Abstract
Identifying microorganisms responsible for recognized environmental processes remains a great challenge in contemporary microbial ecology. Only in the last few years have methodological innovations provided access to the relationship between the function of a microbial community and the phylogeny of the organisms accountable for it. In this study stable-isotope-labeled [13C]phenol was fed into a phenol-degrading community from an aerobic industrial bioreactor, and the 13C-labeled RNA produced was used to identify the bacteria responsible for the process. Stable-isotope-labeled RNA was analyzed by equilibrium density centrifugation in concert with reverse transcription-PCR and denaturing gradient gel electrophoresis. In contradiction with findings from conventional methodologies, this unique approach revealed that phenol degradation in the microbial community under investigation is dominated by a member of the Thauera genus. Our results suggest that this organism is important for the function of this bioreactor.
Our dependence upon laboratory-based studies of culturable bacterial species is acknowledged to be a primary limitation to progress in microbial ecology (2). Traditionally, it has been difficult to attribute a recognized microbially mediated process to the organism(s) responsible for that process in situ (12). Recently, however, culture-independent approaches to linking microbial community function with the genetic identity of key organisms have begun to emerge (7, 18, 23). Among the limited number of elegant methodologies capable of identifying microorganisms responsible for particular biogeochemical processes, stable-isotope probing (SIP) holds considerable promise.
SIP involves the incorporation of stable-isotope-labeled substrates into cellular biomarkers that can be used to identify organisms assimilating the substrate (6). Stable isotopes were first used in this capacity to identify the microbial community component responsible for acetate oxidation in aquatic sediments (7). A 13C-labeled acetate pulse was administered to sediments, with the resulting 13C-enriched polar-lipid-derived fatty acid (PLFA) signature profiles subsequently being preferentially analyzed. Other studies utilizing the same approach have followed suit (9, 24).
The utility of PLFA-based SIP may be limited, because resolving PLFA profiles composed of multiple species can be problematic (26). This limitation has been overcome with the demonstration that stable-isotope-labeled DNA can be recovered from total community nucleic acid extractions on the basis of its increased buoyant density and can be used as an alternative, unambiguous biomarker (21, 26, 32).
Because DNA-based SIP relies on the isolation of labeled DNA by density centrifugation, the degree of isotopic enrichment is crucial. Of the many factors determining the success of an enrichment, including the duration of the pulse and the presence of unlabeled substrate inherent to the system, the rate of DNA synthesis in situ plays a pivotal role. We speculate that DNA synthesis rates in situ, which reflect organism replication rates, limit the enrichment of this biomarker and hence the utility of DNA-based SIP. By comparison, RNA synthesis rates are higher than those of DNA. This is a function of copy number and the continual turnover of RNA, itself a reflection of cellular activity independent of replication. Because RNA also offers the same sequence-based resolution for organism identification as DNA, we hypothesize that RNA could serve as a more responsive biomarker for use in SIP. To establish whether RNA-based SIP (RNA-SIP) is a practicable means of linking a community activity with the organisms responsible we have applied the approach to an industrial bioreactor community responsible for the remediation of phenol-contaminated wastewater.
Phenolic compounds are ubiquitous in the environment, owing to the multitude of human-made and natural processes that produce them. In concert with this ubiquity phenolic compounds are universally toxic, thereby limiting their own biological degradation (27). The resulting accumulation of phenolics from natural sources (plant root exudates, lignins, tannins, etc., in peatland soils) represents a mechanism maintaining a significant sink in the global carbon budget (11). Accumulation from human-made sources (pulp mills, steel manufacturing plants, coal mines, etc.) constitutes an environmental hazard unacceptable to regulatory agencies worldwide (10). Unfortunately, almost all our understanding of industrial and environmental phenol biodegradation is extrapolated from laboratory-based experiments on culturable phenol-degrading bacterial species (31). As a case in point, the identity of the microorganisms responsible for phenol degradation in the bioreactor under investigation and the processes by which they achieve their task are unknown, as are the reasons for the infrequent but costly loss of bioreactor function. This investigation aimed to develop and apply RNA-SIP to the reactor community to demonstrate the utility of the technique and to identify organisms responsible for phenol remediation in the bioreactor.
MATERIALS AND METHODS
Bioreactor characteristics.
The industrial facility generating the waste under investigation produces 1,800 liters of wastewater/min, which is heavily polluted with phenolic compounds (>200 μg/ml). Vitox reactors with a combined volume of 1,700 m3 are employed to support a dense (∼1010 cells/ml) floc-based microbial community responsible for lowering the concentration of total phenolics to less than 10 μg/ml before the wastewater is released into a public waterway (33).
Bacterial strains and culture conditions.
Pseudomonas putida BS564 is a culturable phenol-degrading species previously isolated from the industrial bioreactor under investigation (34). As a pure culture, P. putida BS564 was maintained in a minimal salts medium (M9) supplemented with 200 μg of phenol (BDH laboratory supplies)/ml or phenol-13C6 (Isotech) as a sole carbon source. Pseudomonas chlororaphis BS523 is a phenol-tolerant isolate from the bioreactor unable to utilize phenol as a carbon source (34). Mixed cultures of P. putida BS564 and P. chlororaphis BS523 were maintained in a minimal salts medium (M9) supplemented with 200 μg of phenol-13C6 (Isotech)/ml and 10 μg of glucose/ml. All cultures were incubated at 30°C with shaking (150 rpm).
Assay for phenol concentration.
Phenol was quantified as previously described (16). Briefly, filtered (filter pore size, 0.22 μm) 100-μl aliquots of test sample were treated with 2.5 μl of 0.5 M NH4OH and adjusted to pH 7.9 with 2.25 μl of phosphate buffer (0.5 M KH2PO4, 0.6 M K2HPO4). Samples were then treated with 1 μl of 100 mM 4-aminoantipyrine and 1 μl of 250 mM potassium ferricyanide. Reaction products were measured spectrophotometrically at 500 nm, and phenol concentrations were calculated from standard curves prepared in parallel.
Nucleic acid extraction.
RNA from pure laboratory-grown cultures was extracted by RNeasy protocols (Qiagen). DNA and RNA from bioreactor samples were extracted by washing cells in phosphate-buffered saline and subsequently resuspending 0.25 g of biomass in 0.5 ml of 240 mM potassium phosphate buffer (pH 8.0) and 0.5 ml of phenol:chloroform:isoamyl alcohol (25:24:1) (pH 4.0). Cell suspensions were transferred to Bio-101 Multimix 2 Matrix tubes and were subjected to agitation in a FastPrep FP120 bead beating system for 30 s at 5.5 m/s. The aqueous phase was separated by centrifugation and further purified by RNeasy mini kit protocols (Qiagen). DNA was separated from RNA samples by RNA and DNA mini kit protocols (Qiagen). All samples were stored in RNase-free water at −20°C. DNA and RNA were quantified and checked for purity by using ultramicro volume cuvettes in a GeneQuant pro RNA/DNA calculator (Amersham Pharmacia Biotech).
Centrifugation and fractionation.
Equilibrium (isopycnic) density gradient centrifugation and gradient fractionation were conducted in cesium trifluoroacetate (CsTFA) gradients consisting of 1.86 ml of a 1.99-g/ml CsTFA solution (Amersham Pharmacia Biotech), 375 μl of H2O, and 75 μl of deionized formamide. Gradients were loaded with 500 ng of total RNA in polyallomer bell-top Quick-Seal centrifuge tubes (11 by 32 mm), sealed, and spun in a TLA120.2 rotor in an Optima TLX ultracentrifuge (Beckman Coulter) at 64,000 rpm and 20°C for 36 h. Gradients were fractionated from below by displacement with water by using a syringe pump at a flow rate of 3.3 μl/s. The buoyant density of gradient fractions was determined by weighing known volumes on a four-figure milligram balance. RNA was isolated from gradient fractions by precipitation with isopropanol. Gradient fractions were checked for the presence of RNA by standard agarose gel electrophoresis, ethidium bromide (0.1 μg/ml) staining, and densitometric analysis (Phoretix; Nonlinear Dynamics).
16S rRNA amplification and denaturing gradient gel electrophoresis (DGGE) analysis.
RNA samples from equilibrium density gradient fractions were reverse transcribed with reverse primer 519R (5′-GTATTACCGCGGCTGCTG-3′) (17) and avian myeloblastosis virus reverse transcriptase (Promega). The cDNA produced was used to amplify the V3 region of the 16S rRNA sequence from positions 356 to 519 with an adjoining 39-bp guanine + cytosine (GC) clamp with forward primer GC356F (5′-CGCCCGCCGCGCCCCCGCCCCGGCCCGCCGCCCCCGCCCACTCCTACGGGAGGCAGC-3′) and 519R. The GC-clamped products were separated on 10% (wt/vol) polyacrylamide gels with a 30 to 60% urea/formamide denaturing gradient as previously described (33). Denaturing gradient gels were cast and run by using the Ingeny PhorU2 system at 60°C and 200 V for 8 h. Gels were stained with SYBR gold nucleic acid gel stain (Molecular Probes) and were visualized by UV trans-illumination.
Sequencing DGGE bands.
Organisms represented in DGGE profiles were analyzed by reamplifying, cloning, and sequencing bands of interest. Bands were excised from gels and were eluted in water. Recovered DNA was reamplified by using primers GC356F and 519R as described above. Products were cloned into pCR4-TOPO (Invitrogen) and were sequenced by using M13 forward and reverse primers from three randomly selected clones. To obtain the entire 16S rRNA gene sequence from the dominant species a clone library was constructed from total DNA extracts of sludge cells amplified with primers BF1 [TCAGA(A/T)(C/T)GAACGCTGGCGG] and 1525R (AAGGAGGTGATCCAGCC) (17) in pCR4-TOPO (Invitrogen). Clones were screened for the dominant 16S rRNA gene sequence by DGGE analysis of colony PCR products obtained by using primers GC356F and 519R. Four separate clones from the library with matching DGGE products were sequenced by using internal primer sets. Sequencing was performed with Beckman dye chemistry and a CEQ2000 Beckman sequencer according to the manufacturer's instructions (Beckman Coulter Corporation). Sequence homology was determined by BLASTN searches to determine the most similar sequences in the GenBank and EMBL databases (1).
13C/12C-IRMS analysis of nucleic acids.
Total community DNA and RNA samples were prepared for isotope ratio mass spectrometry (IRMS) analysis as follows. In triplicate, 1-μg samples of DNA or RNA (0.36 μg of C) were cut with 61.6 μg of glucose (24.64 μg of C) to give a minimum C content (25 μg). To assess the localization of 13C-enriched RNA in equilibrium density gradient fractions, samples were prepared by extracting RNA from gradient fractions derived from six separate equilibrium density gradients which were subsequently pooled according to fraction number and were cut with 61.6 μg of glucose. Samples were placed into 6- by 4-mm tin cups (Microanalysis, Okehampton, United Kingdom) and freeze-dried for 16 h (Christ, Osterode, Germany) before being analyzed for 13C content by continuous-flow IRMS at the Natural Environment Research Council (NERC) Stable Isotope Facility located at CEH Merlewood UK by using an Elemental Analyser (Carlo Erba/Fisons UK) linked to a modified isotope ratio mass spectrometer (IRMS) (Dennis Leigh, Merlewood, United Kingdom). The 13C content of DNA and RNA samples are reported as 13C atom percent. The 13C atoms percent values obtained from gradient fractions were adjusted for RNA concentration and contaminating carbon (CsTFA) to reflect relative RNA enrichment by standard mass balance calculations.
RESULTS
RNA labeling relates to metabolic ability.
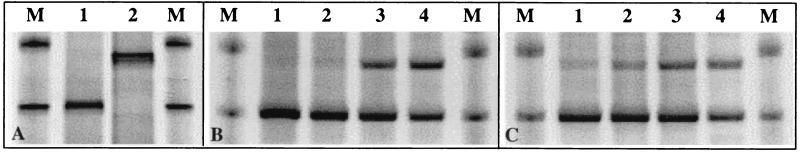
To test whether RNA labels according to specific metabolic activity, a mixed culture consisting of a phenol degrader (P. putida) and a phenol-tolerant nondegrader (P. chlororaphis) was grown in minimal medium supplemented with phenol-13C6 and glucose. RNA was extracted from the mixed culture after 24, 48, and 72 h and was subjected to equilibrium density centrifugation and fractionation. The V3 region of 16S rRNA from fractions 5, 7, 9, and 11 was reverse transcribed and amplified with an adjoining 5′ GC clamp, and products were analyzed by DGGE. Figure 1B reveals that after 24 h the two lower density fractions (9 and 11), associated by buoyant density with unlabeled RNA (1.769 to 1.750 g/ml), contained 16S rRNA templates from both bacterial species present in the mixed culture. In contrast, the two higher-density fractions (5 and 7), associated with enriched RNA (1.805 to 1.787 g/ml), only contained the 16S rRNA template of the phenol-degrading P. putida strain. As at 24 h, there was no detectable assimilation of 13C into the RNA of P. chlororaphis after 48 h (data not shown). After 72 h, however, the 16S rRNA sequence of P. chlororaphis was detected in gradient fractions 5 and 7, indicative of carbon cross-feeding (Fig. 1C). Therefore, over 48 h the assimilation of 13C was specific to the organism expressing the metabolic ability to utilize labeled phenol.
FIG. 1.
RNA-SIP analysis of a defined, mixed bacterial culture. (A) GC-clamped 16S rRNA (V3 region) RT-PCR products from pure cultures of a phenol-degrading isolate (P. putida; lane 1) and a nondegrading isolate (P. chlororaphis; lane 2) on a denaturing gradient gel. (B) RT-PCR products from fractions 5, 7, 9, and 11 (lanes 1 to 4, respectively) of an equilibrium density gradient loaded with RNA extracted at 24 h from a mixed P. putida, P. chlororaphis culture grown on phenol-13C6. RNA from both species was detected in fractions 9 and 11 (associated by buoyant density with unlabeled RNA). The 16S rRNA sequence of P. putida alone was detectable in fractions 5 and 7 (13C-labeled RNA), demonstrating specific access to 13C according to metabolic activity. (C) RT-PCR products from fractions 5, 7, 9, and 11 (lanes 1 to 4, respectively) of a gradient loaded with RNA extracted after 72 h from the mixed culture. RNA from both species was detectable in all four fractions, indicative of 13C cross-feeding between species. These results are representative of two separate experiments. Lanes labeled M were loaded with an unrelated DGGE marker.
RNA labels more efficiently than DNA.
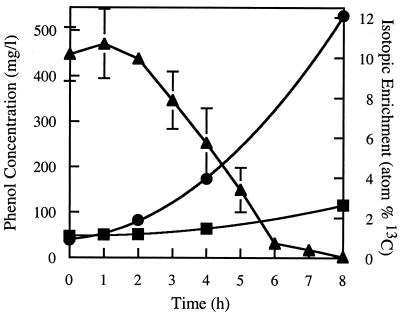
To test the utility of the technique in an industrial bioreactor we added 500 μg of phenol-13C6/ml to sludge samples and assayed hourly for phenol concentration. Native phenol levels in the reactor were undetectable (<1 μg/ml) when the pulse was administered. Figure 2 illustrates the degradation of the labeled phenol pulse over time. The phenol-13C6 concentration remained unchanged for 2 h, after which time it dropped rapidly (101 μg/ml · h−1, R2 = 0.997) until it reached 30 μg/ml at 6 h. The phenol pulse was undetectable at 8 h. An unlabeled phenol pulse displayed degradation kinetics indistinguishable from those of the labeled pulse. The phenol concentration in paraformaldehyde-treated sludge samples (killed control) did not change over time, indicating that the observed degradation was biological.
FIG. 2.
Degradation of phenol-13C6 and increase in 13C atom percent of RNA and DNA in the microbial community from an industrial bioreactor. Bioreactor samples were pulsed with 500 μg of phenol-13C6/ml and were assessed hourly for phenol concentration (triangles). The phenol concentration remained constant over the initial 2 h but then dropped rapidly over the following 5 h. A reduced phenol degradation rate preceded the complete removal of the pulse by 8 h. Samples were assayed for phenol in triplicate. Error bars represent standard errors. These data are representative of three separate experiments. Total community DNA and RNA samples were extracted at 0, 1, 2, 4, and 8 h and were subjected to IRMS analysis. The 13C atom percent of RNA (•) increased more than 10-fold as the labeled phenol pulse was utilized, while that of DNA (▪) increased just over 2-fold. Nucleic acid samples were analyzed in triplicate from a single experiment. Error bars representing standard deviation were too small to be visible.
Total community DNA and RNA was extracted throughout the experiment at 0, 1, 2, 4, and 8 h. The purified nucleic acids were quantified and subject to IRMS analysis to determine the level of 13C enrichment. The 13C atom percent of community DNA remained relatively unchanged throughout the first 4 h of the pulse but had increased over twofold (from 0.98 to 2.63 13C atom%) by the time the pulse was completely removed (Fig. 2). The 13C atom percent of community RNA remained relatively unchanged throughout the first 2 h of the pulse but then increased more than 10-fold (from 0.98 to 12.09 13C atom%) over 6 h (Fig. 2). This confirmed that carbon from the phenol pulse is being incorporated into the microbial community biomass and that the rate of incorporation into RNA exceeds that of DNA.
RNA-SIP reveals phenol assimilation is dominated by a Thauera species.
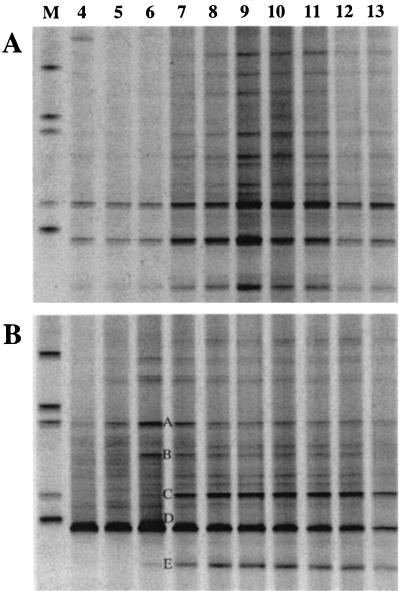
RNA samples taken throughout biodegradation of the phenol pulse were subjected to equilibrium density centrifugation and fractionation. RNA from 10 consecutive fractions (fractions 4 to 13, encompassing buoyant densities of 1.81 to 1.73 g/ml) was reverse transcribed and amplified for analysis by DGGE as described above. Figure 3 shows the reverse transcription (RT)-PCR products from density gradients loaded with RNA extracted at 1 and 8 h into the phenol-13C6 pulse on denaturing gradient gels. These two-dimensional RNA-SIP profiles represent a functional analysis of a microbial community. Each species detected by RT-PCR is separated horizontally according to 13C atom percent (by equilibrium density centrifugation) and vertically according to 16S rRNA melting temperature (by DGGE).
FIG. 3.
RNA-SIP profiles relating community function to specific microbial community components. DGGE profile of community RNA present in fractions 4 to 13 from equilibrium density gradients loaded with RNA extracted 1 (A) and 8 h (B) after the phenol-13C6 pulse was administered. Lane numbers correspond to fraction numbers. Five of the most dominant bands are labeled A to E (B). After 1 h the profile suggests that RNA from all detectable species have the same buoyant density prior to phenol degradation. After 8 h the profile suggests that RNA from particular community members (bands A, B, and D) have increased in buoyant density subsequent to phenol degradation. Lanes labeled M were loaded with an unrelated DGGE marker.
The RNA-SIP profile of the bioreactor community prior to any assimilation of 13C from the phenol pulse (Fig. 3A) revealed RNA templates predominantly in fraction 9 and spanning fractions 7 to 11. The presence of RNA in many fractions of the gradient is a result of the limitations of density gradient centrifugation and the high sensitivity of RT-PCRs. The buoyant density of fraction 9 (1.769 g/liter) corresponds closely to that of unlabeled RNA. The distribution of RT-PCR products illustrated in Fig. 3A (all bands in the profile were most intense around fractions 9 and 10) indicates that RNA from each represented bioreactor species has the same buoyant density 1 h into the pulse. The only exception is a band present in fraction 4 above the first band in the marker. Sequence analysis of this band revealed it to be an Acinetobacter species, a common experimental contaminant (28). Note that the remaining gradient fractions (4 to 6 and 12 to 13) also generated RT-PCR products. This reveals that RNA is present throughout the equilibrium density gradient at concentrations detectable by RT-PCR, thus suggesting that density gradients typically used to isolate RNA according to buoyant density have a limited ability to focus RNA into tightly defined bands.
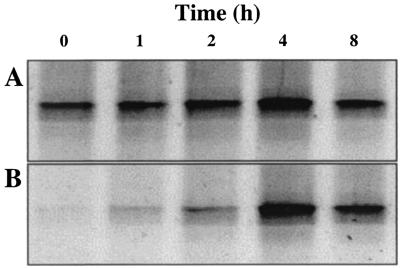
The RNA-SIP profile of the bioreactor community 8 h after the phenol pulse was administered (Fig. 3B) showed marked variation from the profile after 1 h. RT-PCR products derived from RNA from at least three species focused around fraction 6 in the density gradient, indicative of increased buoyant density. The effect is particularly pronounced for the band labeled D in Fig. 3B. In conjunction with the observation that this band is also the most intense in the DGGE profile, suggestive of numerical dominance in the reactor, we believe this result indicates that one bacterial species in particular has dominated the acquisition of carbon from phenol. Figure 4 depicts the changes in intensity of this band over time in gradient fractions 6 and 10, corresponding to buoyant densities of 1.796 and 1.760 g/ml, respectively. This suggests that the rRNA template from which the RT-PCR product is derived increases in concentration in the high-density fraction over time without a concomitant increase in the unlabeled RNA density fraction. This result was observed with bioreactor samples taken on three independent occasions spanning a period of 6 months, suggesting that the ecological processes underlying bioreactor function were stable over this time. Two additional bands (Fig. 3B, bands A and B) also showed a distinct increase in product concentration in gradient fraction 6 after 8 h, suggesting that these species also acquire carbon from phenol. In contrast, the bands labeled C and E in Fig. 3B were dominant in the community profile but did not increase in intensity in the high buoyant density gradient fractions, suggesting that these species do not rely on phenol as a carbon source.
FIG. 4.
Increasing buoyant density of the dominant band in 16S rRNA community profiles throughout the duration of the phenol-13C6 pulse. (A) The intensity of the dominant DGGE band remains constant in fraction 10 over time. (B) The intensity of the dominant DGGE band increases in fraction 6 over time, confirming an increase in the buoyant density of RNA from this species as phenol-13C6 is consumed.
Density gradients were subject to IRMS analysis to confirm that RNA isolated from high-density gradient fractions had become enriched with 13C. The 13C content of RNA was found to be above natural abundance levels in fractions 5 to 9, with RNA in fraction 5 having the highest 13C content with a sequential decrease through fractions 6 to 9. This directly relates the increase in buoyant density of RNA to 13C enrichment.
The five most prominent bands in the 16S rRNA community DGGE profile (Fig. 3B, bands A to E) were excised, reamplified, cloned, and sequenced. The resulting sequences encompassing the V3 region of the 16S rRNA indicated that these organisms belonged to the β-Proteobacteria (bands B, C, and D), the α-Proteobacteria (band A), and the Acidobacteria (band E). The full 16S ribosomal DNA sequence of band D (accession number AY098637) was obtained from a clone library constructed from DNA extracted from untreated bioreactor sludge. BLASTN searches revealed that the organism that dominated the acquisition of carbon from phenol is a member of the Thauera genus (3).
DISCUSSION
SIP provides access to the relationship between environmental functions and the specific microbial community members pivotal to function performance (12). SIP involves pulsing stable-isotope-labeled substrates into microbial communities and subsequently analyzing labeled biomarkers to identify functional organisms (6). To date, the choice of biomarkers has been restricted to PLFAs and DNA, both of which exhibit inherent limitations (26). In this study the use of RNA as a biomarker in SIP has been explored.
The strength of 16S rRNA as a biomarker in SIP lies in the relatively unambiguous taxonomic information it provides (22) and its rapid turnover rate compared to that of DNA. In the densely populated and functionally active bioreactor community investigated here, the total RNA pool was enriched for labeled carbon much more efficiently than the total DNA pool when pulsed with phenol-13C6. This finding is reasonable in light of the continuous-flow biomass recycling architecture of the bioreactor. Biomass generation is expected to be largely dependent on the level of grazing activity of protozoa present in the reactor (unpublished observation). It remains to be demonstrated that the enrichment of RNA will be superior to that of DNA in other microbial communities with different growth dynamics and activities, such as soils. An additional advantage of RNA-SIP stems from the concurrent isolation of entire transcriptomes along with the 16S rRNA of community members utilizing the stable-isotope-labeled substrate. This provides access to the sequence of genes expressed by functional organisms during substrate assimilation, including those encoding the active biochemical pathway associated with biodegradation.
As with DNA-based SIP, RNA-SIP is dependent upon equilibrium density centrifugation for the isolation of stable-isotope-labeled nucleic acids (26). Because of this, an understanding of the migration of RNA on CsTFA equilibrium density gradients is a critical requirement for the successful application of the technique. Analyzing gradient fractions for the presence of RNA on ethidium bromide-stained agarose gels revealed that RNA focuses over a number of fractions in CsTFA gradients. It is important to note that the detection of RNA is dependent on the sensitivity of the detection methods employed. For example, the use of RT-PCR to detect RNA in gradient fractions has revealed that so-called equilibrium density gradient bands of RNA can span the entire length of the gradient. The presence of RNA in high-density gradient fractions, therefore, does not necessarily mean that it is isotopically enriched. Because of this, it becomes essential to analyze each gradient fraction by RT-PCR and DGGE as shown in Fig. 3 and to observe the shifting band intensities throughout the duration of the labeled substrate pulse (Fig. 4). Assurance that RNA from a particular organism has been enriched comes from the demonstration that its template increases in concentration in high-density fractions without a concomitant increase in standard-density fractions. This temporal RT-PCR and DGGE analysis also excludes the possibility that RNA from a particular species is present in high-density fractions because of differences in G+C (5).
Along with its promise, SIP exhibits potential limitations in its use as a tool in molecular microbial ecology, regardless of biomarker choice. The primary assumption inherent to the method is that a microbial community does not distinguish between a stable-isotope-labeled substrate and its natural counterpart. It is well established that bacteria can fractionate rare stable isotopes (13C) from more naturally abundant counterparts (12C) to a small but significant extent (19, 20). The degree to which bacteria have been observed to fractionate against stable-isotope-labeled substrates is unlikely to affect the assimilation of an enrichment pulse. In support of this, there was no detectable difference in the degradation rates of phenol and phenol-13C6 by the bioreactor community under investigation here; however, this does not rule out the possibility that stable-isotope fractionation may manifest itself in less robust processes, such as finely tuned syntrophic relationships. Another potential limitation of the technique concerns the possibility of substrate cross-feeding. Figure 1 illustrates that cross-feeding can influence the results obtained, especially as pulse lengths increase. Temporal analyses may enable the detection of the transition from labeling of specific community members that have access to the carbon in any given substrate to a more generalized scavenging of carbon. The potential limitation substrate dilution confers to the approach has been discussed elsewhere (26). The only potential limitation inherent to RNA-SIP is the ability to extract high-quality RNA from environmental samples. Continuing advances in nucleic acid extraction methodologies suggest this will not impede the broad application of RNA-SIP (13).
Conventional and molecular microbiological approaches have previously been used to characterize the industrial bioreactor community under investigation here (33, 34). Culture-dependent studies suggested that pseudomonad species dominate the reactor community (34), while culture-independent approaches suggested that the γ-Proteobacteria and Cytophaga-Flavobacterium groups were potentially important in the bioremediation process (33). The RNA-SIP approach utilized here revealed that a member of the β-Proteobacteria, of the order Rhodocyclales and belonging to the Thauera genus, dominates the acquisition of carbon from phenol, thus illustrating the pitfalls of assuming functional importance on the basis of culturing and sequencing alone.
Members of the Thauera genus have typically been isolated under denitrifying conditions with aromatic compounds, such as phenol, benzoate, or toluene, as a sole source of carbon and energy (29). Little attention appears to have been paid to the ability of members of this genus and the closely related Azoarcus genus to degrade aromatic compounds aerobically (8, 30). Given that dissolved oxygen is pumped into the vitox reactor to maintain concentrations of 0.1 to 0.3 mM, it is unlikely that the process occurs anaerobically. Fluorescence in situ hybridization employing a probe specific for the Thauera and Azoarcus genera (25) has demonstrated the presence of members of this group on the surface of flocs rather than buried in the floc structure, suggesting that the Thauera species dominating carbon acquisition from phenol does not occupy an anaerobic microenvironment in the reactor (unpublished data). Additionally, unpublished GC-mass spectrometry observations of bioreactor fluid throughout a phenol-13C6 pulse have revealed the generation of 13C-labeled benzendiols, which are formed during the aerobic biodegradation of phenol. Furthermore, the absence of nitrate in the bioreactor as revealed by unpublished ion exchange chromatography analyses confirms that phenol degradation in the reactor is not dependent on denitrification. Finally, while the bioreactor contains 10 mM sulfate, the addition of molybdate does not interfere with phenol degradation, thus ruling out a dependence on sulfate reduction. Taken together, these results suggest that a potentially novel Thauera species is dominating aerobic phenol degradation in the presence of organisms conventionally considered to dictate such processes (31). We propose that this member of the β-Proteobacteria is important for bioreactor function and should be monitored as part of the bioreactor management strategy. Attempts to isolate the organism are presently under way.
Band A in Fig. 3B represents an organism most closely related over the V3 region of the 16S rRNA sequence to the purple nonsulfur bacterium Rhodopseudomonas palustris, which is capable of degrading a range of aromatic compounds both aerobically and anaerobically (14). The precedent for phenol biodegradation in R. palustris supports our finding that the organism represented by band A assimilated 13C from the labeled phenol pulse. This suggests that this member of the α-Proteobacteria also plays a role in bioreactor function. There is also precedent for the degradation of phenolic compounds by Acidovorax species with which the sequence of band B shares homology, again supporting the finding that the organism represented by this band actively catabolized the phenol-13C6 pulse (35). The sequence of band C was most closely related over the V3 region to the polyhydroxyalkanoate-degrading organism Pseudomonas lemoignei (15). Band E was placed within the Acidobacterium division, which contains only three cultivated species and comprises many sequences obtained from cloning and sequencing studies from a diverse range of environments (4). The organisms represented by these bands are not implicated in the degradation of aromatic compounds and were not found to assimilate carbon from the phenol-13C6 pulse administered in this study. The correlation between organisms found to be assimilating phenol by RNA-SIP and organisms known to possess phenol-degrading abilities contributes to the validation of the approach.
The potential applications of a means of unambiguously linking biological processes to specific sequences retrieved in situ are numerous. With respect to biodegradation, a common approach to the understanding and enhancement of environmental bioremediation processes involves the extensive characterization of genes and proteins in biochemical pathways from culturable isolates displaying the activity of interest. RNA-SIP offers a means of identifying organisms dominating bioremediation processes in situ and thus can act to educate the choice of organism for characterization. With such technologies the vagaries of culturability can be avoided and the chances of successful augmentation of desired microbiological phenomena can be greatly enhanced.
Acknowledgments
We thank Phil Ineson, Nick Ostle, and the staff of the NERC/CEH Stable Isotope Facility. Thanks also to Lisa Gieg for the GC-mass spectrometry results.
M.M. was supported by NERC with a CEH nonthematic research grant (GR3/13121).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Scheliefer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders, H.-J., A. Kaetzke, P. Kampfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying Pseudomonad strains K172 and KB740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Intl. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnie, G. D. 1978. Centrifugal methods for characterising macromolecules and their interactions, p. 95-125. In G. D. Birnie and D. Rickwood (ed.), Centrifugal separations in molecular and cell biology. Butterworths, London, United Kingdom.
- 6.Boschker, H. T. S., and J. J. Middelburg. 2002. Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 1334:1-12. [DOI] [PubMed] [Google Scholar]
- 7.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-804. [Google Scholar]
- 8.Breinig, S., E. Schiltz, and G. Fuchs. 2000. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J. Bacteriol. 182:5849-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bull, I. D., N. R. Parekh, G. H. Hall, P. Ineson, and R. P. Evershed. 2000. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature 405:175-178. [DOI] [PubMed] [Google Scholar]
- 10.Environment Agency 1995. Proposed environmental quality standards for phenol in water. Environment Agency, United Kingdom. 475:1. [Google Scholar]
- 11.Freeman, C., N. Ostle, and H. Kang. 2001. An enzymic ‘latch’ on a global carbon store. Nature 409:149. [DOI] [PubMed] [Google Scholar]
- 12.Gray, N. D., and I. M. Head. 2001. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3:481-492. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, C. S., and J. Gibson. 1988. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 54:712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jendrossek, D., A. Frisse, A. Behrends, K. Andermann, H. D. Mratzin, T. Stanislawski, and H. G. Schlegel. 1995. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J. Bacteriol. 177:596-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, R. J., K. A. Short, and R. J. Seidler. 1991. Assay for detection and enumeration of genetically engineered microorganisms which is based on the activity of a deregulated 2,4-dichlorophenoxyacetate monooxygenase. Appl. Environ. Microbiol. 57:1790-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 18.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielson, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meckenstock, R. U., B. Morasch, R. Warthmann, B. Schink, E. Annweiler, W. Michaelis, and H. H. Richnow. 1999. 13C/12C isotope fractionation of aromatic hydrocarbons during microbial degradation. Environ. Microbiol. 1:409-414. [DOI] [PubMed] [Google Scholar]
- 20.Morasch, B., H. H. Richnow, B. Schink, and R. U. Meckenstock. 2001. Stable hydrogen and carbon isotope fractionation during microbial toluene degradation: mechanistic and environmental aspects. Appl. Environ. Microbiol. 67:4842-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 23.Orphan, V. J., C. H. House, K. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-486. [DOI] [PubMed] [Google Scholar]
- 24.Pelz, O., A. Chatzinotas, A. Zarda-Hess, W.-R. Abraham, and J. Zeyer. 2001. Tracing toluene-assimilating sulfate-reducing bacteria using 13C-incorporation in fatty acids and whole-cell hybridisation. FEMS Microbiol. Ecol. 38:123-131. [Google Scholar]
- 25.Rabus, R., H. Wilkes, A. Schramm, G. Harms, A. Behrends, R. Amann, and F. Widdel. 1999. Anaerobic utilisation of alkylbenzenes and n-alkanes from crude oil in an enrichment culture of denitrifying bacteria affiliating with the β-subclass of Proteobacteria. Environ. Microbiol. 1:145-157. [DOI] [PubMed] [Google Scholar]
- 26.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 27.Sparling, G. P., B. G. Ord, and D. Vaughan. 1981. Changes in microbial biomass and activity in soils amended with phenolic acids. Soil Biol. Biochem. 13:455-460. [Google Scholar]
- 28.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tschech, A., and G. Fuchs. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 148:213-217. [DOI] [PubMed] [Google Scholar]
- 30.VanSchie, P., and L. Y. Young. 1998. Isolation and characterisation of phenol-degrading denitrifying bacteria. Appl. Environ. Microbiol. 64:2432-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterisation of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitby, C. B., H. Hall, R. Pickup, J. R. Saunders, P. Ineson, N. R. Parekh, and A. McCarthy. 2001. 13C incorporation into DNA as a means of identifying the active components of ammonia-oxidizer populations. Lett. Appl. Microbiol. 32:398-401. [DOI] [PubMed] [Google Scholar]
- 33.Whiteley, A., and M. J. Bailey. 2000. Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl. Environ. Microbiol. 66:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteley, A. W., S. Wiles, A. K. Lilley, J. Philp, and M. J. Bailey. 2000. Ecological and physiological analyses of Pseudomonad species within a phenol remediation system. J. Microbiol. Methods. 44:79-88. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, J. S., and O. P. Ward. 1999. Microbial degradation of nitrobenzene and mono-nitrophenol by bacteria enriched from municipal activated sludge. Can. J. Microbiol. 45:427-432. [PubMed] [Google Scholar]