Abstract
A molecular characterization of two Mycobacterium marinum genes, 16S rRNA and hsp65, was carried out with a total of 21 isolates from various species of fish from both marine and freshwater environments of Israel, Europe, and the Far East. The nucleotide sequences of both genes revealed that all M. marinum isolates from fish in Israel belonged to two different strains, one infecting marine (cultured and wild) fish and the other infecting freshwater (cultured) fish. A restriction enzyme map based on the nucleotide sequences of both genes confirmed the divergence of the Israeli marine isolates from the freshwater isolates and differentiated the Israeli isolates from the foreign isolates, with the exception of one of three Greek isolates from marine fish which was identical to the Israeli marine isolates. The second isolate from Greece exhibited a single base alteration in the 16S rRNA sequence, whereas the third isolate was most likely a new Mycobacterium species. Isolates from Denmark and Thailand shared high sequence homology to complete identity with reference strain ATCC 927. Combined analysis of the two gene sequences increased the detection of intraspecific variations and was thus of importance in studying the taxonomy and epidemiology of this aquatic pathogen. Whether the Israeli M. marinum strain infecting marine fish is endemic to the Red Sea and found extremely susceptible hosts in the exotic species imported for aquaculture or rather was accidentally introduced with occasional imports of fingerlings from the Mediterranean Sea could not be determined.
Fish mycobacteriosis has been one of the most devastating diseases in Israeli aquaculture in recent years. The disease was first diagnosed in Eilat (Red Sea) in 1990 in cultured sea bass, Dicentrarchus labrax (7), and the causative agent, Mycobacterium marinum, has been isolated from several additional marine fish species since then (9). M. marinum is an acid-fast rod that causes chronic systemic infections in fish (1, 11) and other cold-blooded animals (32) and is capable of causing skin lesions in humans (13, 28).
Traditional diagnosis of this pathogen requires that it be recovered on culture medium and identified by means of a battery of differential biochemical tests (16). This classical method, however, often fails to identify M. marinum conclusively. Not only may the morphology and biosynthetic capabilities of mycobacteria vary depending on culture conditions (20), but also not uncommon are strain varieties that do not quite fit the typical biochemical profile for the species. In fact, the Red Sea M. marinum isolates, by being scotochromogenic and unable to hydrolyze Tween 80 and deamidate pyrazinamide, were atypical (7) and could not be positively identified to the species level until a molecular approach was adopted (17).
Over the last two decades, the 16S rRNA gene has emerged as a good standard for determining phylogenetic relationships of bacteria (35, 36). Knibb et al. (17) identified M. marinum directly from infected fish by using PCR amplification and direct sequencing of 16S rRNA products, allowing at the same time proper taxonomic assignment and opening the way to molecular epidemiologic analysis. This gene is still considered a key standard for bacterial identification due to the wealth of data available from public sequence databases (14, 34). However, as more sequence information accumulated over time, it became evident that the resolution power of 16S rRNA sequences alone is often insufficient when closely related organisms are compared (19, 27). According to Palys et al. (19), protein-encoding genes may be more discriminative than those encoding rRNA, while the analysis of two or more unlinked loci would prevent bacterial misclassification due to possible homologous recombination with other taxa.
The hsp65 gene has been highly conserved during evolution (18), is present in all known Mycobacterium species, contains species-specific variations (21) but, on average, is more variable within the genus than the gene encoding 16S rRNA (15).
In the present work, a molecular characterization of 16S rRNA and hsp65 genes was carried out with M. marinum fish isolates from various geographically distant sources to determine whether combined analysis of the two genes increases the detection of intraspecific variations. We also hoped to shed some light on the sources and epidemiology of the Israeli isolates.
MATERIALS AND METHODS
Fish and bacterial sources.
The origins of all Mycobacterium isolates used in this study and their respective hosts are listed in Table 1. Sixteen isolates originated from cultured and wild fish in Israel; one was isolated from a captive hawksbill sea turtle in Eilat (Red Sea). Six were from Europe and two were from the Far East. Reference strain ATCC 927 was isolated from a marine fish in the United States and was obtained directly from the American Type Culture Collection, Manassas, Va.
TABLE 1.
Mycobacterium isolates and their hosts
| Isolatea | Host | Originb | Groupc
|
Identity to GenBank accession no. for:
|
||
|---|---|---|---|---|---|---|
| 16S rRNA | hsp65 | 16S rRNA | hsp65 | |||
| ATCC 927 | Unspecified fish | United States, marine, captive | C | CII | AF456240 | AF456470 |
| DL240490 | European sea bass (Dicentrarchus labrax) | Israel, marine (RS), cultured | A | A | AF456238 | AF456468 |
| DL150991 | European sea bass (D. labrax) | Israel, marine (MS), cultured | A | A | AF456238 | AF456468 |
| DL045d | European sea bass (D. labrax) | Greece, marine (MS), cultured | A | A | AF456241 | AF456471 |
| DL049d | European sea bass (D. labrax) | Greece, marine (MS), cultured | F | F | AF456242 | AF456472 |
| CF030494 | Butterfly fish (Chaetodon fasciatus) | Israel, marine (RS), captive | A | A | AF456238 | AF456468 |
| PP241194e | Sea bream (Puntazzo puntazzo) | Israel, marine (RS), cultured | ||||
| SO020195e | Red drum (Sciaenops ocellatus) | Israel, marine (RS), cultured | ||||
| SR250195 | Rabbit fish (Siganus rivulatus) | Israel, marine (RS), cultured | A | A | AF456238 | AF456468 |
| DLDKf | European sea bass (D. labrax) | Denmark, marine, cultured | C | CII | AF456240 | AF456470 |
| MC110595e | Grey mullet (Mugil cephalus) | Israel, marine (RS), cultured | ||||
| SA200695e | Gilthead sea bream (Sparus aurata) | Israel, marine (RS), cultured | ||||
| Hybrid 270995g | Hybrid red sea bream (Pagrus major [female] × S. aurata [male]) | Israel, marine (RS), cultured | A | AF456238 | ||
| EA040995g | White grouper (Epinephelus aeneus) | Israel, marine (RS), cultured | A | AF456238 | ||
| SR300397g | Rabbit fish (S. rivulatus) | Israel, marine (RS), wild | A | AF456238 | ||
| SR030597 | Rabbit fish (S. rivulatus) | Israel, marine (RS), wild | A | A | AF456238 | AF456468 |
| EI100398g | Hawksbill sea turtle (Eretmochelys imbricata) | Israel, marine (RS), captive | A | AF456238 | ||
| CC240299 | Koi (Cyprinus carpio) | Israel, freshwater, cultured | B | B | AF456239 | AF456469 |
| EA110499g | White grouper (E. aeneus) | Israel, marine (RS), cultured | A | AF456238 | ||
| BB170200 | Silver perch (Bidyanus bidyanus) | Israel, freshwater, cultured | B | B | AF456239 | AF456469 |
| SV300500 | Lizard fish (Synodus variegatus) | Israel, marine (RS), wild | A | A | AF456238 | AF456468 |
| DL041200 | European sea bass (D. labrax) | Greece, marine (MS), cultured | A | A | AF456238 | AF456468 |
| 185-408h | Golden steaty (Acipenser baeri) | Holland, freshwater | G | G | AF152558j | AF456473 |
| 185-409h | Golden steaty (A. baeri) | Holland, freshwater | G | G | AF152558j | AF456473 |
| S4i | Snakehead (Channa striatus) | Thailand, freshwater | C | CIII | AF251565j | AF456474 |
| S267i | Snakehead (C. striatus) | Thailand, freshwater | C | CIII | AF251565j | AF456474 |
Unless otherwise indicated, isolates were from Israel Oceanographic and Limnological Research Ltd., National Center for Mariculture, Eilat, Israel (the two first letters indicate the initials of the host genus and species; the digits indicate the date of isolation [daymonthyear]).
RS, Red Sea; MS, Mediterranean Sea.
Kindly provided by H. Nousias, Oceanos, Fish Health Center, Preveza, Greece.
Not included in the 16S rRNA and hsp65 gene sequence study.
Kindly provided by I. Dalsgaard, Fish Disease Laboratory, Danish Institute for Fisheries and Marine Research, Frederiksberg, Denmark.
Not included in the hsp65 gene sequence study.
Kindly provided by O. L. M. Haenen and D. Bakker, Laboratory of Fish Diseases, Institute for Animal Science and Health, Lelystad, Holland.
Kindly provided by A. Adams, Institute of Aquaculture, University of Stirling, Stirling, Scotland.
Accession numbers are already available from GenBank.
Bacteriologic analysis.
Mycobacteria were isolated on either Löwenstein-Jensen egg medium or Middlebrook 7H-10 agar medium (Difco) and incubated at 24 ± 0.5°C. Subcultures were made every 3 months by using the same media. Presumptive identification was reached by using standard biochemical tests (16).
DNA extraction.
M. marinum cells from fresh or lyophilized cultures were used. The cells from the cultures were first washed in 10 mM Tris (pH 8.0)-1 mM EDTA (pH 8.0) buffer. The resulting pellet was ground with 300 μl of grinding buffer (100 mM Tris-HCl [pH 9], 100 mM EDTA, 1% sodium dodecyl sulfate). The homogenate was incubated for 30 min at 70°C. Forty-two microliters of 8 M potassium acetate was added, and the mixture was placed on ice for 30 min and then centrifuged for 15 min at 4°C and 12 000 × g. In order to avoid traces of the pellet, the supernatant was transferred to a fresh tube and centrifuged again for 5 min. DNA was precipitated with 1 volume of isopropanol and allowed to stand for 15 min at room temperature. Pelleted DNA was washed in 70% ethanol twice, and the air-dried pellet was dissolved in 50 μl of double-distilled H2O. DNA quality was assessed by electrophoresis in 0.7% agarose and ethidium bromide staining. DNA quantity and purity were estimated with an RNA/DNA calculator (Gene Quant pro; Amersham, Cambridge, England).
PCRs.
PCRs were performed with programmable thermal controllers (PTC-100; MJ Research, San Francisco, Calif.) and a final volume of 50 μl containing 1 U of Taq DNA polymerase (Promega, Madison, Wis.), PCR buffer (Promega), 1.5 mM MgCl2, deoxynucleotide triphosphates each at a final concentration of 0.2 mM, 12.5 pmol of each primer, and 10 to 100 ng of template DNA. Typical cycling parameters were 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 1.5 min of extension at 72°C for 30 cycles. The initial denaturation step was extended to 4 min, and the final extension step was extended to 10 min.
The primers used in this study are listed in Table 2. The primers for the 16S rRNA gene were designed by comparing the 16S rRNA gene sequences of Mycobacterium species with the E. coli 16S rRNA gene sequence (GenBank accession no. J01859). Primers 246, 414, 266, and 1522 were designed by Böddinghaus et al. (2), while primers My1 and 614 were designed by us. The primers for the hsp65 gene were designed on the basis of the sequence of M. tuberculosis published by Shinnick (26). Primers TB11 and TB12 were designed by Telenti et al. (31), primer M2 was designed by Plikaytis et al. (21), and primers HSF1, HS12F, HS4F, and HSR1were designed by us.
TABLE 2.
PCR primers
| Gene | Primer | Positiona | Sequenceb |
|---|---|---|---|
| 16S rRNA | 246c | 1-20 | F 5′-AGA GTT TGA TCC TGG CTC AG-3′ |
| My1 | 61-78 | F 5′-GGA AAG GTC TCT TCG GAG-3′ | |
| 414c | 200-216 | R 5′-CAT CCC ACA CCG CWA AAG-3′ | |
| 266c | 593-612 | R 5′-CAC GCY CAC AGT TAA GCY GT-3′ | |
| 614 | 593-614 | F 5′-CTT AAC TGT GAG CGT GCG-3′ | |
| 1522c | 1503-1522 | R 5′-AAG GAG GTG ATC CAG CCG CA-3′ | |
| hsp65 | HSF1 | 1-21 | F 5′-GAT CCG GAG GAA TCA CTT CGC-3′ |
| TB11d | 167-186 | F 5′-ACC AAC GAT GGT GTG TCC AT-3′ | |
| TB12d | 588-607 | R 5′-CTT GTC GAA CCG CAT ACC CT-3′ | |
| HS12F | 588-607 | F 5′-AGG GTA TGC GGT TCG ACA AG-3′ | |
| HS4F | 1079-1099 | F 5′-ACA GCG ACT CCG ACT ACG ACC-3′ | |
| M2e | 1367-1383 | R 5′-TTG AAG GCG ATC TGC TT-3′ | |
| HSR1 | 1628-1648 | R 5′-TCA GAA ATC CAT GCC ACC CAT-3′ |
According to GenBank accession no. AF456238 for the 16S rRNA gene and accession no. AF456468 for the hsp65 gene.
F, forward, R, reverse. Bold letters in primers sequences represent the following degeneracy: W, A or T; Y, C or T.
Designed by Böddinghaus et al. (2).
Designed by Telenti et al. (31).
Designed by Plikaytis et al. (21).
Restriction mapping.
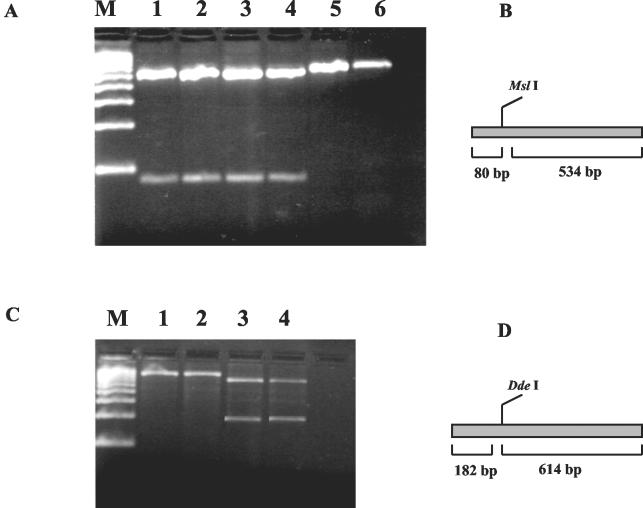
A 614-bp PCR product from the 16S rRNA gene (obtained with primer set 246-266) and a 796-bp PCR product from the hsp65 gene (obtained with primer set HS12F-M2) were purified by using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and digested with restriction enzyme MslI or DdeI (New England Biolabs, Beverly, Mass.), respectively. The digestion was carried out according to the manufacturer's recommendations, except for the MslI reaction, which was extended overnight at room temperature in order to ensure a complete cut. Israeli and foreign isolates were selected as representative of geographically distant regions (see Fig. 3). A 100-bp molecular ruler (Bio-Rad, Hercules, Calif.) was used as a DNA size marker.
FIG. 3.
Enzyme restriction mapping. (A and B) A 614-bp PCR product from the 16S rRNA gene cleaved with restriction enzyme MslI. (A) Gel separation. (B) Restriction map. (C and D) A 796-bp product from the hsp65 gene cleaved with restriction enzyme DdeI. (C) Gel separation. (D) Restriction map. Lanes: M, 100-bp molecular size marker; 1, DL240490; 2, SR030597; 3, CC240299; 4, BB170200; 5, ATCC 927; 6, DLDK.
DNA sequencing.
Double-stranded PCR products were purified by using a QIAquick PCR purification kit. Double-stranded DNA templates (50 ng) were sequenced by using an automated DNA sequencer (Perkin-Elmer model 377) and the Big Dye Deoxy Terminator cycle sequencing enzyme at the DNA Sequencing Biological Services Unit, Weizmann Institute, Rehovot, Israel. Gene-specific primers (Table 2) yielded the complete sequences of both DNA strands and confirmed reading accuracy.
Twenty-one Mycobacterium fish isolates (and the isolate from the sea turtle) were selected for 16S rRNA (1,522 bp) sequencing, and 17 fish isolates were selected for hsp65 (1,648 bp) sequencing.
Phylogenetic analysis.
The sequences of all the isolates used in this study were compared with those of several closely related isolates available in the GenBank database (Table 3).
TABLE 3.
Mycobacterium sp. sequences available in the GenBank database compared to the sequences of our isolates
| Gene study | Isolate | Origin | Clustera | GenBank accession no. |
|---|---|---|---|---|
| 16S rRNA | M. marinum | Unknownb; identical to strain ATCC 927 | C | X52920 |
| M. marinum | Rainbow fish (Melanotaenia praecox); Taiwan | C | AF251565 | |
| Mycobacterium sp. strain M175 | Striped bass (Morone saxatilis); United States | C | AY005147 | |
| M. ulcerans | Human | C | X88926 | |
| M. bovis | Strain BCG | D | M20940 | |
| M. tuberculosis | Human; strain H37Rv | D | Z83862 | |
| M. avium | Human | E | AF306455 | |
| M. avium subsp. paratuberculosis | Strain ATCC 19698 | E | X52934 | |
| M. chlorophenolicum | Freshwater lake sediment | F | X79094 | |
| Mycobacterium sp. strain Fuerth 1999 | Human | G | AF152558 | |
| N. asteroidesc | Strain ATCC 3306 | H | X57949 | |
| hsp65 | M. ulcerans | Human; Australia (isolate 93147172) | CI | AF456475d |
| M. avium subsp. paratuberculosis | Ruminant | E | X74518 | |
| M. avium | Unknownb | E | AF281650 | |
| M. tuberculosis | Human; strain H37Rv | D | AL021932 | |
| M. bovis | Strain BCG P3 | D | M17705 | |
| N. asteroidesc | Strain ATCC 3306 | H | AF352019 |
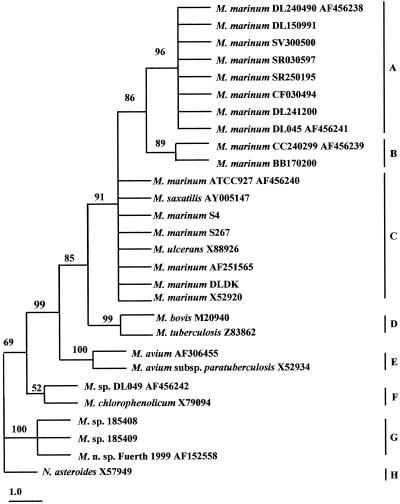
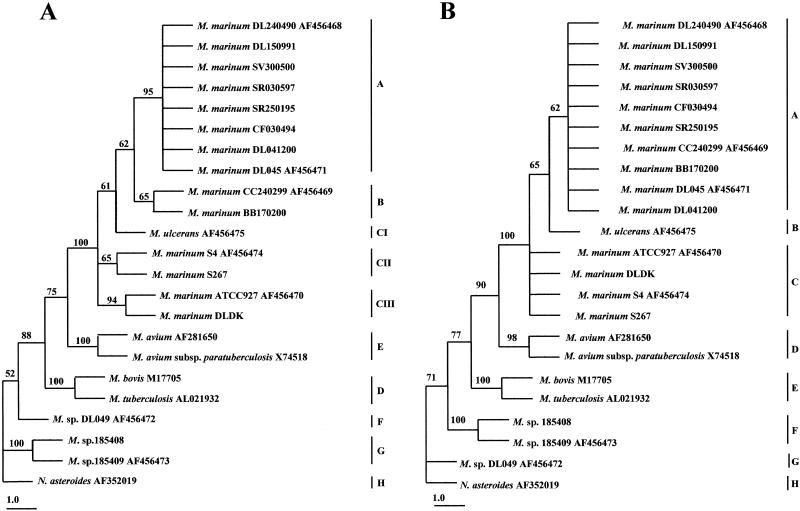
Sequence alignments were obtained by using the ClustalX program (EMBL, Heidelberg, Germany). Phylogenetic trees were constructed by maximum-parsimony analysis with the computer program PAUP 4 (beta7). Bootstrap proportions were used to assess the robustness of the tree with 100 bootstrap replications. Numbers on the branches (Fig. 1 and 2) indicate bootstrap proportions. The Nocardia asteroides 16S rRNA or hsp65 sequences were used as outgroups while constructing the respective phylogenetic trees.
FIG. 1.
Phylogenetic relationships among Mycobacterium species derived from the 16S rRNA gene sequences, as constructed by maximum-parsimony analysis. The tree was rooted by using N. asteroides as the outgroup. Numbers on the branches indicate bootstrap proportions (100 replicates). Bar, 1.0 nucleotide replacement. Available GenBank accession numbers are shown.
FIG. 2.
Phylogenetic relationships among Mycobacterium species derived from the hsp65 gene sequences (A) and the deduced sequences of the amino acids encoded by the Mycobacterium hsp65 gene (B), as constructed by maximum-parsimony analysis. The tree was rooted by using N. asteroides as the outgroup. Numbers on the branches indicate bootstrap proportions (100 replicates). Bar, 1.0 nucleotide or amino acid replacement. Available GenBank accession numbers are shown.
Nucleotide sequence accession numbers.
All the sequences obtained from the 16S rRNA gene and the hsp65 gene were submitted to the GenBank database under the accession numbers listed in Table 1.
RESULTS
16S rRNA phylogenetic analysis.
The phylogenetic tree constructed from the 16S rRNA gene sequences (Fig. 1) revealed eight distinct groups (groups A to H). All the Israeli isolates (marine and freshwater) clustered together in groups A and B and were therefore designated the Israeli type.
M. marinum marine isolates from wild Red Sea fish as well as fish farmed along both the Red Sea and the Mediterranean Sea coasts of Israel all shared an identical nucleotide sequence (group A) different from that of reference strain ATCC 927 (group C) by five bases (99.7% homology). The three Greek isolates from cultured Mediterranean Sea fish were different from each other: the first isolate (DL041200) shared an identical nucleotide sequence with the Israeli marine type, while the second isolate (DL045) was found to differ from it by only one base out of 1,522 bp (99.93% homology). Both clustered in group A. The third Greek isolate (DL049) had only 96.8% homology with the other two isolates and belonged to group F.
The two Israeli isolates from farmed freshwater fish species (koi [Cyprinus carpio] and silver perch [Bidyanus bidyanus]) shared an identical nucleotide sequence (group B), which differed by five bases from the sequence of the Israeli marine isolates in group A (99.7% homology) and by four bases from the sequence of ATCC 927 in group C (99.7% homology).
A sea bass (Dicentrarchus labrax) isolate from Denmark (DLDK), two snakehead (Channa striatus) isolates from Thailand (S4 and S267), one rainbow fish (Melanotaenia praecox) isolate from Taiwan (GenBank accession no. AF251565), one M. marinum isolate from an unspecified source in Germany (GenBank accession no. X52920), and an M. ulcerans isolate from a human infection (GenBank accession no. X88926) all shared high sequence homology to complete identity (99.8 to 100%) with ATCC 927 and clustered in group C.
Group G includes isolates from golden sturgeon (Acipenser baeri) from Holland (isolates 185408 and 185409), which exhibit 100% identity with Mycobacterium sp. strain Fuerth 1999 (GenBank accession no. AF152558), a clinical isolate from a human infection from an unspecified location.
hsp65 phylogenetic analysis.
The tree constructed from the hsp65 gene sequences (Fig. 2A) revealed 10 distinct groups. The marine and freshwater Israeli type isolates (groups A and B) showed the same clustering pattern as in the 16S rRNA tree and differed from each other by four bases (99.7% homology). They differed from strain ATCC 927 by nine bases (99.4% homology) and seven bases (99.6% homology), respectively.
The three Greek isolates showed the same clustering pattern as well: isolates DL041200 and DL045 shared an identical nucleotide sequence with the marine Israeli type isolates and clustered in group A, while the third isolate (DL049) belonged to group F.
In contrast to what was seen with the tree constructed from the 16S rRNA gene sequences, group C was further divided into three subgroups, which included M. ulcerans (subgroup CI) (99.4% homology with ATCC 927), the isolate from sea bass from Denmark (isolate DLDK) and ATCC 927 (subgroup CII) (100% homology), and the isolates from snakehead from Thailand (subgroup CIII) (99.6% homology with ATCC 927). The isolates from golden sturgeon from Holland (isolates 185408 and 185409) clustered in group G.
A comparison of the deduced sequences of the amino acids encoded by the Mycobacterium hsp65 gene is shown in Fig. 2B. All Israeli isolates, marine and freshwater, and two of the Greek isolates clustered in one group (group A). M. ulcerans belonged to group B, while the isolate from sea bass from Denmark (isolate DLDK), snakehead from Thailand (isolates S4 and S267), and ATCC 927 clustered together in group C. The isolates from golden sturgeon from Holland (isolates 185408 and 185409) clustered in group F, while the Greek isolate from sea bass (isolate DL049) belonged to group G.
The sequences of the Israeli type isolates (marine and freshwater) differed from the sequences of all published Mycobacterium spp. in both genes. Consequently, a new name, M. marinum strain Eilaticum DL240490, was proposed for the marine isolates (GenBank accession no. AF456238 for the 16S rRNA gene sequence and AF456468 for the hsp65 gene sequence). The Israeli freshwater isolates were named M. marinum strain Cyprinum CC240299 (GenBank accession no. AF456239 for the 16S rRNA gene sequence and AF456469 for the hsp65 gene sequence).
Restriction enzyme mapping.
Based on the sequence information obtained from both genes, a two-step restriction map was drawn (Fig. 3B and D). MslI digestion of a 614-bp PCR product from the 16S rRNA gene yielded cleavage products of 80 and 534 bp, which separated the Israeli type isolates from the other selected isolates, as they have a unique site for the enzyme (Fig. 3A, lanes 1 to 4). This group was further characterized by DdeI digestion of a 796-bp PCR product from the hsp65 gene, which yielded cleavage products of 182 and 614 bp. Having a unique site for this enzyme, the freshwater isolates could be differentiated from the marine isolates (Fig. 3C).
DISCUSSION
Our results show that the combined analysis of both the 16S rRNA gene and the protein-encoding hsp65 gene of M. marinum increases the detection of intraspecific variations.
Direct sequencing of the 16S rRNA and hsp65 genes confirmed the identities of all the Israeli isolates that were characterized in this study as M. marinum. Phylogenetic analysis based on the 16S rRNA and hsp65 gene sequences clustered the Israeli type Mycobacterium isolates, discriminating this type from all other Mycobacterium isolates found in the literature. Site-specific nucleotide differences revealed that in the last decade, all M. marinum isolates in Israel have belonged to two strains, one that has been infecting marine fish (both cultured and wild) and one that has been infecting freshwater fish (cultured).
Bacterial strains have been considered to be within the same species if they have fewer than 5 to 15 base differences in the 16S rRNA gene (10). For mycobacteria, a difference of less than 5 nucleotides within the complete 16S rRNA gene, along with clear phenotypic differences, is supposed to indicate a genetically unique and distinct taxon (34). Accordingly, M. marinum and members of the M. tuberculosis complex lie on close branches (25, 33). However, M. marinum shares over 99.8% homology with M. ulcerans, with only two nucleotide differences (22, 33). In fact, Tønjum et al. (33) pointed out that the 16S rRNA gene is not useful for discriminating among M. marinum, M. ulcerans, and M. haemophilum. However, the diseases caused by M. marinum and M. ulcerans in humans differ greatly in their clinical, histopathologic, and epidemiologic aspects (3, 29).
The sequences of the hsp65 gene discriminated among M. ulcerans, M. marinum ATCC 927 (reference strain), and M. marinum from the Far East (Thailand; group C; GenBank accession no. AF456474), which could not be distinguished with the 16S rRNA gene sequences. Whole-genome techniques, such as IS2404 restriction fragment length polymorphism analysis and amplified fragment length polymorphism analysis, clustered M. ulcerans and M. marinum into separate groups as well (5).
In this study, the restriction enzyme map based on the combination of the two gene (16S rRNA and hsp65) sequences enabled us to differentiate not only the Israeli type isolates from all other M. marinum isolates but also the Israeli marine isolates from the Israeli freshwater isolates. PCR-restriction fragment length polymorphism analysis of the hsp65 gene (31), although an established discriminating tool used with mycobacteria by many laboratories (4, 8, 24), is not applicable to the M. marinum complex, as different strains have the same restriction sites. This problem was thus overcome with our method.
Our sequence of the 16S rRNA gene of M. marinum ATCC 927 (reference strain) (GenBank accession no. AF456240) is identical to that published by Rogall et al. (25) (GenBank accession no. X52920) and is in contrast (93 to 98% homology) to the partial sequence published by Talaat et al. (30) (GenBank accession no. U92088). Also, the 16S rRNA gene sequence of the isolates from golden sturgeon (A. baeri) from Holland (isolates 185408 and 185409) exhibited 100% identity with that of Mycobacterium sp. strain Fuerth 1999 (GenBank accession no. AF152558), whose sequence was in turn found by Turenne et al. (34) to be identical to that of M. chelonae ATCC 19237.
Because there were no records of M. marinum in the Red Sea before the report by Colorni (7) for sea bass (D. labrax) and because of the active fish trade between Israel and other Mediterranean countries in recent years (in particular, for fingerlings hatched in one region to be reared in another), it had been assumed that the pathogen was introduced into Israel via mariculture operations and had then spread to other farmed and native species in the Gulf of Eilat (A. Diamant and A. Colorni, Abstr. Proc. 7th Int. Conf. Eur. Assoc. Fish Pathol., p. 66, 1995). This hypothesis was supported by the facts that acid-fast bacteria, some of which were specifically identified as M. marinum, had already been detected in D. labrax in Europe (12; W. Verdonck, L. Lambrechts, and F. Ollevier, Abstr. Proc. 2nd Int. Colloq. Pathol. Mar. Aquacult., p. 149, 1986; S. Mellergaard and I. Dalsgaard, Abstr. 6th Int. Conf. Eur. Assoc. Fish Pathol., p. 60, 1993) and that disease in Israel was first detected in offspring of D. labrax which originated in France. While our results show that no linkage exists between the Israeli type and the northern European M. marinum isolates, the hypothesis that Israeli marine M. marinum was nevertheless imported from Europe received support by the identity of one of the Greek isolates to the Israeli marine isolates. On the other hand, the fact that in the last decade, the same strain of M. marinum, different from the freshwater isolates, infected all the Israeli marine fish species as well as a captive hawksbill sea turtle in the Red Sea may indicate that a local, Red Sea strain of M. marinum exists and found extremely susceptible hosts in the exotic species (mainly sea bass) imported for aquaculture. In fact, our results suggest that every isolate of M. marinum examined is endemic, with a unique genotype specific and circumscribed to its geographic region. Interestingly, Portaels et al. (23) and Chemlal et al. (6) have similarly shown for M. ulcerans that the polymorphisms in the 3′ end of the 16S rRNA gene sequence and in the IS2404 restriction fragment length polymorphism pattern, respectively, are related to their geographic origins (Australia, Africa, and Central America). Further characterization of M. marinum isolates from additional geographic regions may clarify whether this is indeed the case.
Acknowledgments
This work was supported by the Israeli Ministry of National Infrastructures (grant 4328-3-96) and Yad haNadiv (grant 5245.12).
REFERENCES
- 1.Austin, B., and D. A. Austin. 1999. Mycobacterium spp., p. 52-54. In L. Laird (ed.), Bacterial fish pathogens: disease of farmed and wild fish, 3rd ed. Springer, London, England.
- 2.Böddinghaus, B., T. Rogall, T. Flohr, H. Blöcker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9:456-458. [DOI] [PubMed] [Google Scholar]
- 4.Brunello, F., M. Ligozzi, E. Cristelli, S. Bonora, E. Tortoli, and R. Fontana. 2001. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 39:2799-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemlal, K., G. Huys, P.-A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chemlal, K., K. de Ridder, P.-A. Fonteyne, W. M. Meyers, J. Swings, and F. Portaels. 2001. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographical areas. Am. J. Trop. Med. Hyg. 64:270-273. [DOI] [PubMed] [Google Scholar]
- 7.Colorni, A. 1992. A systemic mycobacteriosis in the European sea bass Dicentrarchus labrax cultured in Eilat (Red Sea). Isr. J. Aquacult. Bamidgeh 44:75-81. [Google Scholar]
- 8.Devallois, A., K. S. Goh, and N. Rastogi. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 35:2969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamant, A., A. Banet, M. Ucko, A. Colorni, W. Knibb, and H. Kvitt. 2000. Mycobacteriosis in wild rabbitfish Siganus rivulatus associated with cage farming in the Gulf of Eilat, Red Sea. Dis. Aquat. Org. 39:211-219. [DOI] [PubMed] [Google Scholar]
- 10.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 11.Frerichs, G. N. 1993. Acid-fast fish pathogens, p. 217-233. In V. Inglis, R. J. Roberts, and N. R. Bromage (ed.), Bacterial diseases of fish. Blackwell Scientific Publications Ltd., Oxford, England.
- 12.Ghittino, P. 1970. Piscicoltura e ittiopatologia. Riv. Zootec. 2:131-139. [Google Scholar]
- 13.Giavenni, R. 1979. Alcuni aspetti zoonosici delle micobatteriosi di origine ittica. Riv. Ital. Piscicolt. Ittiopatol. 4:123-126. [Google Scholar]
- 14.Gillman, L. M., J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2001. Identification of Mycobacterium species by multiple-fluorescence PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 39:3085-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapur, V., L. L. Li, M. R. Hamrick, B. B. Plikaytis, T. M. Shinnick, A. Telenti, W. R. Jacobs, Jr., A. Banerjee, S. Cole, K. Y. Yuen, J. E. Clarridge III, B. N. Kreiswirth, and J. M. Musser. 1995. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119:131-138. [PubMed] [Google Scholar]
- 16.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology. A guide for the level III laboratory. Centers for Disease Control, Atlanta, Ga.
- 17.Knibb, W., A. Colorni, M. Ankaoua, D. Lindell, A. Diamant, and H. Gordin. 1993. Detection and identification of a pathogenic marine mycobacterium from the European seabass Dicentrarchus labrax using polymerase chain reaction and direct sequencing of 16 rDNA sequences. Mol. Mar. Biol. Biotechnol. 2:225-232. [PubMed] [Google Scholar]
- 18.Lindquist, S. 1986. The heat-shock response. Annu. Rev. Biochem. 55:1151-1191. [DOI] [PubMed] [Google Scholar]
- 19.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 20.Patel, R., J. T. Kvach, and P. Mounts. 1986. Isolation and restriction endonuclease analysis of mycobacterial DNA. J. Gen. Microbiol. 132:541-551. [DOI] [PubMed] [Google Scholar]
- 21.Plikaytis, B. B., B. D. Plikaytis, M. A. Yakrus, W. R. Butler, C. L. Woodley, V. A. Silcox, and T. M. Shinnick. 1992. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 30:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portaels, F., J. Aguiar, K. Fissette, P. A. Fonteyne, H. de Beenhouwer, P. de Rijk, A. Guédénon, R. Lemans, C. Steunou, C. Zinsou, J. M. Dumonceau, and W. M. Meyers. 1997. Direct detection and identification of Mycobacterium ulcerans in clinical specimens by PCR and oligonucleotide-specific capture plate hybridization. J. Clin. Microbiol. 35:1097-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portaels, F., P. A. Fonteyne, H. de Beenhouwer, P. de Rijk, A. Guédénon, J. Hayman, and W. M. Meyers. 1996. Variability in the 3′ end of the 16S rRNA sequence of the species Mycobacterium ulcerans is related to geographic origins of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rastogi, N., K. S. Goh, and M. Berchel. 1999. Species-specific identification of Mycobacterium leprae by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 37:2016-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogall, T., J. Wolters, T. Flohr, and E. C. Böttger. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 26.Shinnick, T. M. 1987. The 65-kilodalton antigen of Mycobacterium tuberculosis. J. Clin. Microbiol. 169:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 28.Steitz, A., A. Feddersen, C. Freytag, S. Daniello, R. E. Schopf, W. O. Böcher, S. Bhakdi, and M. Husmann. 1997. Rapid identification of Mycobacterium marinum by comparative 16S-rRNA-gene analysis in five cases of progredient cutaneous infections. Eur. J. Dermatol. 7:295-299. [Google Scholar]
- 29.Stinear, T. P., G. A. Jenkin, P. D. R. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talaat, A. M., R. Reimschuessel, and M. Trucksis. 1997. Identification of mycobacteria infecting fish to the species level using polymerase chain reaction and restriction enzyme analysis. Vet. Microbiol. 58:229-237. [DOI] [PubMed] [Google Scholar]
- 31.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoen, C. O., and T. A. Schliesser. 1984. Mycobacterial infections in cold-blooded animals, p. 1297-1311. In G. P. Kubica and L. G. Wayne (ed.), The mycobacteria, a sourcebook. Part B. Marcel Dekker, Inc., New York, N.Y.
- 33.Tønjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, J. P. 1998. Bacterial evolution and the nature of species, p. 119-131. In G. R. Carvalho (ed.), Advances in molecular biology, vol. 306. IOS Press, Amsterdam, The Netherlands.