Abstract
In the filamentous fungus Aspergillus nidulans, iron homeostasis is regulated at the transcriptional level by the negative-acting GATA factor SREA. In this study the expression of a putative heme-containing metalloreductase-encoding gene, freA, was found to be upregulated by iron limitation independently of SREA, demonstrating the existence of an iron-regulatory mechanism which does not involve SREA. In contrast to freA, various other genes encoding proteins in need of iron-containing cofactors—acoA, lysF, and cycA—were downregulated in response to iron depletion. Remarkably, SREA deficiency led to increased expression of acoA, lysF, and cycA under iron-replete growth conditions.
Virtually all organisms require iron for their growth. The electron transfer ability of the iron atom makes it essential for redox reactions ranging from respiration to ribonucleotide synthesis. Despite the fact that iron is the fourth most abundant element in the earth's crust, the amount of bioavailable iron is very limited since this metal is most commonly found as insoluble Fe(III)-hydroxide. Thus, microorganisms need specialized iron mobilization systems (14). On the other hand, an excess of iron in the cell can be detrimental, because iron can catalyze the production of cell-damaging hydroxyl radicals in the presence of oxygen. Therefore, the concentration of iron in biological fluids is tightly regulated, and control is accomplished primarily by the rate of uptake.
Under iron starvation, most fungi synthesize and excrete low-molecular-weight, Fe(III)-specific chelators, termed siderophores, in order to solubilize environmental iron. Subsequently, cells recover the iron from the ferrisiderophore complexes via specific uptake mechanisms (17). Furthermore, most fungi possess intracellular siderophores as an iron storage compound. In this respect Saccharomyces cerevisiae is an exception since it lacks the ability to synthesize siderophores, although it can utilize siderophores produced by other species. This yeast employs two distinct high-affinity iron uptake systems which are both regulated by the paralogous transcriptional activators Aft1p and Aft2p (2, 32). The first mechanism—termed reductive iron assimilation—requires the action of surface metalloreductases with different substrate specificities (Fre1p to Fre4p) to reduce Fe(III) to Fe(II), which is subsequently transported into the cell by the permease-oxidase complex Ftr1p/Fet3p (1, 5, 27, 34). This system allows the uptake of both siderophore-bound and unbound iron (33). The second iron uptake system—called nonreductive iron assimilation—is specialized for the uptake of siderophore-bound iron and depends on members of the major facilitator superfamily (16, 18, 33).
In ascomycetes and basidiomycetes, siderophore biosynthesis and siderophore-mediated iron uptake are controlled by orthologous, negative-acting GATA transcription factors, e.g., Aspergillus nidulans SREA, Neurospora crassa SRE, and Ustilago maydis URBS1 (15, 29, 35). In A. nidulans, deletion of sreA results in derepressed intracellular and extracellular siderophore biosynthesis as well as increased accumulation of iron under sufficient iron supply due to derepressed siderophore uptake (21). Recently various members of the SREA regulon which are presumably involved in biosynthesis, transport, and utilization of siderophores have been identified, e.g., mirA, which encodes an orthologue of the S. cerevisiae siderophore permeases (21, 22). Notably, neither the available A. nidulans cDNA and genomic sequences nor the publicly accessible complete genomes of the close relatives Aspergillus fumigatus (http://www.sanger.ac.uk/Projects/A_fumigatus/) and N. crassa (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/) seem to contain orthologues of S. cerevisiae AFT1 or AFT2. Furthermore, S. cerevisiae does not possess an orthologue of A. nidulans SREA. Thus, the question remains if SREA represents the major iron regulator or if it is specific for control of siderophore metabolism.
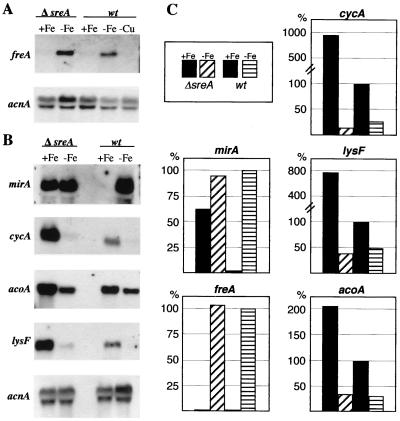
Up to now, it was not known if A. nidulans has the ability for reductive iron uptake. Searches for putative components of this system in various A. nidulans sequence databases led to the identification of expressed sequence tag clone o5f06a1, whose translation product displayed significant similarity to metalloreductases. The sequence information was used to isolate corresponding genomic clones from a cosmid library provided by the Fungal Genetic Stock Center (4). The five hybridizing clones, L4F02, L28H11, L25F03, L23A09, and L32A010, localized freA to chromosome IV, and the entire sequence of freA was sequenced directly from cosmid L23A09. Comparison of the genomic and cDNA sequences, obtained by 5′ and 3′ rapid amplification of cDNA ends according to the protocols of Frohman et al. (8), revealed an open reading frame of 1,797 bp interrupted by five introns, 53, 50, 43, 47, and 45 nucleotides (nt) in length (Fig. 1A). Additionally, two introns, 66 and 58 nt in length, are present in the 827-bp 5′ untranslated region. The 3′ untranslated region was found to be 84 nt in length. The deduced FREA protein has a calculated molecular mass of 67.2 kDa and shows significant similarity to various metalloreductases, e.g., 24% identity (blastp E-value of 8e−32) to S. cerevisiae Fre2p. An alignment of A. nidulans FREA, S. cerevisiae Fre2p (10), Arabidopsis thaliana FRO2 (24), and the gp91phox subunit of the NADPH oxidase (25), which is critical for production of microbicidal oxidants in human neutrophils, is shown in Fig. 1B. FREA possesses all typical features of metalloreductases (7, 12): a flavin adenine dinucleotide cofactor binding site, an NADPH binding motif, and four typically spaced histidine residues predicted to coordinate a bis-heme structure between transmembrane domains of the protein (Fig. 1B). S. cerevisiae possesses nine paralogous, metalloreductase-encoding genes which display different expression profiles: FRE1 is upregulated by iron and copper depletion, FRE2 to FRE6 are upregulated by iron starvation only, FRE7 is specifically upregulated by copper limitation, and YGL160w and YLR047c are regulated by neither copper nor iron availability (12, 19). Iron regulation of these genes is mediated by Aft1p, and copper regulation is mediated by Mac1p. Fre1p to Fre4p are involved in reduction of siderophore-bound and unbound iron (5, 34), Fre1p and Fre2p additionally function in copper uptake (11), and the function of Fre5p to Fre7p is unknown. To study the expression pattern of freA, A. nidulans wild-type and sreA deletion strains were grown for 24 h at 37°C under standard conditions, iron limitation, and copper starvation as described previously (20). Northern blot analysis revealed that the A. nidulans freA expression pattern resembles that of Saccharomyces FRE2 to FRE4 by being iron but not copper regulated (Fig. 2). These data indicate that Aspergillus FREA is involved in securing iron homeostasis. It might be a component of a possible reductive iron assimilation system or function as an intracellular metalloreductase. In contrast to typical members of the SREA regulon, e.g., mirA, SREA deficiency did not lead to derepressed freA expression under iron-replete conditions. These data show that in A. nidulans an iron-regulatory mechanism exists which does not involve SREA.
FIG. 1.
A. nidulans freA. (A) Intron-exon structure of freA. (B) Alignment of A. nidulans FREA, S. cerevisiae Fre2p (P36033), A. thaliana FRO2 (CAA70770), and human gp91phox (NP_000388). Amino acid residues identical in three of the four proteins are in boldface, and amino acid residues potentially involved in the bis-heme binding (H), NADPH binding (HPFT), and flavin adenine dinucleotide binding (GPYG) are shaded in gray.
FIG. 2.
Expression of freA, mirA, cycA, acoA, and lysF under standard (+Fe), iron depletion (−Fe), and copper depletion (−Cu) conditions in A. nidulans wild-type (wt) and SREA-deficient (ΔsreA) strains. Fungal strains were grown for 24 h in minimal medium containing 10 μm FeSO4 and 10 μm CuSO4 as described previously (20); for iron- and copper-depleted growth the addition of the respective metal was omitted. As a control for loading and RNA quality, blots were hybridized with the γ-actin-encoding acnA gene (6). (A and B) Northern blot analysis was performed with 1 μg of mRNA (A) or 10 μg of total RNA (B), respectively. (C) Quantification of mRNA levels normalized to acnA levels with a PhosphorImager. Bars represent mean values of two independent experiments; standard deviations did not exceed 20%.
Furthermore, SREA-independent expression of freA confirms that SREA indeed acts as a direct repressor of extracellular siderophore biosynthesis and uptake. SREA deficiency results in 20-fold-increased accumulation of the intracellular siderophore ferricrocin during iron-replete growth (21). Therefore, it could have been alternatively hypothesized that SREA acts only as a repressor of ferricrocin biosynthesis and that SREA deficiency causes iron deprivation via sequestration of intracellular iron. But in this case, the expression of all iron starvation-induced genes, including freA, would be expected to be upregulated under iron-replete conditions in an sreA deletion strain.
Iron depletion can lead to upregulation of expression, as in the case of genes involved in high-affinity iron uptake. But the opposite regulatory pattern can also be found: expression of catB, encoding a heme-containing catalase, is downregulated at the transcript level under iron starvation (21). To investigate if this regulatory pattern is specific for catB or holds for other proteins in need of iron-containing cofactors, the expression of the genes encoding the iron-sulfur cluster containing aconitase (acoA) and homoaconitase (lysF), as well as the heme-containing cytochrome c (cycA), was studied (23, 30). For partial analysis of the putative A. nidulans aconitase gene acoC, the expressed sequence tag clone c8d09 was sequenced. It contains the C-terminal 398 amino acids of ACOC displaying 88 and 73% identity to the aconitases of Aspergillus terreus and S. cerevisiae, respectively. Northern blot analysis proved that expression of genes involved in pathways as distinct as the citric acid cycle (acoA) and respiration (cycA), as well as lysine and penicillin biosynthesis (lysF), is downregulated between two- and eightfold under iron limitation in the wild-type and the SREA-deficient strains (Fig. 2). With an eightfold-decreased transcript level, cycA was the gene most dramatically affected by iron depletion. Notably, CYCA-deficient A. nidulans mutants are viable, and it was suggested previously that this is due to the ability of Aspergillus to ferment and to use alternative respiratory pathways (3). Taken together, these data suggest that, during iron depletion, decreased expression of cycA saves energy and iron for other processes essential for survival under iron limitation. Assuming that FREA is involved in iron homeostasis, as has been shown previously for four of the six iron-regulated S. cerevisiae paralogues (5, 10, 34), the opposite regulation of freA versus acoA, lysF, and cycA by iron availability suggests that under iron depletion the flow of this limiting metal might be directed from various metabolic pathways to systems needed to secure iron homeostasis.
Interestingly, the transcript levels of acoA, lysF, and cycA were elevated between two- (acoA) and ninefold (cycA) under iron-replete conditions in the sreA deletion strain (Fig. 2). Therefore, expression of these genes might be subject to SREA regulation. Alternatively, upregulation of these genes might be caused indirectly since SREA deficiency leads to increased iron accumulation and increased oxidative stress (21): (i) it may reflect the increased bioavailability of iron within SREA-deficient cells, or (ii) it may represent an oxidative stress response. In the latter case, the increased expression of these genes could represent a compensatory response invoked to maintain cellular enzyme activities because, e.g., iron-sulfur cluster-containing enzymes are particularly sensitive to inactivation by oxidative attack (9). In this respect it is noteworthy that, in Escherichia coli, expression of aconitase-encoding acnA is specifically induced by iron and oxidative stress (13), and it was suggested previously that the aconitase proteins serve as a protective buffer against oxidative stress by acting as a sink for reactive oxygen species (28). The upregulation of cycA expression might also be a response to oxidative stress because cytochrome c plays an important role in the antioxidant system of mitochondria (26). Remarkably, the promoter region of lysF contains several GATA motifs which potentially represent SREA binding sites. But since mutational analysis showed that at least two of these GATA sites mediate a positive effect on lysF expression, a direct involvement of the repressor SREA seems to be doubtful (31).
Hybridization probes.
The hybridization probes used in this study were generated by PCR with oligonucleotides 5′-AGCCCGGTGTGAAAAGAG and 5′-AACAGGAGGAGGATTGCGCC for mirA, 5′-AGATCATGGGAGTTGACCTG and 5′-AGACGGATTGTATGGCGATGAG for freA, 5′-ACCCTTTCTCTCTACCTC and 5′-CGCGATTAGACGAGATAA for cycA, 5′-TATCCATGTAGTCCGCCC and 5′-GGTCCCACTGTCCAATGC for acoA, 5′-GCTGACGAACGAAGAAG and 5′-GCGTTCTTAACCCATTTC for lysF, and 5′-CGGTGATGAGGCACAGT and 5′-CGGACGTCGACATCACA for γ-actin-encoding acnA.
Nucleotide sequence accession number.
The freA and acnA sequences were assigned GenBank accession no. AF515629 and AF515630, respectively.
Acknowledgments
We are grateful to Bruce A. Roe et al. for the information supplied by the A. nidulans cDNA sequencing project and to the Whitehead Institute/MIT Center for Genome Research for access to the N. crassa genome sequence, as well as to the Sanger Institute and its collaborators David Denning and Andrew Brass at the University of Manchester for access to the A. fumigatus genome sequence. We thank Axel Brakhage for a plasmid containing a lysF fragment.
This project was supported by Austrian Science Foundation grant FWF-P13202-MOB (to H.H.).
REFERENCES
- 1.Askwith, C. C., D. de Silva, and J. Kaplan. 1994. Molecular biology of iron acquisition in Saccharomyces cerevisiae. Mol. Microbiol. 20:27-34. [DOI] [PubMed] [Google Scholar]
- 2.Blaiseau, P. L., E. Lesuisse, and J. M. Camadro. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276:34221-34226. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw, R. E., D. M. Bird, S. Brown, R. E. Gardiner, and P. Hirst. 2001. Cytochrome c is not essential for viability of the fungus Aspergillus nidulans. Mol. Genet. Genomics 266:48-55. [DOI] [PubMed] [Google Scholar]
- 4.Brody, H., J. Griffith, A. J. Cuticchia, J. Arnold, and W. E. Timberlake. 1991. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 19:3105-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fidel, S., J. H. Doonan, and N. R. Morris. 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene 70:283-293. [DOI] [PubMed] [Google Scholar]
- 7.Finegold, A. A., K. P. Shatwell, A. W. Segal, R. D. Klausner, and A. Dancis. 1996. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 271:31021-31024. [DOI] [PubMed] [Google Scholar]
- 8.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, P. R., and I. Fridovich. 1992. Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J. Biol. Chem. 267:8757-8763. [PubMed] [Google Scholar]
- 10.Georgatsou, E., and D. Alexandraki. 1994. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgatsou, E., L. A. Mavrogiannis, G. S. Fragiadakis, and D. Alexandraki. 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272:13786-13792. [DOI] [PubMed] [Google Scholar]
- 12.Georgatsou, E., and D. Alexandraki. 1999. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 15:573-584. [DOI] [PubMed] [Google Scholar]
- 13.Gruer, M. J., and J. R. Guest. 1994. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140:2531-2541. [DOI] [PubMed] [Google Scholar]
- 14.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 15.Haas, H., I. Zadra, G. Stöffler, and K. Angermayr. 1999. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274:4613-4619. [DOI] [PubMed] [Google Scholar]
- 16.Heymann, P., J. F. Ernst, and G. Winkelmann. 2000. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186:221-227. [DOI] [PubMed] [Google Scholar]
- 17.Leong, S. A., and G. Winkelmann. 1998. Molecular biology of iron transport in fungi. Met. Ions Biol. Syst. 35:147-186. [PubMed] [Google Scholar]
- 18.Lesuisse, E., M. Simon-Casteras, and P. Labbe. 1998. Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144:3455-3462. [DOI] [PubMed] [Google Scholar]
- 19.Martins, L. J., L. T. Jensen, J. R. Simon, G. L. Keller, D. R. Winge, and J. R. Simons. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 20.Oberegger, H., I. Zadra, M. Schoeser, and H. Haas. 2000. Iron starvation leads to increased expression of Cu/Zn-superoxide dismutase in Aspergillus. FEBS Lett. 485:113-116. [DOI] [PubMed] [Google Scholar]
- 21.Oberegger, H., M. Schoeser, I. Zadra, B. Abt, and H. Haas. 2001. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 41:1077-1089. [DOI] [PubMed] [Google Scholar]
- 22.Oberegger, H., I. Zadra, M. Schoeser, B. Abt, W. Parson, and H. Haas. 2002. Identification of members of the Aspergillus nidulans SREA regulon: genes involved in siderophore biosynthesis and utilization. Biochem. Soc. Trans. 30:781-783. [DOI] [PubMed] [Google Scholar]
- 23.Raitt, D. C., R. E. Bradshaw, and T. M. Pillar. 1994. Cloning and characterisation of the cytochrome C gene of Aspergillus nidulans. Mol. Gen. Genet. 242:17-22. [DOI] [PubMed] [Google Scholar]
- 24.Robinson, N. J., C. M. Procter, E. L. Connolly, and M. L. Guerinot. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397:694-697. [DOI] [PubMed] [Google Scholar]
- 25.Royer-Pokora, B., L. M. Kunkel, A. P. Monaco, S. C. Goff, P. E. Newburger, R. L. Baehner, F. S. Cole, J. T. Curnutte, and S. H. Orkin. 1986. Cloning the gene for an inherited human disorder—chronic granulomatous disease—on the basis of its chromosomal location. Nature 322:32-38. [DOI] [PubMed] [Google Scholar]
- 26.Skulachev, V. P. 1998. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 423:275-280. [DOI] [PubMed] [Google Scholar]
- 27.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 28.Tang, Y., M. A. Quail, P. J. Artymiuk, J. R. Guest, and J. Green. 2002. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology 148:1027-1037. [DOI] [PubMed] [Google Scholar]
- 29.Voisard, C., J. Wang, J. L. McEvoy, P. Xu, and S. A. Leong. 1993. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol. Cell. Biol. 13:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidner, G., B. Steffan, and A. A. Brakhage. 1997. The Aspergillus nidulans lysF gene encodes homoaconitase, an enzyme involved in the fungus-specific lysine biosynthesis pathway. Mol. Gen. Genet. 255:237-247. [DOI] [PubMed] [Google Scholar]
- 31.Weidner, G., S. Steidl, and A. A. Brakhage. 2001. The Aspergillus nidulans homoaconitase gene lysF is negatively regulated by the multimeric CCAAT-binding complex AnCF and positively regulated by GATA sites. Arch. Microbiol. 175:122-132. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi-Iwai, Y., A. Dancis, and R. D. Klausner. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun, C. W., T. Ferea, J. Rashford, O. Ardon, P. O. Brown, D. Botstein, J. Kaplan, and C. C. Philpott. 2000. Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J. Biol. Chem. 275:10709-10715. [DOI] [PubMed] [Google Scholar]
- 34.Yun, C. W., M. Bauler, R. E. Moore, P. E. Klebba, and C. C. Philpott. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:10218-10223. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, L. W., H. Haas, and G. A. Marzluf. 1998. Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol. Gen. Genet. 259:532-540. [DOI] [PubMed] [Google Scholar]