Abstract
Chromogranin A (CgA) is transported restrictedly to secretory granules in neuroendocrine cells. In addition to pH- and Ca2+-dependent aggregation, CgA is known to bind to a number of vesicle matrix proteins. Because the binding-prone property of CgA with secretory proteins may be essential for its targeting to secretory granules, we screened its binding partner proteins using a yeast two-hybrid system. We found that CgA bound to secretogranin III (SgIII) by specific interaction both in vitro and in endocrine cells. Localization analysis showed that CgA and SgIII were coexpressed in pituitary and pancreatic endocrine cell lines, whereas SgIII was not expressed in the adrenal glands and PC12 cells. Immunoelectron microscopy demonstrated that CgA and SgIII were specifically colocalized in large secretory granules in male rat gonadotropes, which possess large-type and small-type granules. An immunocytochemical analysis revealed that deletion of the binding domain (CgA 48–111) for SgIII missorted CgA to the constitutive pathway, whereas deletion of the binding domain (SgIII 214–373) for CgA did not affect the sorting of SgIII to the secretory granules in AtT-20 cells. These findings suggest that CgA localizes with SgIII by specific binding in secretory granules in SgIII-expressing pituitary and pancreatic endocrine cells, whereas other mechanisms are likely to be responsible for CgA localization in secretory granules of SgIII-lacking adrenal chromaffin cells and PC12 cells.
INTRODUCTION
Chromogranin A (CgA) is a granin family protein, which includes chromogranin B (CgB), secretogranin II (SgII), secretogranin III (SgIII), and secretogranin V (7B2) (Huttner et al., 1991; Winkler and Fischer-Colbrie, 1992). They are rich in acidic amino acids, exhibit aggregation at low pH, and possess a high capacity for calcium binding. Because granins are localized restrictedly to secretory granules of neuroendocrine cells, two characteristics of their sorting mechanisms have been studied extensively (Tooze, 1998; Tooze et al., 2001). One is low pH/high calcium–induced aggregation, with which regulated secretory proteins such as prolactin and growth hormone are coaggregated, whereas constitutively secreted proteins such as IgG and albumin are excluded from the aggregate in vitro (Colomer et al., 1996). Thus, granins are thought to be essential for the sorting of secretory proteins at the trans-Golgi network (TGN) (Tooze, 1998).
The second characteristic is an N-terminal disulfide loop of CgA and CgB. In rat pheochromocytoma-derived PC12 cells, mutant CgB lacking the N-terminal disulfide loop is missorted to the constitutive secretory vesicles under conditions in which endogenous protein synthesis was shut down (Krömer et al., 1998). In rat pituitary tumor–derived GH4C1 cells, by contrast, mutated CgA lacking the N-terminal loop is properly sorted to the regulated secretory pathway (Cowley et al., 2000). However, removal of the C-terminal 90 amino acids caused rerouting to the constitutive secretory pathway and impaired the aggregation properties. Surprisingly, in PC12 cells, this mutant CgA is sorted to the regulated secretory pathway. Thus, the N-terminal loop functions differently between CgA and CgB and between PC12 and GH4C1 (Tooze et al., 2001).
Concerning sorting receptors between secretory proteins and granule membranes, it was suggested that the membrane-associated form of carboxypeptidase E (CPE) sorts regulated secretory proteins to secretory granules (Cool et al., 1997). The sorting of proopiomelanocortin (POMC) to secretory granules is impaired in the pituitary intermediate lobe endocrine cells of CPE-deficient mice (Cool et al., 1997). Thus, CPE is thought to act as a sorting receptor for POMC. CPE has been shown to bind to lipid rafts within secretory granules (Dhanvantari and Loh, 2000). Although CgA, like POMC, is sorted to secretory granules, the sorting of CgA was not disturbed in Neuro-2a cells depleting CPE (Normant and Loh, 1998). CgA may use CPE-independent mechanisms for targeting to secretory granules. Furthermore, CgA appears to act as a key molecule not only for the sorting of secretory proteins but also for the biogenesis of secretory granules. The biogenesis of secretory granules was impaired in PC12 cells stably transfected with CgA antisense RNA (Kim et al., 2001). Other secretory proteins, including CgB, CPE, and synaptotagmin, were also diminished in their mRNA expression, whereas transfection of bovine CgA into CgA-deficient cell lines restored secretory granule formation.
Some granins colocalize, whereas others distribute distinctly in the same cells. Colocalization of CgA and CgB was demonstrated in the same secretory granule in rat atrial cardiocytes (Steiner et al., 1990). Rat pituitary male gonadotropes contain large and small types of secretory granules. Large granules contain CgA, whereas small ones contain SgII (Watanabe et al., 1991). Thus, the expression of each granin appears to be tissue-specific; furthermore, the sorting of granins is not always common to each secretory granule-type. Some granins may have a potential to generate their specific granules.
CgA has been shown to bind to the inositol (1,4,5)-triphosphate (IP3) receptor and a number of vesicle matrix proteins (Yoo, 1996, 2000). Binding partner proteins for each granin may participate in the biogenesis of mature secretory granules, because IP3 receptor is essential for secretory function (Yoo, 2000). In seeking CgA binding partners, we considered that the yeast two-hybrid screen is sensitive, although we noted that the intranuclear pH of yeast is neutral, i.e., it is not acidic. Even in the neutral condition, CgA binds weakly to vesicle matrix proteins (Yoo, 1996).
In the present study, we demonstrated that CgA binds to SgIII in secretory granules of endocrine cells. CgA appears to target to secretory granules by specific binding with SgIII in pituitary corticotropes, gonadotropes, and perhaps pancreatic β-cells.
MATERIALS AND METHODS
Yeast Two-Hybrid Screens
Rat CgA cDNA was cloned from a PC12 cDNA library (Iacangelo et al., 1988). For the yeast two-hybrid screens, we used a rat brain cDNA library of postnatal day 8 in pVP16–3 as described previously (Vojtek et al., 1993; Hosaka and Südhof, 1999). A bait vector, pLexN, contained a full-length rat CgA (CgA 1–448) fused to LexA without a putative signal sequence. The rat CgA amino acid sequence is numbered by counting the first methionine as the −18th residue (Iacangelo et al., 1988).
Rat SgIII cDNA was obtained by PCR with rat brain RNA. The sequence was identical to that in the previously reported data (Dopazo et al., 1993). The first methionine of rat SgIII was numbered as 1. Rat granin cDNAs were used in most experiments except where otherwise specified.
Quantitative β-Galactosidase Assays of Yeast Two-Hybrid Interactions
Full-length or partial fragments of rat CgA, rat SgIII, or lamin (as a negative control) were cloned into pLexN or pVP16–3. Bait vector pLexN contained 1) rat CgA 1–448; 2) rat CgA 1–111; 3) rat CgA 112–448; 4) rat CgA 1–54; 5) rat CgA 48–111; 6) mouse CgA 48–115 (Wu et al., 1991); 7) human CgA 48–95 (Konecki et al., 1987); and 8) rat CgA 48–111ΔQ, excluding 20 polyglutamine residues CgA 75–94. Prey vector pVP16–3 contained 1) rat SgIII 187–214; 2) rat SgIII 214–373; 3) rat SgIII 214–277; and 4) rat SgIII 214–373. Yeast strain L40 was cotransfected with bait and prey vectors by use of lithium acetate. The transformants were plated on selection plates lacking uracil, tryptophane, and leucine. After 3 d of incubation at 30°C, colonies were inoculated into the selection medium and incubated for 48 h at 30°C. β-Galactosidase assay was performed on yeast extracts with a protein concentration of 20–40 mg/l per assay (Rose et al., 1990).
Construction of Bacterial Expression Vectors for Recombinant Proteins
SgIII cDNA fragments were placed in the bacterial expression vector pGEX-KG (Guan and Dixon, 1991). SgIII 187–373 was used for coprecipitation experiments, and SgIII 23–186 was used for antibody preparation. Glutathione S-transferase (GST)-fusion proteins were expressed in BL21(DE3) and were purified on glutathione beads.
Antibodies
The antibodies used were mouse monoclonal antibody (mAb) to human LHβ (Immunotech S.A., Marseille, France), mouse mAb to FLAG (Sigma, St. Louis, MO), rat mAb to HA (Roche, Hertfordshire, UK), rabbit polyclonal antiserum to ACTH (Chemicon, Temecula, CA), FITC-labeled antimouse IgG, and Texas Red–labeled anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA). We raised antiserum to CgA (CgA-C#101) against a peptide covering rat CgA 430–442 and antiserum to SgIII (SgIII-N#1) against a GST-rat SgIII 23–186 in rabbits.
Cell Culture
Mouse corticotrope-derived AtT-20 cells and rat insulinoma-derived INS-1 cells (Asfari et al., 1992) were maintained in DMEM supplemented with 10% fetal bovine serum.
Immunoprecipitations
AtT-20 and INS-1 cells grown in 100-mm dishes were solubilized in 1 ml of 50 mM 2-[N-morpholino]ethanesulfonic acid (MES), pH 5.5, containing 0.15 M NaCl, 10 mM CaCl2, 0.1% NP-40, and a protease inhibitor cocktail. After removal of insoluble materials by centrifugation, an aliquot (1 ml) of soluble extracts was incubated for 1 h at 4°C with 5–10 μl of antisera (diluted 1:100–200), after which 30 μl of a 50% (wt/vol) slurry of protein A or G-Sepharose was added, and the mixture was further incubated for 12 h at 4°C under continuous rotation. The immunocomplexes were run on SDS-PAGE, followed by immunoblotting.
In Vitro Binding between CgA and SgIII
For solubilization of the AtT-20 cells and INS-1 cells, we used a buffer containing 0.1 M NaCl, 1% Triton X-100, 2 mM EGTA, and a protease inhibitor cocktail. The buffer was adjusted to pH 5.5 by 50 mM MES or to pH 7.4 by 20 mM HEPES. The high-calcium solution was made by supplementing CaCl2 at a final concentration of 10 mM. Soluble extracts were incubated for 12 h at 4°C with either GST or GST-SgIII 187–373 immobilized on glutathione beads under continuous rotation. The beads were then pelleted by centrifugation. The proteins bound to the GST or GST-SgIII 187–373 were run on SDS-PAGE, followed by immunoblotting.
Northern Blot Analysis
Total RNAs (10 μg) from rat tissues and culture cells were electrophoresed on an agarose gel and then blotted onto a membrane. The blot was hybridized with radiolabeled cDNAs encoding rat CgA 300–448 or rat SgIII 1–471.
Immunoelectron Microscopy
AtT-20 cells grown in 100-mm dishes were fixed first with 0.2% glutaraldehyde–4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2 (10 min, 4°C), and subsequently with 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2) containing 3% sucrose (1 h, 4°C). The cells were gently scraped from dishes and microcentrifuged to pellets, which were dehydrated with 70% ethanol and infiltrated into pure LR White resin (London Resin, Hampshire, UK) for 12 h at 4°C. The pellets were placed in gelatin capsules with fresh LR White resin and polymerized for 24 h at 55°C.
For preparing tissue samples from pituitary glands, three male rats were anesthetized with pentobarbital, perfused with 50 ml of physiological saline, and then perfused with 250 ml of 0.2% glutaraldehyde–4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2. The pituitaries were quickly excised, cut into small pieces, and immersed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2) containing 3% sucrose for 2 h at 4°C. The samples were subjected to 7.5% sucrose replacement and ethanol dehydration, then placed into LR White resin for polymerization.
Immunogold labeling was performed as described previously (Watanabe et al., 1998). Briefly, ultrathin sections from tissue blocks were incubated for 12 h at 4°C with the rabbit first antisera to CgA (diluted 1:500), SgII (diluted 1:4000), or SgIII (diluted 1:500). For double immunostaining, the two-face technique (Bendayan, 1982) was applied. Four colloidal gold particles (5, 10, 15, or 20 nm in diameter) conjugated to goat anti-rabbit IgG (Amersham Pharmacia Biotech) were used. To identify gonadotropes, the sections were simultaneously immunostained with the mouse mAb to LHβ (diluted 1:2000) and colloidal gold particles (5 nm in diameter) conjugated to goat antimouse IgG. The sections were contrasted with saturated aqueous solutions of uranyl acetate and lead citrate and examined with a Hitachi H-7100 electron microscope (Hitachi, Tokyo, Japan).
Expression of CgA or SgIII Constructs
CgA and SgIII constructs fused to FLAG (Stratagene) or HA (Roche) were made in the pcDNA3 (Invitrogen) with a putative signal sequence. The following constructs with FLAG were made: 1) CgA 1–448, 2) CgA 1–112, 3) CgA Δ(41–109) deleting CgA 41–109, 4) CgA Δ(17–38) deleting CgA 17–38, 5) SgIII 1–471, and 6) SgIII Δ(187–373) deleting SgIII 187–373. The following constructs with HA were made: 1) CgA 1–448, 2) CgA Δ(41–109) deleting CgA 41–109, 3) SgIII 1–471, and 4) SgIII Δ(187–373) deleting SgIII 187–373. AtT-20 cells were cultured on eight-well Lab-Tek chamber slides and then expressed constructs with LipofectAMINE 2000 reagents (Invitrogen).
Laser Confocal Microscopy
AtT-20 cells were fixed with 4% paraformaldehyde and then permeabilized with high-salt TPBS (0.01 M sodium phosphate buffer, 0.5 M NaCl, 0.1% Tween 20, pH 7.3) containing 0.1% Triton X-100. The cells were incubated with a mixture of rabbit anti-ACTH (1:500) and mouse monoclonal anti-FLAG (1:2500) antibodies for 18 h at 4°C. For the second antibody reaction, the cells were incubated for 1 h at 20°C with a mixture of FITC-labeled antimouse IgG and Texas Red–labeled anti-rabbit IgG. The coverslips were mounted in 90% glycerol (vol/vol in PBS) containing 0.1% p-phenylenediamine dihydrochloride (Sigma). Stained cells were observed with a laser scanning confocal microscope (Olympus, Tokyo, Japan).
Radiolabeling and Immunoprecipitation
After transfection with the CgA or SgIII constructs, the AtT-20 cells were radiolabeled with 0.2 mCi of [35S] methionine (Amersham Pharmacia Biotech) for 2 h. After radiolabeling, the medium was changed to DMEM with or without 5 mM 8-Br-cAMP for 1 h. The cell extracts and culture medium were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE for fluorography. The 35S signals of blots were recorded in BAS2000 (Fujifilm, Tokyo, Japan). An automatic integration method was used to calculate the intensity of each band. The relative signals were obtained for each cell extract and medium.
RESULTS
Yeast Two-Hybrid Screen Identified SgIII as a Binding Partner for CgA
To identify partner proteins for CgA, we screened a rat brain cDNA library using a yeast two-hybrid system with a bait vector containing the full-length rat CgA. Thirty million transformants were screened, yielding 128 positive clones, of which 43 were positive. The positive clones were retested for interaction with the bait CgA and were then sequenced. Twenty-four of the 43 prey clones encoded SgIII, which was selected frequently as the independent overlapping clones SgIII 187–373 and SgIII 177–471. Another eight clones encoded activating transcription factor 4 (Nehring et al., 2000), and three clones encoded exo70 (Kee et al., 1997). The rest of the sequenced clones could not be identified in the data banks of cDNA. Among the proteins identified, SgIII is a residential protein in the secretory granules of neuroendocrine cells. The latter two are cytosol proteins, which are less likely to encounter CgA at the TGN in vivo. We consider that SgIII is a physiological binding partner for CgA.
Determination of Specific Interaction Domains between CgA and SgIII
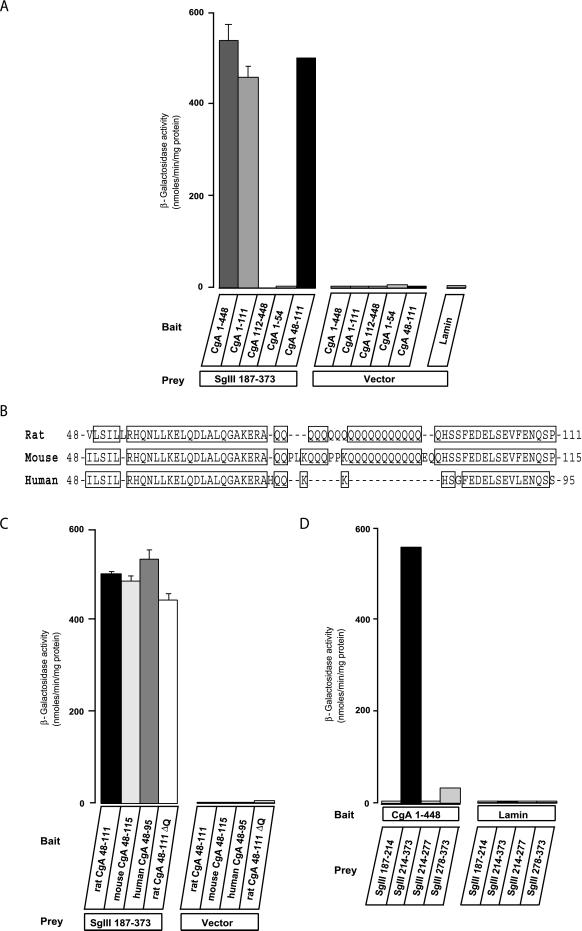
We examined a combination of bait and prey vectors containing various CgA and SgIII regions for their specific interaction. The interaction of SgIII 187–373 with CgA 48–111 was as strong as that with CgA 1–448, whereas CgA 1–54 and CgA 112–448, lacking 48–111, did not bind to the SgIII 187–373 (Figure 1A). This finding suggests that CgA 48–111 constitutes a binding domain for SgIII. Because the rat CgA 48–111 contains a cluster of polyglutamine (polyQ), which exists in rat and mouse CgAs but not in human CgA (Figure 1B) (Winkler and Fischer-Colbrie, 1992), we suspected that polyQ is required for the binding. The SgIII 187–373 efficiently bound to the polyQ-deleting rat CgA 48–111ΔQ and human CgA 48–95, as well as to the polyQ-containing rat CgA 48–111 and mouse CgA 48–115 (Figure 1C). Thus, the polyQ region is not required for binding to SgIII.
Figure 1.
Quantification of the interactions between CgA and SgIII by a yeast two-hybrid assay. (A) Yeast cells cotransfected with bait vectors containing rat CgA 1–448, 1–111, 112–448, 1–54, 48–111, or lamin and prey vectors containing rat SgIII 187–373 or vector only were grown and lysed for β-galactosidase assay. Data are shown in nmol/min per mg protein as the mean ± SE (n = 5). (B) Alignment of putative binding domains to SgIII from the rat, mouse, and human CgA sequences. Human CgA does not have a cluster of polyglutamine. (C) Yeast cells cotransfected with bait vectors containing rat CgA 48–111, mouse CgA 48–115, human CgA 48–95, or rat CgA 48–111ΔQ and prey vectors containing rat SgIII 187–373 or vector only were grown and lysed for β-galactosidase assay. Data are shown in nmol/min per mg protein as the mean ± SE (n = 5). (D) Prey vectors containing rat SgIII 187–214, 214–373, 214–277, or 278–373 and bait vectors containing rat CgA 1–448 or vector only were grown and lysed for β-galactosidase assay. Data are shown in nmol/min per mg protein as the mean ± SE (n = 5).
We next determined the binding domains of SgIII for CgA using the four constructs SgIII 187–214, 214–373, 214–277, and 278–373. Only SgIII 214–373 reacted strongly with the CgA 1–448, whereas the other three did not (Figure 1D). These data suggest that the SgIII 214–373 is required, although its half-split fragments, SgIII 214–277 and 278–373, are insufficient for binding with CgA.
CgA and SgIII Form a Complex in Culture Cell Lines
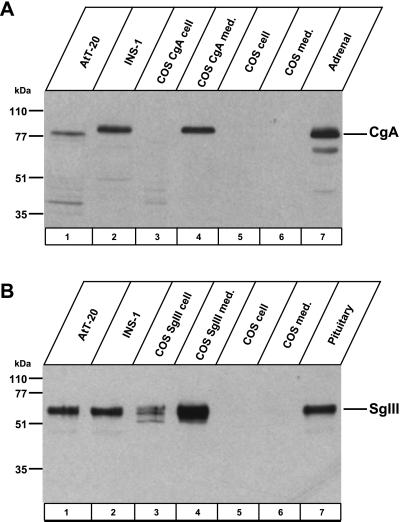
We confirmed the specific binding of CgA and SgIII with a combination of immunoprecipitation and immunoblot analyses in endocrine cell lines, AtT-20 cells, and INS-1 cells. We initially immuno-detected endogenous CgA in AtT-20 cells, INS-1 cells, cell extracts and culture medium from COS-7 cells expressing CgA, and rat adrenal glands at the expected molecular size positions (Figure 2A). Endogenous SgIII was also confirmed in AtT-20 cells, INS-1 cells, cell extracts and culture medium from COS-7 cells expressing SgIII, and rat pituitary gland extracts (Figure 2B). Both CgA and SgIII were extensively released to the culture medium and remained a negligible amount within the granin-expressing COS-7 cells. No bands of both CgA and SgIII appeared in cell extracts and medium from nontransfected COS-7 cells.
Figure 2.
Immunoblot of CgA and SgIII. Cell extracts were from AtT-20 cells, INS-1 cells, COS cells expressing CgA or SgIII, control COS cells, rat adrenal glands, or rat pituitary glands. Culture medium was collected from COS cells expressing CgA or SgIII, and from control COS cells. Samples were immunoblotted with antibody to CgA (A) and to SgIII (B).
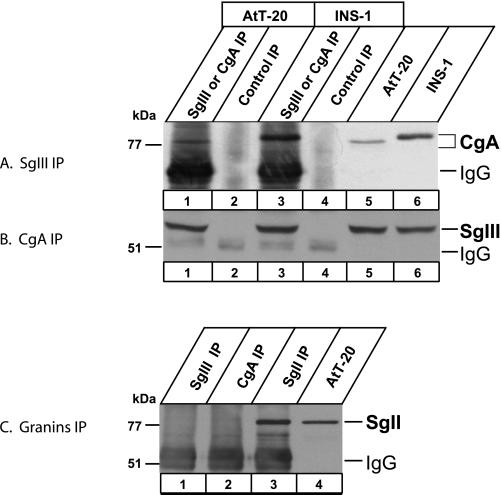
When AtT-20 and INS-1 cell extracts were precipitated with the SgIII-specific antiserum, CgA was recovered in the precipitates and was detected with the CgA-specific antiserum (Figure 3A). A similar pull-down experiment using the CgA-specific antiserum revealed the presence of SgIII in the precipitates (Figure 3B). To prove the specificity of CgA or SgIII immunoblot, we used the SgII-specific antiserum for the similar pull-down experiment. The SgII-specific antiserum pulled down SgII alone without SgIII and CgA (Figure 3C, lane 3); furthermore, neither SgIII- nor CgA-specific antiserum pulled down SgII (Figure 3C, lanes 1 and 2). These findings suggest that CgA forms a complex with SgIII but not with SgII in AtT-20 and INS-1 cells.
Figure 3.
Coimmunoprecipitation of CgA and SgIII from AtT-20 and INS-1 cells. AtT-20 and INS-1 cell lysates were subjected to immunoprecipitations using rabbit antibody to SgIII (A) and to CgA (B) (lanes 1 and 3, respectively). Control immunoprecipitations were performed without the primary antibody (lanes 2 and 4). One-fifth of starting cell lysates (lanes 5 and 6) were used for immunoblotting without immunoprecipitations. Immunoprecipitates (lanes 1–4) were run on an SDS-PAGE for immunoblotting using rabbit antibody to CgA (A) and to SgIII (B). (C) Immunoblot with the antibody to SgII. AtT-20 cell lysates were immunoprecipitated with the antibody to SgIII (lane 1), to CgA (lane 2), and to SgII (lane 3) for immunoblotting with the antibody to SgII. One-fifth of the cell lysate was run on lane 4.
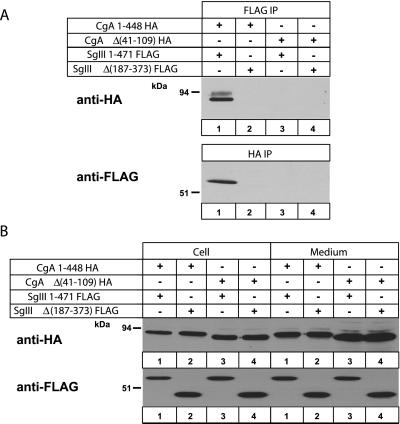
To examine specific binding domains between CgA and SgIII, we made two CgA constructs: CgA 1–448 and Δ(41–109) fused to HA; and two SgIII constructs: SgIII 1–471 and Δ(187–373) fused to FLAG. These constructs were expressed in AtT-20 cells with four combinations: CgA 1–448 and SgIII 1–471 (Figure 4, lane 1), CgA 1–448 and SgIII Δ(187–373) (lane 2), CgA Δ(41–109) and SgIII 1–471 (lane 3), and CgA Δ(41–109) and SgIII Δ(187–373) (lane 4). With anti-FLAG antibody, CgA 1–448-HA was pulled down with SgIII 1–471-FLAG, but CgA Δ(41–109)-HA was not (Figure 4A, top). Likewise, with the anti-HA antibody, SgIII 1–471-FLAG was detected, but not SgIII Δ(187–373)-FLAG (Figure 4A, bottom). Thus, for CgA and SgIII binding, CgA 41–109 and SgIII 187–373 domains appear to be essential. As an input control, Figure 4B shows that all constructs were expressed in AtT-20 cells and secreted to culture media.
Figure 4.
Coimmunoprecipitation of epitope-tagged CgA and SgIII constructs from the AtT-20 cell lysates. (A) AtT-20 cell lysates transfected with combinations of CgA 1–448-HA and SgIII 1–471-FLAG (lane 1), CgA 1–448-HA and SgIII Δ(187–373)-FLAG (lane 2), CgA Δ(41–109)-HA and SgIII 1–471-FLAG (lane 3), or CgA Δ(41–109)-HA and SgIII Δ(187–373)-FLAG (lane 4) were precipitated with mouse mAb to FLAG (A, top) and rat mAb to HA (A, bottom). Immunoprecipitates were run on an SDS-PAGE for immunoblotting with anti-HA (top) and anti-FLAG (bottom) antibodies. (B) One-fifth of the cell lysates and the corresponding media were immunoblotted with HA and FLAG antibodies to confirm their expression in AtT-20 cells.
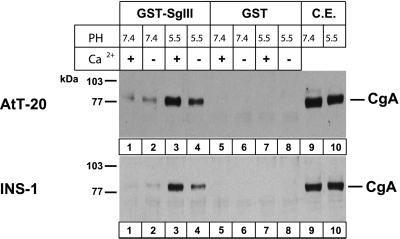
Granin aggregation is induced under low pH/high-calcium conditions that mimic the luminal milieu at the TGN (Chanat and Huttner, 1991; Colomer et al., 1996). To investigate the intensity of CgA binding to GST-SgIII 187–373, the GST-SgIII 187–373 immobilized on the glutathione beads was incubated with AtT-20 or INS-1 cell extracts at pH 5.5 or pH 7.4. SgIII 187–373 bound to CgA at pH 5.5/10 mM Ca2+ (Figure 5, lane 3) and less strongly even without Ca2+ at pH 7.4 in either AtT-20 or INS-1 cell extracts (Figure 5, lane 2). At pH 5.5, SgIII bound to CgA much more strongly with 10 mM Ca2+ than without Ca2+, whereas at pH 7.4 the difference with or without 10 mM Ca2+ was minimal. Thus, CgA binding to SgIII was enhanced by pH 5.5 and 10 mM Ca2+.
Figure 5.
Binding of CgA to SgIII at pH 7.4 or 5.5 with or without 10 mM Ca2+. GST-SgIII (lanes 1–4) and GST (lanes 5–8) were incubated with AtT-20 and INS-1 cell extracts at either pH 7.4 or 5.5 and 10 mM of either Ca2+ (+) or Ca2+ (−). Cell lysates with GST-SgIII (lanes 1–4) or GST alone (lanes 5–8) or one-tenth of cell lysate alone (lanes 9–10) were run on an SDS-PAGE and were immunoblotted with the antibody to CgA.
SgIII and CgA are Colocalized in the Same Secretory Granules
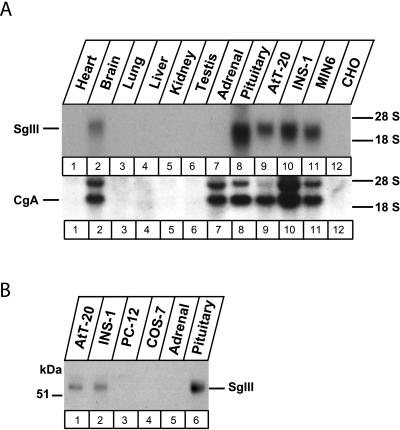
The codistribution of CgA and SgIII was examined using rat tissues and culture cell lines. On Northern blot analyses, they were coexpressed in the brain and pituitary gland but not in nonneuroendocrine tissues, such as heart, lung, liver, kidney, and testis. Their coexpression was also observed in neuroendocrine cell lines, AtT-20, INS-1, and MIN6, but not in CHO cells (Figure 6A). In contrast, only CgA was detected and no SgIII message was detected in adrenal glands (Figure 6A). On Western blot analysis, no SgIII bands were detected in adrenal glands and rat pheochromocytoma-derived PC12 cells (Figure 6B).
Figure 6.
Tissue distribution of SgIII and CgA analyzed by Northern blot (A) and Western blot (B). Total RNA (10 μg) from the indicated tissues and cell lines were run on an agarose gel; this blot was hybridized with a 32P-labeled cDNA probe for SgIII and CgA (A). Cell lysates (25 μg protein) from the indicated tissues and cell lines were run on an SDS-PAGE for immunoblotting using the antibody to SgIII (B).
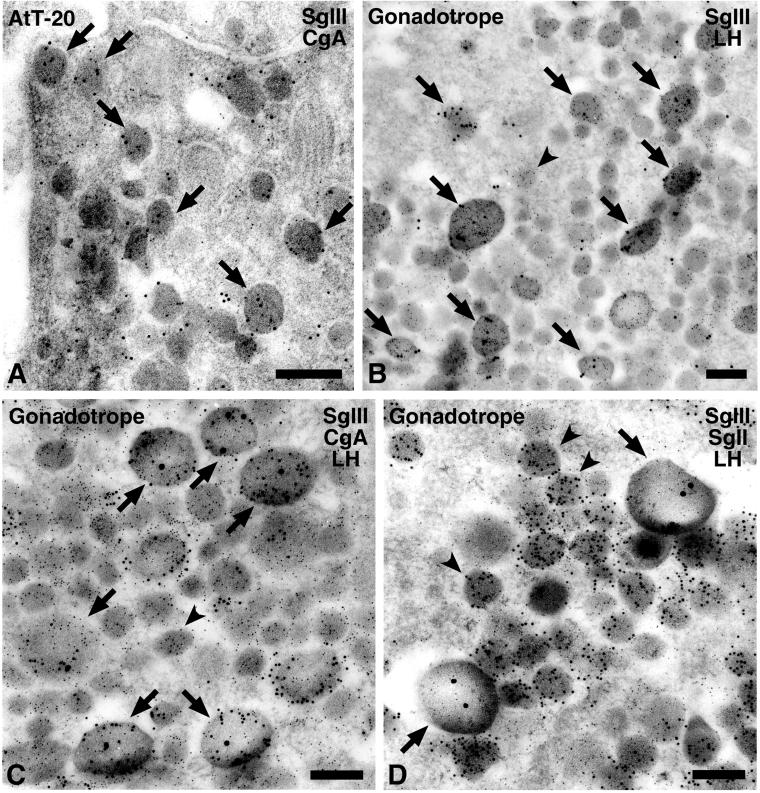
Because CgA and SgIII are coexpressed in the AtT-20, their intracellular localization was examined by immunoelectron microscopy. As shown in Figure 7A, distinct colloidal gold particles for CgA and for SgIII were colocalized on most secretory granules. To further confirm their specific association, we selected the male rat gonadotropes as ideal subjects because they possess two distinct secretory granules: large ones containing CgA and small ones containing SgII (Watanabe et al., 1991). LH (5 nm gold) is contained in the two types of granules (Figure 7, B–D). SgIII was expected to localize with CgA in the large granules. Indeed, SgIII (15 nm gold) was colocalized with LH in large granules (Figure 7B, arrows). Furthermore, SgIII (20 nm gold) was also colocalized with CgA (10 nm gold) along with LH (Figure 7C). However, SgII (10 nm gold) was localized in small granules distinctly from SgIII (20 nm gold) (Figure 7D). These findings suggest that SgIII is specifically associated with CgA in the large secretory granules, and SgII has a distinct characteristic from CgA and SgIII in association with other secretory proteins.
Figure 7.
Immunoelectron microscopy of SgIII and CgA in (A) AtT-20 cells and (B–D) male rat pituitary gonadotropes. (A) Two sizes of colloidal gold particles indicative of CgA (5 nm gold) and SgIII (10 nm gold) were colocalized in most secretory granules in the AtT-20 cell (arrows). (B) Gold particles indicative of SgIII (15 nm gold) were observed restrictedly on the large-type secretory granules (arrows) in the male rat gonadotropes. (C) Gold particles indicative of SgIII (20 nm gold) were localized in the granules labeled with anti-CgA (10 nm gold; arrows) but (D) not in those labeled with anti-SgII (10 nm gold; arrowheads). (B–D) LH was simultaneously labeled with smaller gold particles (5 nm gold) in these ultrathin sections. (B and C) Small granules with LH were indicated by an arrowhead. Bars, 200 nm.
CgA Constructs Lacking the SgIII-binding Domain Could Not Target to Secretory Granules
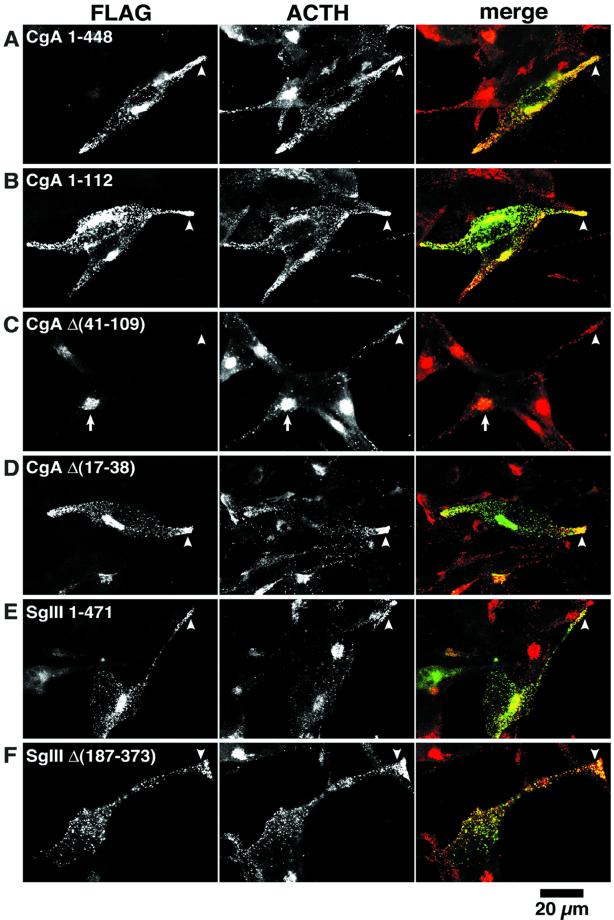
To examine the functional significance of CgA binding to SgIII in their sorting to secretory granules, we made a series of deletion constructs of CgA and SgIII tagged with a FLAG epitope, as shown in Figure 8, left. Their targeting to secretory granules was evaluated in AtT-20 cells with immunocytochemistry and pulse-chase labeling experiments. By immunostaining, CgA 1–448 and CgA 1–112, containing the binding domain for SgIII, were localized densely at the tips of cell processes and a Golgi area near the nucleus (Figure 8, A and B). Both of them were colocalized virtually with ACTH, suggesting that CgA containing the binding domain 41–109 is properly targeted to secretory granules. To test the functional significance of the domain 41–109, we made two additional constructs, CgA Δ(41–109) and CgA Δ(17–38), lacking the disulfide loop. Although these constructs were comparable in length, their intracellular localizations were strikingly distinct: CgA Δ(41–109) was virtually restricted to the Golgi area and failed to target to secretory granules (Figure 8C). In contrast, CgA Δ(17–38) was properly targeted to secretory granules at the tips of cell processes (Figure 8D). These findings suggest that the CgA-binding domain 41–109 for SgIII is indispensable for CgA targeting to secretory granules and that the N-terminal disulfide loop region 17–38 of CgA is not required for its targeting to secretory granules in AtT-20 cells.
Figure 8.
Localization of CgA and SgIII constructs in AtT-20 cells. AtT-20 cells transiently expressing (A) CgA 1–448-FLAG, (B) CgA 1–112-FLAG, (C) CgA Δ(41–109)-FLAG, (D) CgA Δ(17–38)-FLAG, (E) SgIII 1–471-FLAG, and (F) SgIII Δ(187–373)-FLAG were first immunoreacted with mouse anti-FLAG antibody followed by FITC- labeled antimouse IgG (left) and rabbit anti-ACTH antiserum followed by Texas Red–labeled anti-rabbit IgG (middle). Merged images of FLAG (green) and ACTH (red) staining are demonstrated at right. Note that (A–D) CgA constructs with the SgIII-binding domain (CgA 41–109) are punctately stained with ACTH granules at the tip of cell process (arrowhead). In contrast, CgA Δ(41–109) lacking the SgIII-binding domain is restricted to the Golgi area (arrow) (C). Both SgIII 1–471 (E) and SgIII Δ(187–373) (F) are localized in the ACTH granules at the tip of the cell process (arrowheads). Bar, 20 μm.
In contrast to the results of the CgA constructs, the binding domain 187–373 of SgIII for CgA did not affect its targeting to secretory granules; both SgIII 1–471 and SgIII Δ(187–373) were targeted to secretory granules at the tips of cell processes (Figure 8, E and F). These observations suggest that the targeting of SgIII to secretory granules is independent of its binding to CgA in AtT-20 cells. The experiments with use of a FLAG epitope were reproduced by those with HA-epitope tagged granins, and similar results were obtained (our unpublished data).
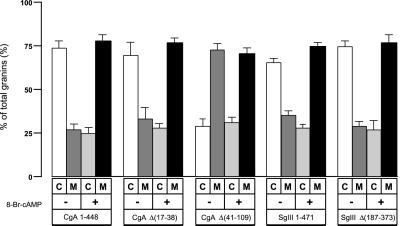
We then confirmed these morphological data by 1 h of pulse labeling with [35S] methionine and 1 h of chasing with or without 8-Br-cAMP, a stimulant of ACTH secretion. As shown in Figure 9, radiolabeled CgA 1–448 and CgA Δ(17–38) were detected two to three times as often in the cell extract as in the culture medium in a nonstimulating state. In an 8-Br-cAMP–stimulating state, their radio content in the cell extract decreased to one-third of that in the culture medium, indicating that it was secreted more to the culture medium. In contrast, ∼2.5-fold more radiolabeled CgA Δ(41–109) was secreted into the culture medium than in the cell extract, even in a nonstimulating state. Thus, it appears that the SgIII-binding domain CgA 41–109 is essential for the retention of CgA inside the cell extract. Together with the immunocytochemical results shown in Figures 8, it is apparent that CgA requires the SgIII-binding domain 41–109 for its targeting to secretory granules in AtT-20 cells. In contrast, both SgIII 1–471 and SgIII Δ(187–373) were detected at the same ratio in the cell extract and in the culture medium in either the nonstimulating or 8-Br-cAMP-stimulating state. Although SgIII 187–373 acts as a binding domain for CgA, we suggest that in AtT-20 cells, SgIII does not need the CgA-binding domain to target to secretory granules.
Figure 9.
Distribution of CgA and SgIII in AtT-20 cells and media. AtT-20 cells were transfected with CgA 1–448, CgA Δ(17–38), CgA Δ(41–109), SgIII 1–471, or SgIII Δ(187–373) tagged with FLAG. The transfected cells were pulse-labeled with [35S] methionine for 1 h. The cells were further chased for 1 h in DMEM with or without 5 mM 8-Br-cAMP. The radiolabeled cell extracts (C) and culture media (M) were immunoprecipitated with the anti-FLAG antibody. The precipitates were run on an SDS-PAGE for fluorography. The radioactivity ratio in the cell extract and culture medium was quantified by BAS 2000 (Fujifilm) detection. All experiments were independently repeated at least four times.
DISCUSSION
Granins are known as residential proteins in secretory granules of neuroendocrine cells. Granins display two properties, aggregation and binding to specific partner proteins exemplified by CgA to IP3 receptor (Yoo, 2000). In the present study, using CgA as a bait, we screened a rat brain cDNA library with a yeast two-hybrid system to find other binding partner proteins with CgA in secretory granules. Although membrane-bound proteins such as IP3 receptor were initially expected, a soluble secretory protein, SgIII, was identified. Biochemical and immunocytochemical analyses clearly demonstrated that SgIII is one of the physiological binding partners of CgA. Consistently, in AtT-20 cells, binding of CgA with SgIII is involved in the targeting of CgA to secretory granules, because deletion of the SgIII-binding domain CgA 41–109 impairs its targeting to secretory granules. In this study, for examining specific binding and colocalization of CgA and SgIII, we used FLAG and HA tags instead of the readily available green fluorescent protein, because green fluorescent protein–tagged protein was shown to localize in secretory granules in neuroendocrine cells in PC12 cells, AtT-20 cells, and INS-1 cells (Molinete et al., 2000; El Meskini et al., 2001).
CgA binds to SgIII via the short primary sequence CgA 48–111 (Figure 1). This sequence contains 20 residues of polyglutamine (CgA 75–94), which turned out not to be essential for binding to SgIII. Thus, the binding determinant was limited to <50 residues on the CgA 48–111. In contrast, SgIII needs a wider primary sequence, SgIII 214–373, for CgA binding, and a half-split of this region (214–277 and 278–373) eliminated the SgIII binding to CgA. The CgA 48–111 does not contain a disulfide loop between Cys17 and Cys38, which is common in CgA from many species, including human, bovine, and rat (Winkler and Fischer-Colbrie, 1992). The N-terminal disulfide loop is thought to be essential for the sorting of CgA and CgB to regulated secretory granules in adrenal chromaffin cells and PC12 cells (Thiele and Huttner, 1998; Yoo and Lewis, 2000). However, in AtT-20 cells, the N-terminal loop–deleted CgA Δ(17–38) was correctly targeted to secretory granules, whereas the N-terminal loop–containing CgA Δ(41–109) did not in the present study (Figure 8). Moreover, when this CgA Δ(41–109) construct tagged with FLAG was expressed in AtT-20 cells, more CgA Δ(41–109) was recovered in the culture medium than in the cell lysate either in a nonstimulating or in an 8-Br-cAMP–stimulating condition (Figure 9). These data indicate that the targeting of CgA Δ(41–109) to the secretory granules was inefficient in AtT-20 cells. However, we could not demonstrate that CgA 41–109 sequence– tagged angiotensinogen, a nongranule secretory protein, did target to secretory granules in AtT-20 cells (our unpublished data). This is in contrast with the data by Glombik et al. (1999), who demonstrated the secretory granule localization of a constitutive secretory protein, α1-antitrypsin, in PC12 cells when α1-antitrypsin was tagged with two N-terminal CgB fragments. In GH4C1 cells, CgA lacking a disulfide loop showed multimeric aggregation properties and normal targeting to secretory granules (Gorr et al., 1999; Cowley et al., 2000). Further study should be performed on whether CgA Δ(41–109) displays aggregation properties like those of disulfide loop–lacking CgA.
SgIII is a product of the mouse 1B1075 gene (Kingsley et al., 1990). SgIII mRNA is expressed in particular subsets of CNS neurons and pituitary cells (Ottiger et al., 1990). Mice missing the 1B1075 gene revealed no major obvious effects on viability, fertility, or locomotor behavior, so that the product of the 1B1075 gene, SgIII, may not be required for their survival (Kingsley et al., 1990). Because SgIII was not required for visible behavioral changes in mice lacking the SgIII gene, it has not been as extensively studied as other granins, such as CgA, CgB, SgII, and 7B2. However, because SgIII is a residential protein in secretory granules, Martens and colleagues discussed its secretory function, using the Xenopus pituitary intermediate lobe. The mRNA levels of both SgIII and POMC increased more than 30-fold in the intermediate pituitary when Xenopus was placed on a black background from a white background, resulting in an increase in melanophore-stimulating hormone for color adaptation (Holthuis and Martens, 1996). The coordinated induction of SgIII and POMC messages and resultant rise in melanophore-stimulating hormone may imply a finely integrated secretory system for biological adaption. In AtT-20 cells, SgIII 214–373 serves as a binding domain for CgA, but its half-split fragments, SgIII 214–277 and 278–373, failed to bind to CgA. Furthermore, the SgIII fragment lacking 187–373 was sorted to secretory granules. Thus, SgIII appears to be stored to secretory granules independently of binding to CgA. Because SgIII may take hold of CgA by specific binding in AtT-20 cells, it is interesting to see how CgA targets to secretory granules in SgIII-deleted endocrine cells. In natural SgIII-lacking cells such as adrenal chromaffin cells and PC12 cells, CgA homodimerization through the N-terminal disulfide loop appears to be essential for its targeting to secretory granules (Thiele and Huttner, 1998).
Currently, two different models of granule protein targeting, “sorting for entry” and “sorting by retention,” are controversial (Arvan and Castle, 1998; Glombik and Gerdes, 2000). In the sorting for entry model, sorting takes place at the TGN, and secretory granule proteins are selected for immature secretory granules (ISGs), which become mature secretory granules (MSGs) without losing cargo proteins. In the sorting by retention model, sorting takes place in the ISGs. ISGs become MSGs after nongranule proteins are removed, resulting in retention of only secretory granule proteins. Although CgA is a residential protein in MSGs, CgA Δ(41–109) did not target to MSGs in AtT-20 cells, and more CgA Δ(41–109) was recovered in the culture medium than in the cell lysate in a nonstimulating condition. Further studies are required in which CgA binds to SgIII and in which CgA Δ(41–109) is removed at the TGN or at the ISGs.
CgA was found to bind to secretory granule membranes via cholesterol-rich regions (Blàzquez et al., 2000). It should be examined whether CgA Δ(41–109) binds to secretory granule membranes like native CgA. CPE was also reported to bind to secretory granule membranes by utilizing cholesterol-sphingolipid–rich lipid rafts (Dhanvantari et al., 2000). Depletion of cholesterol in lipid rafts by a cholesterol biosynthesis inhibitor prevented sorting of POMC and CPE to regulated secretory granules (Dhanvantari et al., 2000). Thus, the sorting to the regulated secretory pathway might require association of regulated secretory proteins with cholesterol-sphingolipid–rich lipid rafts in the TGN. Further studies should be performed to ascertain whether SgIII binds to membrane lipids or to membrane-bound proteins such as IP3 receptor. In pancreatic β-cells, which highly express CgA, SgII, SgIII, and IP3 receptor, CgA oligomers are shown to form a complex with IP3 receptor (Yoo, 2000).
In conclusion, we have demonstrated that two distinct granins, CgA and SgIII, form a heterophilic complex in the secretory granules of AtT-20 and INS-1 cells via a pair of novel domains. Although SgIII properties of targeting to secretory granules require further study, we suggest that CgA targets to secretory granules in association with SgIII in pituitary and pancreatic endocrine cells.
ACKNOWLEDGMENTS
We thank Dr. Claes B. Wollheim for INS-1 cells; Drs. T.C. Südhof, T. Izumi, and K. Nakayama for their helpful discussions; and Y. Ohsawa, M. Suda, M. Hosoi, and K. Tomizawa for their valuable technical support. This work was supported by Grants-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
Abbreviations used:
- POMC
proopiomelanocortin
- CgA
chromogranin A
- CgB
chromogranin B
- CPE
carboxypeptidase E
- ISG
immature secretory granules
- MES
2-[N-morpholino]ethanesulfonic acid
- MSG
mature secretory granules
- polyQ
polyglutamine
- SgII
secretogranin II
- SgIII
secretogranin III
- TGN
trans-Golgi network
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–03–0040. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–03–0040.
REFERENCES
- Arvan P, Castle D. Sorting and storage during secretory granule biogenesis: looking back and looking forward. Biochem J. 1998;332:593–610. doi: 10.1042/bj3320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- Bendayan M. Double immunocytochemical labeling applying the protein A-gold technique. J Histochem Cytochem. 1982;30:81–85. doi: 10.1177/30.1.6172469. [DOI] [PubMed] [Google Scholar]
- Blàzquez M, Thiele C, Huttner WB, Docherty K, Shennan KI. Involvement of the membrane lipid bilayer in sorting prohormone convertase 2 into the regulated secretory pathway. Biochem J. 2000;349:843–852. doi: 10.1042/bj3490843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanat E, Huttner WB. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J Cell Biol. 1991;115:1505–1519. doi: 10.1083/jcb.115.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer V, Kicska GA, Rindler MJ. Secretory granule content proteins and the luminal domains of granule membrane proteins aggregate in vitro at mildly acidic pH. J Biol Chem. 1996;271:48–55. doi: 10.1074/jbc.271.1.48. [DOI] [PubMed] [Google Scholar]
- Cool DR, Normant E, Shen F, Chen HC, Pannell L, Zhang Y, Loh YP. Carboxypeptidase E is a regulated secretory pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpe (fat) mice. Cell. 1997;88:73–83. doi: 10.1016/s0092-8674(00)81860-7. [DOI] [PubMed] [Google Scholar]
- Cowley DJ, Moore YR, Darling DS, Joyce PBM, Gorr S-U. N- and C-terminal domains direct cell type-specific sorting of chromogranin A to secretory granules. J Biol Chem. 2000;275:7743–7748. doi: 10.1074/jbc.275.11.7743. [DOI] [PubMed] [Google Scholar]
- Dhanvantari S, Loh YP. Lipid raft association of carboxypeptidase E is necessary for its function as a regulated secretory pathway sorting receptor. J Biol Chem. 2000;275:29887–29893. doi: 10.1074/jbc.M005364200. [DOI] [PubMed] [Google Scholar]
- Dopazo A, Lovenberg TW, Danielson PE, Ottiger H-P, Sutcliffe JG. Primary structure of mouse secretogranin III and its absence from deficient mice. J Mol Neurosci. 1993;4:225–233. doi: 10.1007/BF02821554. [DOI] [PubMed] [Google Scholar]
- El Meskini R, Jin L, Marx R, Bruzzaniti A, Lee J, Emerson R, Mains R. A signal sequence is sufficient for green fluorescent protein to be routed to regulated secretory granules. Endocrinology. 2001;142:864–873. doi: 10.1210/endo.142.2.7929. [DOI] [PubMed] [Google Scholar]
- Glombik MM, Gerdes H-H. Signal-mediated sorting of neuropeptides and prohormones: secretory granule biogenesis revisited. Biochimie. 2000;82:315–326. doi: 10.1016/s0300-9084(00)00195-4. [DOI] [PubMed] [Google Scholar]
- Glombik MM, Krömer A, Salm T, Huttner WB, Gerdes H-H. The disulfide-bonded loop of chromogranin B mediates membrane binding and directs sorting from the trans-Golgi network to secretory granules. EMBO J. 1999;18:1059–1070. doi: 10.1093/emboj/18.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr S-U, Huang XF, Cowley DJ, Kuliawat R, Arvan P. Disruption of disulfide bonds exhibits differential effects on trafficking of regulated secretory proteins. Am J Physiol. 1999;277:C121–C131. doi: 10.1152/ajpcell.1999.277.1.C121. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Martens GJ. The neuroendocrine proteins secretogranin II and III are regionally conserved and coordinately expressed with proopiomelanocortin in Xenopus intermediate pituitary. J Neurochem. 1996;66:2248–2256. doi: 10.1046/j.1471-4159.1996.66062248.x. [DOI] [PubMed] [Google Scholar]
- Hosaka M, Südhof TC. Homo- and heterodimerization of synapsins. J Biol Chem. 1999;274:16747–16753. doi: 10.1074/jbc.274.24.16747. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Gerdes H-H, Rosa P. The granin (chromogranin/secretogranin) family. Trends Biochem Sci. 1991;16:27–30. doi: 10.1016/0968-0004(91)90012-k. [DOI] [PubMed] [Google Scholar]
- Iacangelo A, Okayama H, Eiden LE. Primary structure of rat chromogranin A and distribution of its mRNA. FEBS Lett. 1988;227:115–121. doi: 10.1016/0014-5793(88)80880-9. [DOI] [PubMed] [Google Scholar]
- Kee Y, Yoo JS, Hazuka CD, Peterson KE, Hsu SC, Scheller RH. Subunit structure of the mammalian exocyst complex. Proc Natl Acad Sci USA. 1997;94:14438–14443. doi: 10.1073/pnas.94.26.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Tao-Cheng JH, Eiden LE, Loh YP. Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell. 2001;106:499–509. doi: 10.1016/s0092-8674(01)00459-7. [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Rinchik EM, Russell LB, Ottiger H-P, Sutcliffe JG, Copeland NG, Jenkins NA. Genetic ablation of a mouse gene expressed specifically in brain. EMBO J. 1990;9:395–399. doi: 10.1002/j.1460-2075.1990.tb08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecki DS, Benedum UM, Gerdes H-H, Huttner WB. The primary structure of human chromogranin A and pancreastatin. J Biol Chem. 1987;262:17026–17030. [PubMed] [Google Scholar]
- Krömer A, Glombik MM, Huttner WB, Gerdes H-H. Essential role of the disulfide-bonded loop of chromogranin B for sorting to secretory granules is revealed by expression of a deletion mutant in the absence of endogenous granin synthesis. J Cell Biol. 1998;140:1331–1346. doi: 10.1083/jcb.140.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinete M, Lilla V, Jain R, Joyce PB, Gorr SU, Ravazzola M, Halban PA. Trafficking of non-regulated secretory proteins in insulin secreting (INS-1) cells. Diabetologia. 2000;43:1157–1164. doi: 10.1007/s001250051507. [DOI] [PubMed] [Google Scholar]
- Nehring RB, Horikawa HPM, El Far O, Kneussel M, Brandstatter JH, Stamm S, Wischmeyer E, Betz H, Karschin A. The metabotropic GABAB receptor directly interacts with the activating transcription factor 4. J Biol Chem. 2000;275:35185–35191. doi: 10.1074/jbc.M002727200. [DOI] [PubMed] [Google Scholar]
- Normant E, Loh YP. Depletion of carboxypeptidase E, a regulated secretory pathway sorting receptor, causes misrouting and constitutive secretion of proinsulin and proenkephalin, but not chromogranin A. Endocrinology. 1998;139:2137–2145. doi: 10.1210/endo.139.4.5951. [DOI] [PubMed] [Google Scholar]
- Ottiger H-P, Battenberg EF, Tsou AP, Bloom FE, Sutcliffe JG. 1B1075: a brain- and pituitary-specific mRNA that encodes a novel chromogranin/secretogranin-like component of intracellular vesicles. J Neurosci. 1990;10:3135–3147. doi: 10.1523/JNEUROSCI.10-09-03135.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Steiner HJ, Weiler R, Ludescher C, Schmid KW, Winkler H. Chromogranins A and B are co-localized with atrial natriuretic peptides in secretory granules of rat heart. J Histochem Cytochem. 1990;38:845–850. doi: 10.1177/38.6.2139887. [DOI] [PubMed] [Google Scholar]
- Thiele C, Huttner WB. The disulfide-bonded loop of chromogranins, which is essential for sorting to secretory granules, mediates homodimerization. J Biol Chem. 1998;273:1223–1231. doi: 10.1074/jbc.273.2.1223. [DOI] [PubMed] [Google Scholar]
- Tooze SA. Biogenesis of secretory granules in the trans-Golgi network of neuroendocrine and endocrine cells. Biochim Biophys Acta. 1998;1404:231–244. doi: 10.1016/S0167-4889(98)00059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Martens GJ, Huttner WB. Secretory granule biogenesis: rafting to the SNARE. Trends Cell Biol. 2001;11:116–122. doi: 10.1016/s0962-8924(00)01907-3. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Banno T, Jeziorowski T, Ohsawa Y, Waguri S, Grube D, Uchiyama Y. Effects of sex steroids on secretory granule formation in gonadotropes of castrated male rats with respect to granin expression. Endocrinology. 1998;139:2765–2773. doi: 10.1210/endo.139.6.6059. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Uchiyama Y, Grube D. Topology of chromogranin A and secretogranin II in the rat anterior pituitary: potential marker proteins for distinct secretory pathways in gonadotrophs. Histochemistry. 1991;96:285–293. doi: 10.1007/BF00271348. [DOI] [PubMed] [Google Scholar]
- Winkler H, Fischer-Colbrie R. The chromogranins A and B: the first 25 years and future perspectives. Neuroscience. 1992;49:497–528. doi: 10.1016/0306-4522(92)90222-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Rozansky DJ, Parmer RJ, Gill BM, O'Connor DT. Structure and function of the chromogranin A gene: clues to evolution and tissue-specific expression. J Biol Chem. 1991;266:13130–13134. [PubMed] [Google Scholar]
- Yoo SH. pH- and Ca2+-dependent aggregation property of secretory vesicle matrix proteins and the potential role of chromogranins A and B in secretory vesicle biogenesis. J Biol Chem. 1996;271:1558–1565. [PubMed] [Google Scholar]
- Yoo SH. Coupling of the IP3 receptor/Ca2+ channel with Ca2+ storage proteins chromogranins A and B in secretory granules. Trends Neurosci. 2000;23:424–428. doi: 10.1016/s0166-2236(00)01621-0. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Lewis MS. Interaction of chromogranin B and the near N-terminal region of chromogranin B with an intraluminal loop peptide of the inositol 1,4,5-trisphosphate receptor. J Biol Chem. 2000;275:30293–30300. doi: 10.1074/jbc.M001204200. [DOI] [PubMed] [Google Scholar]