [DOI] [PubMed] [Google Scholar]
- 41.Obst, M., E. R. Meding, R. F. Vogel, and W. P. Hammes. 1995. Two genes encoding the beta-galactosidase of Lactobacillus sake. Microbiology 141:3059-3066. [DOI] [PubMed] [Google Scholar]
- 42.Okazaki, N., X. X. Jue, H. Miyake, M. Kuroda, T. Shimamoto, and T. Tsuchiya. 1997. A melibiose transporter and an operon containing its gene in Enterobacter cloacae. J. Bacteriol. 179:4443-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poolman, B., R. Modderman, and J. Reizer. 1992. Lactose transport system of Streptococcus thermophilus. The role of histidine residues. J. Biol. Chem. 267:9150-9157. [PubMed] [Google Scholar]
- 44.Poolman, B., T. J. Royer, S. E. Mainzer, and B. F. Schmidt. 1989. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J. Bacteriol. 171:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 46.Rackis, J. J. 1981. Flatulence caused by soya and its control through processing. J. Am. Oil Chem. Soc. 58:503-510. [Google Scholar]
- 47.Raya, R., J. Bardowski, P. S. Andersen, S. D. Ehrlich, and A. Chopin. 1998. Multiple transcriptional control of the Lactococcus lactis trp operon. J. Bacteriol. 180:3174-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reese, M. G. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51-56. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Shimamoto, T., T. Shimamoto, X.-J. Xu, N. Okazaki, H. Kawakami, and T. Tsuchiya. 2001. A cryptic melibiose transporter gene possessing a frameshift from Citrobacter freundii. J. Biochem. (Tokyo) 129:607-613. [DOI] [PubMed] [Google Scholar]
- 51.Slominski, B. A. 1994. Hydrolysis of galactooligosaccharides by commercial preparations of α-galactosidase and β-fructofuranosidase: potential for use as dietary additives. J. Sci. Food Agric. 65:323-330. [Google Scholar]
- 52.Tamura, C., and O. Matsushita. 1992. Melibiose transport system in Lactobacillus plantarum. Microbiol. Immunol. 36:1119-1128. [DOI] [PubMed] [Google Scholar]
- 53.Vaillancourt, K., S. Moineau, M. Frenette, C. Lessard, and C. Vadeboncoeur. 2001. Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products. J. Bacteriol. 184:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaughan, E. E., S. David, and W. M. de Vos. 1996. The lactose transporter in Leuconostoc lactis is a new member of the LacS subfamily of galactoside-pentose-hexuronide translocators. Appl. Environ. Microbiol. 62:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan, E. E., P. T. van den Bogaard, P. Catzeddu, O. P. Kuipers, and W. M. de Vos. 2001. Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkinson, M., J. Doskow, and S. Lindsey. 1990. RNA blots: staining procedures and optimization of conditions. Nucleic Acid Res. 19:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanagida, F., K.-I. Suzuki, T. Kaneko, M. Kozaki, and K. Komagata. 1987. Morphological, biochemical, and physiological characteristics of spore-forming lactic acid bacteria. J. Gen. Appl. Microbiol. 33:33-45. [Google Scholar]