Abstract
We engineered Saccharomyces cerevisiae cells that produce large amounts of fungal glucoamylase (GAI) from Aspergillus awamori var. kawachi. To do this, we used the δ-sequence-mediated integration vector system and the heat-induced endomitotic diploidization method. δ-Sequence-mediated integration is known to occur mainly in a particular chromosome, and the copy number of the integration is variable. In order to construct transformants carrying the GAI gene on several chromosomes, haploid cells carrying the GAI gene on different chromosomes were crossed with each other. The cells were then allowed to form spores, which was followed by dissection. Haploid cells containing GAI genes on multiple chromosomes were obtained in this way. One such haploid cell contained the GAI gene on five chromosomes and exhibited the highest GAI activity (5.93 U/ml), which was about sixfold higher than the activity of a cell containing one gene on a single chromosome. Furthermore, we performed heat-induced endomitotic diploidization for haploid transformants to obtain polyploid mater cells carrying multiple GAI genes. The copy number of the GAI gene increased in proportion to the ploidy level, and larger amounts of GAI were secreted.
The yeast Saccharomyces cerevisiae has often been used as a host organism for heterologous gene expression and protein secretion (1, 9, 10, 16, 20, 21). This is because S. cerevisiae is safe and handling and cultivation of this yeast are easy and simple. In addition, the expressed proteins can be secreted into the medium. The expression of heterologous genes in yeast is influenced by many factors, such as the strength of the promoter, mRNA stability, and translation efficiency. In addition, the copy number of the gene and mitotic stability are also very important. The characteristics of expression plasmids strongly depend on the type of replication, the selectable marker, and the growth conditions. In S. cerevisiae, the high-copy-number episomal plasmids (YEp) are commonly used as expression vectors. However, these plasmids are unstably maintained if they have no selective pressure (6, 15). On the other hand, the integrative plasmids (YIp) are stably maintained with a low copy number. The intrinsic genetic stability of integrated constructs is well established. The way to improve heterologous gene expression is to construct YEp plasmids that are maintained stably or to construct YIp plasmids that are present at a high copy number. S. cerevisiae, an organism widely used for ethanol fermentation, lacks hydrolytic enzymes for starch so that raw starch must be liquefied and saccharified by koji mold or other sources. Therefore, application of GAI, a fungal glucoamylase from Aspergillus awamori var. kawachi, to recombinant yeast is useful for improving ethanol fermentation and reducing the use of koji mold or other sources. In this study, we used a novel approach for expression of GAI in polyploid S. cerevisiae cells. In a previous study, we developed a method to obtain homozygous polyploid cells (22). Endomitotic diploidization is readily induced by heat treatment of spores during germination. The diploid cells obtained in this way are homozygous and retain the ability to mate. By using the same method, tetraploid cells were obtained by crossing the diploid mater cells and then heat treating the spores. We tried to use these polyploid mater cells as host cells for gene engineering of S. cerevisiae. Here we describe high-level expression of glucoamylase by polyploid cells of S. cerevisiae in which multiple GAI genes are integrated into the δ-sequence.
MATERIALS AND METHODS
Microorganisms and cultivation.
Escherichia coli JM109 was used for general genetic manipulations. S. cerevisiae CG378 (MATa ura3 leu2 trp1) and CG379 (MATα ura3 leu2 trp1 his3) were used for determination of the mating type. CG379 was used for expression of GAI and for polyploidization. S. cerevisiae was grown at 30°C in YPD medium (1% yeast extract, 2% polypeptone, 2% glucose) and SD medium (2% glucose, 0.67% yeast nitrogen base without amino acids) in the presence of appropriate amino acids. For sporulation, the cells were grown at 30°C for 3 days on sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% glucose, 2% agar). In order to detect the secreted glucoamylase, YPD plates containing 0.5% soluble starch were used.
Construction of plasmids.
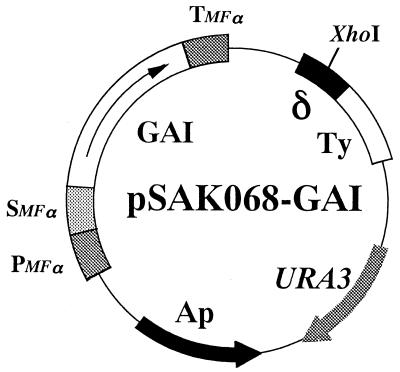
The δ-integrative plasmid pSAK068 was obtained from the Mitsubishi Kasei Institute of Life Science. YEUp-GAs is a YEUp plasmid carrying the GAI cDNA and was previously constructed by introducing the promoter, signal sequence, and terminator of the MFα1 gene (7, 13). YEp13 43A and YCpHIS4 were obtained from S. Harashima (Osaka University) (23). The 3.4-kb EcoRI fragment from YEUp-GAs containing the GAI cDNA along with the promoter, signal sequence, and terminator was introduced into the EcoRI site of pSAK068 to generate plasmid pSAK068-GAI (Fig. 1). The 2.2-kb SalI-XhoI fragment containing the LEU2 gene from YEp13 43A was introduced into the XhoI site of pHSG299 to generate pHSG299-LEU2, which was then digested at the vector-borne SphI and SmaI sites. The 2.2-kb SphI-SmaI fragment was inserted into the SphI-SmaI site of pSAK068-GAI. The pSAKL068-GAI plasmid constructed in this way possesses the LEU2 gene instead of the URA3 gene. The 2.5-kb HindIII fragment containing the HO gene from YEp13 43A was introduced into the HindIII site of yeast episomal plasmid YEUp3 to generate YEUp3-HO. The 1.5-kb EcoRI fragment containing the TRP1 gene from YCpHIS4 was introduced into the EcoRI site of YEUp3-HO to generate YEUTp3-HO.
FIG. 1.
Structure of pSAK068-GAI. pSAK068-GAI is a δ-integrative plasmid constructed for GAI secretion as described in Materials and Methods. Pmfα, Smfα, and Tmfα indicate promoter, signal, and terminator sequences of mating factor α, respectively.
Transformation.
S. cerevisiae was transformed by the lithium acetate method (11). The δ-integrative plasmids were digested at the XhoI site within the δ-sequence. E. coli was transformed by electroporation (model BTX600 system; Biotechnologies & Experimental Research Inc.).
Construction of polyploid mater cells.
Endomitotic diploidization was induced as a result of heat treatment of spores at an early germination stage. a/α diploid cells were obtained by transformation of α haploid cells with YEUTp3-HO. The transformed HO gene was lost by successive cultivation in YPD liquid medium. Moreover, a/a/α/α tetraploid cells were obtained by crossing a/a diploid mater cells with α/α diploid mater cells. Spores from the a/α diploid cells or the a/a/α/α tetraploid cells were incubated in YPD liquid medium to induce germination at 30°C for 30 min. The culture broth was incubated at 55°C for 10 min, and then the cells were harvested by centrifugation. After the cells were washed in sterilized water, they were incubated at 37°C for 1 h in a Zymolyase solution (0.6 mg of Zymolyase 20-T [Seikagaku Co., Tokyo, Japan] per ml, 0.01 M 2-mercaptoethanol, 0.6 M KCl, 1/15 M phosphate buffer [pH 7.5]) to digest the ascus wall. The cells were then washed twice with sterilized water and resuspended in 0.05% Tween 80. When these procedures were used, almost none of the vegetative cells survived. The suspensions were sonicated and spread on a YPD medium plate. The viable cells obtained in this way were examined for mating ability, and then the ploidy of the cells was examined based on the DNA content. The DNA content was determined by the method of Burton (2).
Pulsed-field gel electrophoresis and Southern blot analysis.
Chromosomal DNA was prepared as described by Mochizuki et al. (14). Electrophoresis was performed with an LKB2015 Pulsaphor electrophoresis unit (Pharmacia LKB Biotechnology). The electrophoresis conditions were based on the manufacturer's instructions. Southern blot analysis was performed with a digoxigenin DNA labeling and detection kit used according to the manufacturer's instructions (Boehringer Mannheim).
Plate assay for secreted glucoamylase.
Transformants with the GAI genes were streaked on YPD medium plates containing starch and incubated at 30°C for 3 days. The plates were then inverted onto plates containing solid iodine and left for several minutes.
Enzyme assay.
Glucoamylase activity was assayed by the 3,5-dinitrosalicylic acid method as previously described (8). The transformants were grown aerobically on YPD medium at 30°C for 3 days, after which the cells were in the stationary phase. The glucoamylase activity of the culture supernatant was measured. One unit of glucoamylase activity was defined as the amount of enzyme that liberated 1 μmol of glucose per ml of reaction mixture per min. All values shown below are averages of data from three independent experiments.
RESULTS
Transformation of strain CG379 with δ-integrative plasmids containing the GAI cDNA.
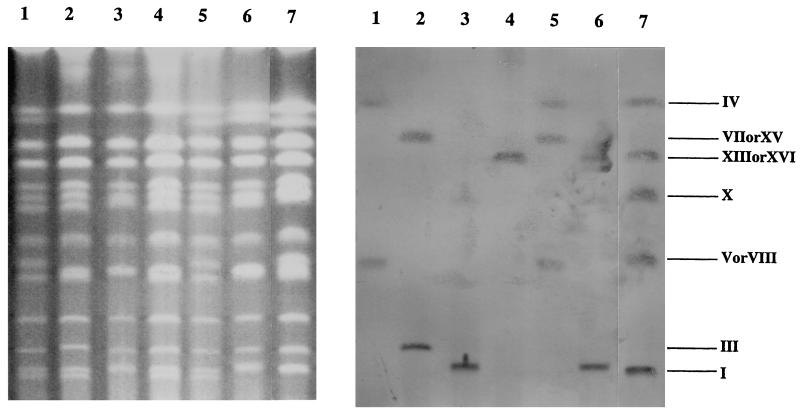
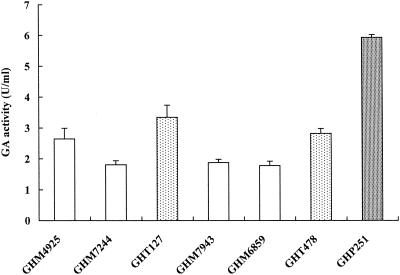
Heterothallic strain CG379 was transformed with the pSAK068-GAI plasmid linearized at the XhoI site within the δ-sequence. Production of glucoamylase in the transformants was measured. First, the secretion of glucoamylase was examined on YPD medium plates containing starch by determining the formation of clear zones (halos) on the dark iodine-stained background. Several URA transformants that produced the largest halos were selected from 100 transformants. These URA transformants were analyzed to determine how the GAI genes were integrated into the δ-sequences in the chromosomes. The chromosomes prepared from the transformants were separated by pulsed-field gel electrophoresis and were analyzed by Southern blot hybridization by using the GAI gene as a probe. Four transformants (GHM-49, GHM-68, GHM-72, and GHM-79) containing the GAI gene on different chromosomes were selected. The secreted glucoamylase activities of GHM-49, GHM-68, GHM-72, and GHM-79 were 0.91, 0.89, 0.90, and 0.98 U/ml, respectively. These clones were further transformed with XhoI-digested pSAKL068-GAI. The four URA and LEU transformants obtained produced large halos and contained the GAI gene on different chromosomes; GHM-4925 derived from GHM-49, GHM-6859 derived from GHM-68, GHM-7244 derived from GHM-72, and GHM-7943 derived from GHM-79 were selected in the same way. The δ-integrations occurred on chromosomes IV and V or VIII in GHM-4925, on chromosomes IV and XIII or XVI in GHM-6859, on chromosomes III and VII or XV in GHM-7244, and on chromosomes I and X in GHM-7943 (Fig. 2). The chromosomes with the GAI genes tended to migrate more slowly than chromosomes with unintegrated chromosomes, suggesting that the decreased mobilities were due to high numbers of copies that were integrated. The secreted glucoamylase activities of GHM-4925, GHM-6859, GHM-7244, and GHM-7943 were 2.65, 1.81, 1.78, and 1.88 U/ml, respectively (Fig. 3).
FIG. 2.
Localization of δ-integrated GAI genes on the chromosomes in various haploid cells. The chromosomal DNAs separated by pulsed-field gel electrophoresis were hybridized with GAI cDNA as a probe. GHM-4925, GHM-7244, GHM-7943, and GHM-6859 were obtained by transformation of strain CG379 with pSAK068-GAI and pSAKL068-GAI. GHT-127 was generated from diploid crosses between GHM-4925 and GHM-7244a. GHT-478 was generated from diploid crosses between GHM-6859 and GHM-7943a. GHP-251 was generated from diploid crosses between GHT-127 and GHT-478a. Lanes 1, GHM-4925; lanes 2, GHM-7244; lanes 3, GHM-7943; lanes 4, GHM-6859; lanes 5, GHT-127; lanes 6, GHT-478; lanes 7, GHP-251.
FIG. 3.
Glucoamylase activities of haploid cells carrying multiple GAI genes on chromosomes. Glucoamylase (GA) activities of various haploid cells are shown. GHM-4925, GHM-7244, GHM-7943, and GHM-6859 contain GAI genes on two chromosomes. GHT-127 and GHT-478 contain GAI genes on three chromosomes. GHP-251 contains GAI genes on five chromosomes. The error bars indicate the standard deviations of three separate experiments.
Construction of haploid cells with more glucoamylase activity.
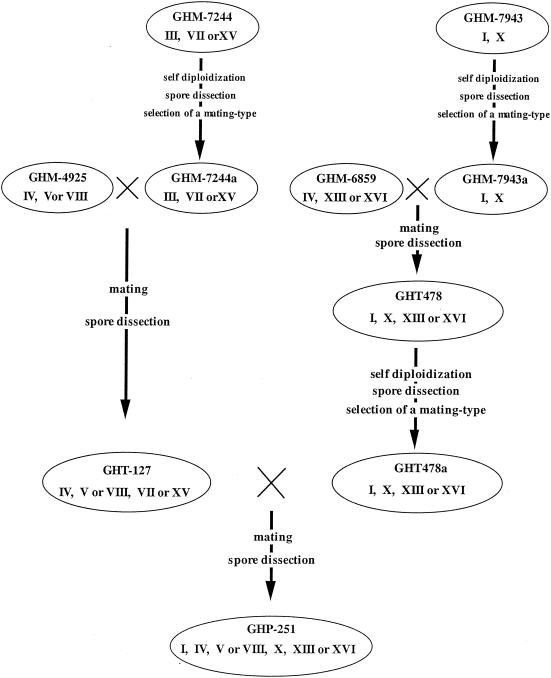
To obtain haploid cells expressing a higher level of glucoamylase, a/α diploid cells were first constructed by crossing the haploid cells containing the GAI genes. To do this, α-mating-type haploid cells of GHM-7244 and GHM-7943 were transformed with YEUTp3-HO containing the HO gene to induce self-diploidization. The diploid cells obtained were grown in rich medium, which resulted in loss of the YEUTp3-HO plasmid. The diploid cells which had lost the HO gene were allowed to form spores on the sporulation medium. The spores were separated by random spore dissection and were examined to determine the mating type by using the standard strains CG378 (MATa) and CG379 (MATα). The a haploid cells from GHM-7244 and GHM-7943 (designated GHM-7244a and GHM-7943a, respectively) were crossed with the α haploid cells of GHM-4925 and GHM-6859, respectively. Again, spores were formed and dissected in the same manner. The haploid cells obtained were examined for glucoamylase activity by halo formation. The locations of the GAI genes on the chromosomes were also determined by pulsed-field gel electrophoresis followed by Southern blot analyses. Two types of α haploid cells were obtained by these procedures. These were GHT-127 cells derived from GHM-4925 and GHM-7244a and GHT-478 cells derived from GHM-6859 and GHM-7943a (Fig. 2). These organisms contained the GAI gene on three different chromosomes. The glucoamylase activities of GHT-127 and GHT-478 were 3.34 and 2.81 U/ml, respectively (Fig. 3). Finally, α haploid cells containing GAI genes on five chromosomes were constructed by crossing and spore dissection. α haploid GHP-251 cells were constructed from the diploid crosses between GHT-127 cells and GHT-478a cells, which were the a haploid cells generated from GHT-478 (Fig. 2). The strategy used to construct these polyploid cells is illustrated in Fig. 4. The glucoamylase activity secreted from GHP-251 was 5.93 U/ml, which was about six times higher than the activities of the transformants containing the GAI genes on only a single chromosome (Fig. 3).
FIG. 4.
Construction of the haploid yeast cells carrying GAI genes on multiple chromosomes.
Polyploidization of transformants.
By using heat treatment of spores during germination, we constructed α/α diploid mater cells and α/α/α/α tetraploid mater cells from GHM-4925 and GHT-127. In GHM-4925, the GAI genes were integrated on chromosomes IV and V or VIII. In GHT-127, the GAI genes were integrated on chromosomes IV, V, or VIII and VII or XV (Fig. 2). The diploid and tetraploid mater cells obtained from GHM-4925 were GDM-94 and GTM-240, respectively. Diploid GDT-104 mater cells and tetraploid GTT-270 mater cells were obtained from GHT-127 (Table 1). The glucoamylase activity increased linearly in proportion to ploidy only when the values were expressed per cell, not when they were expressed per milliliter of culture. Thus, the tetraploid GTM-240 cells derived from GHM-4925 exhibited activities of 3.93 U/ml and 4.03 U/108 cells. The tetraploid GTT-270 cells exhibited higher GAI activities (5.28 U/ml and 5.56 U/108 cells). In both cases, the number of cells decreased as the ploidy level increased.
TABLE 1.
Glucoamylase activities of polyploid mater cells from GHM-4925 and GHT-127a
| Strain | Glucoamylase activity
|
No. of cells (108 cells/ml) | |
|---|---|---|---|
| U/ml | U/108 cells | ||
| GHM-4925 (MATα) | 2.65 ± 0.34 | 0.89 ± 0.05 | 2.95 ± 0.22 |
| GDM-94 (MATα/α) | 3.35 ± 0.16 | 1.98 ± 0.45 | 1.76 ± 0.47 |
| GTM-240 (MATα/α/α/α) | 3.93 ± 0.09 | 4.03 ± 0.41 | 0.98 ± 0.12 |
| GHT-127 (MATα) | 3.34 ± 0.40 | 1.16 ± 0.12 | 2.89 ± 0.13 |
| GDT-104 (MATα/α) | 4.46 ± 0.17 | 2.61 ± 0.24 | 1.72 ± 0.22 |
| GTT-270 (MATα/α/α/α) | 5.28 ± 0.29 | 5.56 ± 0.09 | 0.95 ± 0.07 |
All values are means ± standard deviations of results from three independent experiments.
DISCUSSION
A number of reports on heterologous protein production in S. cerevisiae have been published; these reports include descriptions of breeding of supersecretion mutants (20), construction of a multicopy plasmid with a strong promoter (17), improvement in culture conditions (16), and so on. In this study, a high level of production of heterologous protein was obtained in S. cerevisiae by using the endomitotic diploidization method. The efficiency of protein secretion in a yeast cell can be affected by how the heterologous gene is maintained in the cell. When a gene fused to a yeast promoter and secretion signal sequence was integrated into a yeast chromosome, a greater proportion of the protein was secreted than when the same construction was introduced on a multicopy plasmid vector (21). The lower efficiency of secretion of multicopy plasmids might be due to the higher copy number and correspondingly higher level of gene expression and larger reservoir of unsecretable protein. However, this hypothesis is not correct. Smith et al. (21) constructed two types of strains, one containing several integrated copies of invertase-prochymosin fusion genes and the other containing the same construction on a multicopy plasmid. These strains produced approximately the same absolute amount of prochymosin, but the strain with the integrated genes secreted at least four times as much. Thus, we suggest that chromosomal integration is a useful method for introducing heterologous genes into a yeast, especially if secretion of the products is desired. However, the genes integrated into chromosomes in the host cells are present at levels of only one to several copies per cell. To improve the copy number of the YIp-type plasmids, δ-integrative plasmids were constructed (18). δ-Integration occurred on various chromosomes due to the presence of multicopy δ-sequences on the chromosomes (3). In most transformants, however, the integration occurred on a single chromosome, although the reasons remain to be elucidated (18). Previously, the leu2-d allele and PDR4 were used as the selective makers of a δ-integrative plasmid to improve the copy number for integration (12, 19). The leu2-d allele is a poorly expressed allele of LEU2; therefore, the copy number and the stability of YEp-type plasmids were expected to increase if the transformants carrying leu2-d could grow without added leucine. PDR4 is the dominant selective marker and confers resistance to the antifungal agent cerulenin at a high copy number. In each case, the δ-integrative plasmids could increase the copy number. However, it seems that δ-sequence-mediated integration occurred only on a single chromosome in one transformation. Furthermore, in order to increase the number of δ-integrated chromosomes, Sakai et al. constructed recombinant haploid yeasts carrying human nerve growth factor genes that were δ-integrated on several chromosomes by crosses between the transformants, followed by dissection of the spores (19). In addition to the characteristics of the integrated heterologous genes revealed by the δ-sequence system, the expression level of these genes is affected by the cell type of the host strain. Since expression of the δ-sequence is governed by haploid-specific transcriptional activation, the expression level of a δ-integrated heterologous gene in an a/α diploid cell is much lower than that in a haploid cell (4, 5). The heat-induced endomitotic diploidization method is simple and very efficient for obtaining polyploid mater cells which have haploid-cell-specific characteristics. It is particularly suitable for increasing the copy number of the δ-sequence-integrated heterologous gene when a strain has only a few auxotrophic selectable markers. Thus, the chromosomal copy number of a heterologous gene expression cassette can be amplified in proportion to the ploidy level. One tetraploid mater strain which was obtained, GTT-270, carried the GAI genes on the 12 chromosomes and exhibited the highest glucoamylase activity. Interestingly, the glucoamylase activity increased in proportion to the ploidy level, indicating that the GAI gene on each chromosome seems to be independently expressed without any regulatory interactions. By repeating these methods, we should be able to construct mater cells with higher degrees of polyploidy and higher GAI activities. However, it is also true that the size of the population of cells decreased as the level of ploidy increased. The reason for this is not clear, but we believe that this method is very useful for heterologous protein production in S. cerevisiae.
REFERENCES
- 1.Bitter, G. A., K. M. Egan, E. R. Koski, M. D. Jones, S. G. Eliotto, and J. C. Giffin. 1987. Expression and secretion vector for yeasts. Methods Enzymol. 153:516-544. [DOI] [PubMed] [Google Scholar]
- 2.Burton, K. 1956. Diphenylamine reaction with nucleic acid. Biochem. J. 62:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron, J. R., E. Y. Loh, and R. W. Davis. 1979. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell 16:739-751. [DOI] [PubMed] [Google Scholar]
- 4.Company, M., and B. Errede. 1987. Cell-type-dependent gene activation by yeast transposon Ty1 involves multiple regulatory determinants. Mol. Cell. Biol. 7:3205-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Errede, B., M. Company, and C. A. Huchinson III. 1987. Ty1 sequence with enhancer and mating-type-dependent regulatory activities. Mol. Cell. Biol. 7:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Futcher, A. B., and B. S. Cox. 1984. Copy number and the stability of 2-μm circle-based artificial plasmids of Saccharomyces cerevisiae. J. Bacteriol. 157:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto, M., T. Semimaru, K. Furukawa, and S. Hayashida. 1994. Analysis of the raw starch-binding domain by mutation of a glucoamylase from Aspergillus awamori var. kawachi expressed in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 60:3926-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto, M., K. Tanigawa, W. Kanlayakrit, and S. Hayashida. 1994. The mechanism of binding of glucoamylase I from Aspergillus awamori var. kawachi to cyclodextrins and raw starch. Biosci. Biotechnol. Biochem. 58:49-54. [DOI] [PubMed] [Google Scholar]
- 9.Hitzeman, R. A., D. W. Leung, L. J. Perry, W. J. Kohr, H. L. Levine, and D. V. Goeddel. 1983. Secretion of human interferons by yeast. Science 219:620-625. [DOI] [PubMed] [Google Scholar]
- 10.Innis, M. A., M. J. Holland, P. C. McCabe, G. E. Cole, V. P. Wittman, R. Tal, K. W. K. Watt, D. H. Gelfand, J. P. Holland, and J. H. Meade. 1985. Expression, glycosylation, and secretion of an Aspergillus glucoamylase by Saccharomyces cerevisiae. Science 228:21-26. [DOI] [PubMed] [Google Scholar]
- 11.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura, T., N. Kitamoto, Y. Iimura, and Y. Kito. 1995. Production of HM-1 killer toxin in Saccharomyces cerevisiae transformed with the PDR4 gene and δ-sequence-mediated multi-integration system. J. Ferment. Bioeng. 80:423-428. [Google Scholar]
- 13.Kitada, K., and F. Hishinuma. 1988. Evidence for preferential multiplication of the internal unit in tandem repeats of the mating factor a genes in Saccharomyces cerevisiae. Curr. Genet. 13:1-5. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki, D., K. Miyahara, D. Hirata, H. Matsuzaki, T. Hatana, S. Fukui, and T. Miyakawa. 1994. Overexpression and secretion of cellulolytic enzymes by δ-sequence-mediated multicopy integration of heterologous DNA sequences into the chromosomes of Saccharomyces cerevisiae. J. Ferment. Bioeng. 77:468-473. [Google Scholar]
- 15.Murry, A. W., and J. W. Szostak. 1983. Pedigree analysis of plasmid segregation in yeast. Cell 34:961-970. [DOI] [PubMed] [Google Scholar]
- 16.Nagashima, T., Y. Yamamoto, K. Gomi, K. Kitamoto, and C. Kumagai. 1994. A novel culture method for high level production of heterologous protein in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 7:1292-1296. [DOI] [PubMed] [Google Scholar]
- 17.Romanos, M. A., C. A. Scorer, and J. J. Clare. 1992. Foreign gene expression in yeast: a review. Yeast 6:423-488. [DOI] [PubMed] [Google Scholar]
- 18.Sakai, A., Y. Shimizu, and F. Hishinuma. 1990. Integration of heterologous genes into chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Appl. Microbiol. Biotechnol. 33:302-306. [DOI] [PubMed] [Google Scholar]
- 19.Sakai, A., F. Ozawa, T. Hishigaki, Y. Shimizu, and F. Hishinuma. 1991. Enhanced secretion of human nerve growth factor from Saccharomyces cerevisiae using an advanced δ-integration system. Bio/Technology 9:1382-1385. [DOI] [PubMed] [Google Scholar]
- 20.Sato, T., S. Tsunasawa, Y. Nakamura, E. Mituura, F. Sakiyama, and F. Matsubara. 1986. Expression of the human salivary α-amylase gene in yeast and characterization of the secreted protein. Gene 50:247-257. [DOI] [PubMed] [Google Scholar]
- 21.Smith, R. A., M. J. Duncan, and D. T. Moir. 1985. Heterologous protein secretion from yeast. Science 229:1219-1224. [DOI] [PubMed] [Google Scholar]
- 22.Tani, Y., Y. Tomohiro, A. Miyata, K. Furukawa, and S. Hayashida. 1993. Endomitotic diploidization of Saccharomyces cerevisiae by heat treatment during spore germination. Yeast 9:519-521. [DOI] [PubMed] [Google Scholar]
- 23.Watari, J., Y. Takata, M. Ogawa, N. Nishikawa, and M. Kimura. 1989. Molecular cloning of a flocculation gene in Saccharomyces cerevisiae. Agric. Biol. Chem. 53:901-903. [Google Scholar]