Abstract
The ability of the pathogenic fungus Candida albicans to switch from a yeast to a hyphal morphology in response to external signals is implicated in its pathogenicity. We used glass DNA microarrays to investigate the transcription profiles of 6333 predicted ORFs in cells undergoing this transition and their responses to changes in temperature and culture medium. We have identified several genes whose transcriptional profiles are similar to those of known virulence factors that are modulated by the switch to hyphal growth caused by addition of serum and a 37°C growth temperature. Time course analysis of this transition identified transcripts that are induced before germ tube initiation and shut off later in the developmental process. A strain deleted for the Efg1p and Cph1p transcription factors is defective in hyphae formation, and its response to serum and increased temperature is almost identical to the response of a wild-type strain grown at 37°C in the absence of serum. Thus Efg1p and Cph1p are needed for the activation of the transcriptional program that is induced by the presence of serum.
INTRODUCTION
Candida albicans is an important pathogen, causing the majority of fungal infections in humans. These can range from relatively minor surface infections, such as thrush and vaginal yeast infections, to more serious and life-threatening systemic infections, particularly in immunocompromised individuals. Cancer chemotherapy, tissue transplantation, and HIV infection are generating a growing pool of individuals susceptible to such systemic infections (Corner and Magee, 1997).
Candida is usually a relatively benign commensal of humans, and the ability to become virulent is thus primarily determined by the immune state of the host (Lortholary and Dupont, 1997; Ashman, 1998). However, there are characteristics of C. albicans that contribute to its ability to cause disease in susceptible individuals. One of these is the ability to switch from a yeast form of growth to a filamentous form characterized either by pseudohyphae or true hyphae. Morphogenesis appears to be important for pathogenesis, because cells that are trapped in either the yeast (Lo et al., 1997; Rocha et al., 2001) or pseudohyphal states (Braun and Johnson, 1997) are less virulent in murine systemic infection models. Thus, the determinants of the morphological yeast-to-hyphal switch appear important for virulence.
By several approaches, genes have been identified that are expressed exclusively or primarily in the hyphal state. Signal transduction cascades, modulated by elements such as cAMP, mitogen-activated protein kinases, and pH-responsive modules, appear to regulate this yeast-to-hyphal transition (Whiteway, 2000). Transcription factors important in the ability to filament have been identified (for recent reviews, see Ernst, 2000; Liu, 2001). However, the draft sequence of the C. albicans genome (Tzung et al., 2001; Scherer, 2002) has now also made the powerful technology of DNA microarrays available to investigate the transcriptional profiles of C. albicans cells. Initial efforts to apply this technology have used filters arrays containing 700 (Lane et al., 2001a) or 2002 genes (Murad et al., 2001a). Glass microarrays have also been used by our group and others to study the response of C. albicans to antifungals (De Backer et al., 2001; Cowen et al., 2002). In the current study, we have investigated the behavior of over 6300 genes under a variety of conditions that include two different stimuli that induce a yeast-to-hyphal switch, the individual effects of serum or increased temperature, and the response of transcription factor mutants that are deficient in hyphal development.
MATERIALS AND METHODS
Strains
We used SC5314 (Gillum et al., 1984) as a wild-type strain. Strains containing the Δefg1 (HLC52) or Δefg1/ΔcphI (HLC54) deletions have already been described (Lo et al., 1997)
Construction of the C. albicans Microarrays
The C. albicans genome has been sequenced using a shotgun approach by the Stanford Genome Technology Center to a 7.5-fold redundancy (Version 4; http://www-sequence.stanford.edu/group/candida). We identified a total of 6580 potential open reading frames (ORFs) greater than 250 base pairs and produced a preliminary annotation. Until the publication of a unified nomenclature, we have used the following priority to name Candida genes: published genes in GenBank, mapping elements as reported by the University of Minnesota (http://alces.med.umn.edu/candida/), orf6.#### reference numbers as reported by the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida/), or our own Contig4-###_### nomenclature (see Supplementary Material). Some novel genes described in this report have been given a common name and deposited in GenBank. A more comprehensive annotation will be provided by a recently established international consortium. For details on the production of the microarrays, please see our web page (http://www.bri.nrc.ca/microarraylab) or the Supplementary Material. Version 5.2 of the array consists of 6333 amplicons printed in duplicates arranged in 48 subarrays (20 × 17 spots) including a row of exogenous control spots. Previous versions contained 55, 70, and 85% of the 6580 ORFs.
Growth Media and Conditions
Cultures were grown in Lee's medium (Lee et al., 1975) or in 1% yeast extract, 2% peptone 2% dextrose (YPD)-based medium. Overnight cultures were inoculated from a fresh colony and were grown in YPD (pH 6.0–6.5) at 30°C. These overnight cultures were diluted to an OD600 of 0.05–0.1 in YPD or YPD + 10% FBS (fetal bovine serum) from Invitrogen (Carlsbad, CA), which has been previously incubated at 56°C for 30 min) and grown at 30 and 37°C, respectively, to an OD600 of 0.6–0.8 (∼3 generations). Cultures grown in Lee's medium containing 10% glucose were started from an overnight culture grown in Lee's medium at 25°C. A 10-ml aliquot of this overnight culture was used to inoculate 1 liter of temperature-adjusted medium. Cultures were grown for ∼3 generations at either 37°C to induce hyphae or 25°C to maintain yeast growth. For the time course analysis, cells were grown in YPD medium overnight to stationary phase, diluted to OD600 0.05 in separate 500-ml flasks containing 250 ml of fresh YPD, and allowed to grow to OD600 0.4 in a 30°C shaker. Half of the flasks were then inoculated with heat-inactivated FBS to a final concentration of 10% and incubated at 37°C for either 30 or 60 min. Cultures were harvested by filtration (0.45-μm filters, cat. no. schvu10re; Millipore, Bedford, MA) and were quick-frozen in an ethanol/dry-ice bath.
Isolation of RNA
Total RNA was extracted with the hot phenol protocol (Kohrer and Domdey, 1991) with the following minor modifications. The cells from a 300-1000-ml culture (OD600 = 0.8) were processed separately in 50-ml tubes and were extracted three times for 10 min. For Lee's medium cultures, glass beads (425–600 μm; Sigma, St. Louis, MO, cat. no. G-8772) were added for the extraction. PolyA(+) mRNA was isolated with the MicroFastTrack 2.0 kit (Invitrogen, cat. no. K1520–03). Quantification was performed by fluorescence with the RiboGreen kit (Molecular Probes, Eugene, OR, cat. no. R-11490) on a CytoFluor 2300 (Millipore).
RNA Labeling
A mixture of 3 μg of polyA(+) mRNA, 1 μl control RNA (2 ng/μl; in vitro transcribed Arabidopsis thaliana G4 gene), 1.5 μl oligo(dT)21 (100 pmol/μl), 3 μl dNTP-minus dCTP (6.67 mM each), 1 μl dCTP (2 mM), 4 μl DTT (100 mM), 8 μl 5× First Strand Buffer (Invitrogen) and water to a volume of 36 μl was denatured at 65°C for 10 min and cooled to room temperature for 5 min. The reverse transcription reaction was done at 42°C for 2 h after addition of 2 μl of cyanine 3-dCTP (1 mM) or cyanine 5-dCTP (1 mM; Perkin Elmer-Cetus/NEN, Boston MA, cat.. no. NEL999) and 2 μl of SuperScript II (Invitrogen, cat. no. 18064–014 : reverse transcriptase, DTT and 5× First Strand Buffer). The reaction was stopped and RNA degraded by addition of 5 μl EDTA (50 mM, pH 8.0), 2 μl NaOH (10 N) and incubation at 70°C for 10 min. The reaction was neutralized with 4 μl of acetic acid (5 M). Purification was done by isopropanol precipitation (1 volume) at −20°C for 1.5 h, followed by centrifugation (1 h at 12,000 rpm). The pellet was washed twice with cold 70% ethanol or, alternatively, with a Qiagen (Valencia, CA) column.
Hybridization
Solutions were made using standard saline-citrate buffer (1× SSC is 0.15 M NaCl and 0.015 M Na-citrate). Slides were prehybridized at 42°C for at least 1 h, with 50 μl of a solution containing 5× SSC, 0.1% SDS, 50× Denhardt's solution (1% Ficoll, 1% BSA, 1% PVP), and 1.5 μl tRNA (10 mg/ml; Baker's yeast, Roche Applied Science, http://biochem.roche.com, cat. no. 109517) and 1.5 μl of denatured genomic DNA (10 mg/ml; herring testes, Invitrogen, cat. no. s0277). The microarray slides were covered with a 24 × 60-mm glass coverslip (Fisher Scientific, Nepean, ON, Canada, cat. no. 12–545m) during all hybridization steps, and the hybridization chamber was kept at high humidity level with wet pieces of paper towels placed in the lower part of the chamber. Just before the hybridization, the DNA microarray slide was washed twice with 0.1× SSC at room temperature for ∼2 min and centrifuged at 800 rpm for 3 min. The DNA microarray slide was kept dry for a minimal amount of time just before hybridization. The hybridization was as follows: the two cDNA targets were resuspended with 10 μl water, pooled together, and mixed with the hybridization buffer to a volume of 50 μl at a final concentration of 25% formamide, 5× SSC, 0.1% SDS containing 1.5 μl tRNA (10 mg/ml) and 1.5 μl denatured genomic DNA (10 mg/ml). This hybridization solution was heat denatured at 95°C for 3 min, cooled to room temperature, and applied onto the DNA microarray slide for overnight hybridization at 42°C. Afterward, slides were completely immersed in a large volume chamber (∼250 ml buffer), and the coverslip was carefully removed before washing for 10 min at 42°C with 1× SSC, 0.2% SDS, and for two times 10 min at 37°C with 0.1× SSC, 0.2% SDS, and, finally, rinsing four times at room temperature in 0.1× SSC for ∼3 min per rinse. Slides were spin-dried (800 rpm, 8 min) and stored protected from light until scanning.
Data Analysis
The DNA microarray slides were scanned with a ScanArray 5000 scanner (GSI Lumonics, then Packard BioScience, now Perkin Elmer-Cetus, Wellesley, CA; version 2.11) at a 10-μm resolution. The resulting 16-bit TIFF files were quantified with QuantArray software (Perkin Elmer-Cetus; versions 2.0 and 3.0). Quality control and normalization of the data were performed in Microsoft Excel using standardized spreadsheets. To be included in the normalization and analysis, each spot had to satisfy three quality control criteria: (1) the signal intensity had to be significantly greater than local background (namely, in one of the two color channels, the signal intensity minus half of the SD had to be greater than the local background plus half of the SD); (2) the signal intensity had to be within the dynamic range of the photomultiplier tube as determined by the user with the help of a scatter plot of the log10 of background-substracted intensities; and (3) the raw intensities of the duplicate spots for each gene had to be within 50% of one another. For spots that met these criteria, the ratio of intensity of the two channels was normalized by the median ratio for the entire subarray consisting of 400 spots that had passed quality control. Finally, the log2 values of the ratios for each duplicate spot were averaged. Statistical analysis and visualization were performed with GeneSpring software (Silicon Genetics, Redwood City, CA). Using the available statistical tools (Student's t test of replicate samples showing a variation different from 1), we selected a list of 742 genes that showed a statistically significant (p < 0.02) variation of at least 1.5-fold under one of 18 studied conditions. Hierarchical clustering (Eisen et al., 1998) of these 742 genes was done in GeneSpring using their standard conditions. K-means clustering (Calinski and Harabasz, 1974) of genes modulated after 30 min, 60 min, or 6 h of treatment with FBS/37°C was also done in GeneSpring but used the Pearson algorithm in which the shape of the expression profiles are more significant than their amplitude.
RESULTS
DNA Microarrays
The C. albicans genome sequence produced by the Stanford Genome Technology Center (release 4.0; http://www-sequence.stanford.edu/group/candida/) was used as the source for the ORFs required for the analysis. The available contigs were scanned by the Magpie sequence analysis software (Gaasterland and Sensen, 1996), and 6580 ORFs larger than 250 base pairs were selected for PCR amplification. Several of the reported experiments were performed while we were developing this technology; thus, hybridizations were performed on slides containing 55, 71, 86, or 91% (6333 ORFs) coverage (see Table 1). Details of the PCR amplification, quality control, and spotting procedures, as well as a preliminary genome annotation are available at our website (http://www.bri.nrc.ca/microarraylab), and figures and the complete dataset are located at http://www.cbr.nrc.ca/genetics/MBC2002/.
Table 1.
Number of replicates per experiment
| Number | Cell line, treatment
|
Presence of elongated cells
|
No. of microarrays (no. of ORFs per array)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Experiment | Reference | Experiment | Reference | 3609 | 4651 | 5668 | 6333 | |
| C | 30°C | 30°C | − | − | 2 | 1 | 6 | |
| 1 | FBS 37°C 30 min | 30°C | − | − | 2 | 4 | ||
| 2 | FBS 37°C 60 min | 30°C | +/− | − | 3 | 4 | ||
| 3 | FBS 37°C 6 h | 30°C | +++ | − | 2 | 4 | 4 | |
| 4 | FBS 37°C 6 h | 25°C 6 h | +++ | − | 2 | 1 | ||
| 5 | FBS 37°C 6 h | FBS 30°C 6 h | +++ | + | 3 | 2 | ||
| 6 | FBS 37°C 6 h | FBS 25°C 6 h | +++ | − | 2 | 1 | ||
| 7 | FBS 37°C 6 h | HLC52 FBS 37°C 6 h | +++ | +/− | 3 | 2 | ||
| 8 | FBS 37°C 6 h | HLC54 FBS 37°C 6 h | +++ | − | 3 | 1 | ||
| 9 | FBS 37°C 6 h | 37°C 6 h | +++ | +/− | 4 | 2 | ||
| 10 | Lee's 37°C 24 h | Lee's 25°C | ++++ | + | 1 | 2 | ||
| 11 | FBS 25°C 6 h | 25°C | − | − | 2 | 1 | ||
| 12 | HLC52, 30°C | 30°C | − | − | 4 | 2 | ||
| 13 | HLC54, 30°C | 30°C | − | − | 3 | 2 | ||
| 14 | FBS 30°C 6 h | 30°C | + | − | 3 | |||
| 15 | HLC52 FBS 37°C | HLC52, 30°C | +/− | − | 4 | |||
| 16 | HLC54 FBS 37°C | HLC54, 30°C | − | − | 4 | |||
| 17 | 37°C | 25°C | +/− | − | 1 | 2 | ||
| 18 | 37°C | 30°C | +/− | − | 2 | 1 | ||
Unless noted, experiments were performed on SC5314 (wt). Other strains used were HLC52 (Δegf1), and HLC54 (Δefg1Δcph1). Culture medium is YPD-based in all experiments except for No. 10. FBS, Addition of 10% fetal bovine serum.
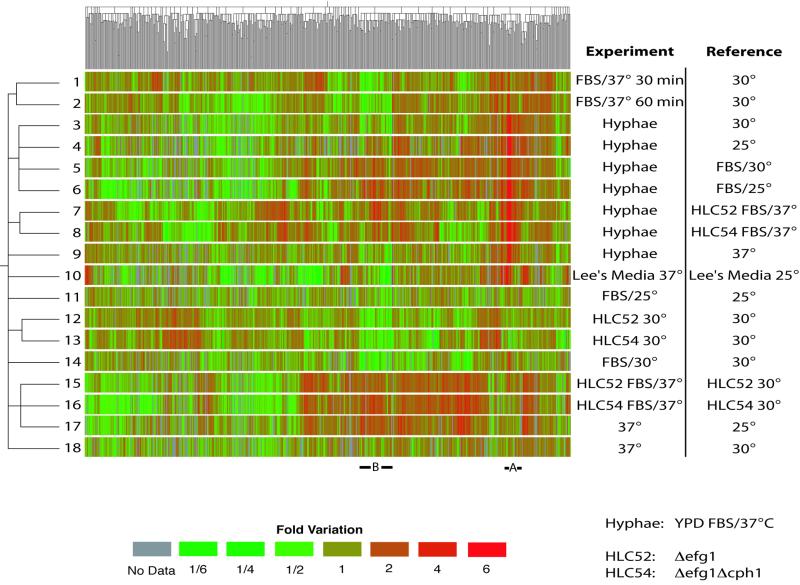
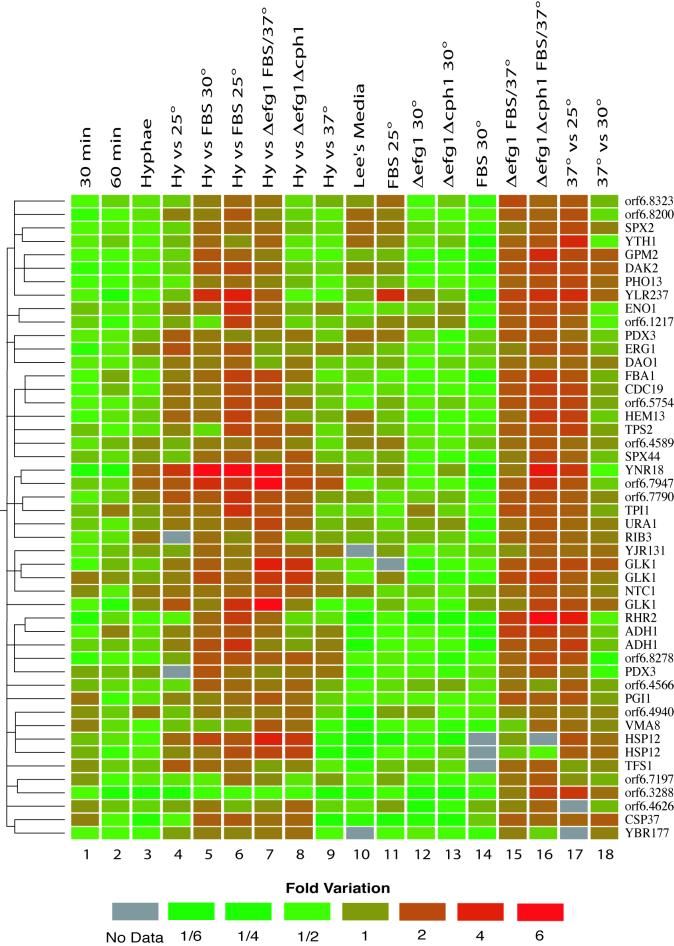
The analyzed data included a significant number of biological and technical replicates (n = 3–10, see Table 1). These include nine control hybridization experiments in which we compared the transcription profiles of independent cultures of Candida cells grown in YPD at 30°C. From 18 additional experimental regimes, we selected 742 ORFs that qualified as “significantly modulated” by passing both a statistical (t test, p < 0.02) and a fold-variation (1.5-fold up or down) cutoff. In the control experiments, only 7 ORFs (0.12%) would have been identified by such stringent criteria. As shown in Figure 1, these results were organized by two-dimensional hierarchical clustering (Eisen et al., 1998). On the X axis, the 742 ORFs were clustered according to the similarity in their expression profiles, with each gene colored according to its change in transcript abundance (downregulated genes in green, upregulated genes in red). In the absence of reliable data, the genes are colored in gray. On the Y axis, the transcriptional responses observed in 18 different experimental regimes were clustered according to the similarities in the resulting transcriptional profiles. A detailed description of each experiment is shown in Table 1. The experiments include an evaluation of the transcriptional changes during a time course of yeast-to-hyphae transition induced by serum and high temperature in YPD (lanes 1–3), an alternative hyphal induction model in Lee's Medium (lane 10), the individual effects of serum (lanes 13 and 14) or high temperature (lanes 17 and 18), a comparison of fully developed hyphae with yeast or elongated cells grown under partially inductive conditions (lanes 3–6 and 9), and the responses of cells missing one or two genes encoding transcription factors (lanes 7, 8, 11, 12, 15, and 16). Horizontal and vertical dendrograms are used to represent the similarity between the expression profiles of each gene and experiment, respectively. For example, in experiments 3 to 6, we compared cells with the hyphal morphology with cells with the yeast morphology. The resulting profiles are thus very similar, and the vertical dendrogram shows that these four experiments form a single subcluster. The same will be true of groups of genes with similar responses to stimuli, two of which (subclusters A and B) will be examined in more detail.
Figure 1.
Two-dimensional clustering of gene expression data. The analysis was performed on 742 genes that showed a statistically significant variation in at least one of 18 experiments (see Table 1). Ratios of gene expression obtained by dividing the experimental by the reference samples are represented as a green-to-red color scale. Similarity between gene expression patterns is represented by the horizontal dendrogram. The vertical dendrogram represents the similarity between experiments. Bars and labels (A and B) represent subclusters examined in more detail in Figures 3 and 5.
Serum and Elevated Temperature Induction of the Yeast-to-Hyphal Switch
The yeast-to-hyphal transition is induced by transfer of C. albicans cells from growth in liquid YPD at 30°C to YPD + 10% serum at 37°C (compare Figure 2, a and d). The transcriptional profile of hyphae formed after 6 h of culturing in serum-containing medium at 37°C was compared with that of yeast control cells. The expression of 18 genes at least doubled upon the formation of hyphae, whereas the expression of an additional 56 genes reproducibly increased by ≥50%. (It should be noted that our use of experimental replicates tends to reduce the amplitude of fold-variations.) In addition, there were 46 genes whose expression was consistently reduced in the hyphal cells compared with the yeast cells. A partial list of these hyphae-modulated genes is presented in Table 2.
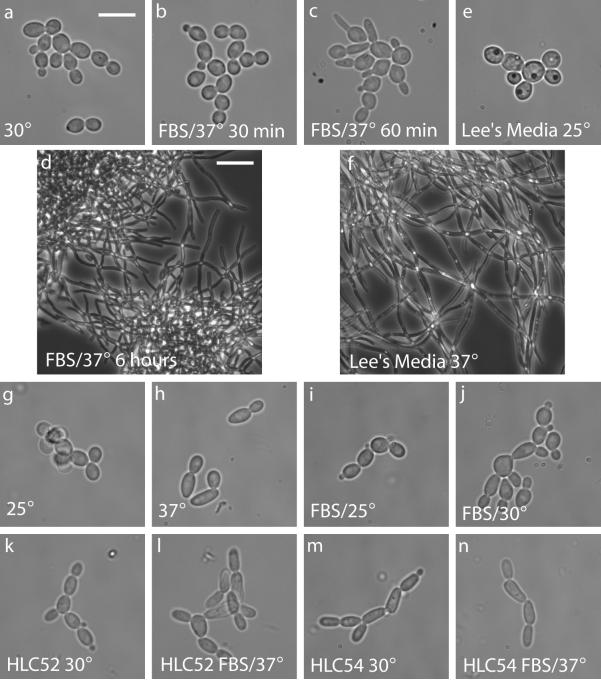
Figure 2.
Morphology of Candida albicans. Yeast phenotype observed in YPD at 30°C (a). Hyphal development after growth at 37°C in YPD + 10% FBS for 30 min (b), 60 min (c), or 6 h (d). In Lee's medium, cells have a yeast-like morphology at 25°C (e) but develop into hyphae by 24 h after an increase in temperature to 37°C (f). Growth in YPD at 25°C (g) is indistinguishable from growth at 30°C, but an increase to 37°C induces cell elongation (h). Addition of 10% FBS in YPD has fewer effects at reduced temperatures of 25°C (i) or 30°C (j). Appearance of the hyphae-defective strains HLC52 (k and l) and HLC54 (m and n) grown in YPD at 30°C (k and m) or under hyphal-inducing conditions at 37°C in YPD + 10% FBS (l and n). The white bars represent a length of either 20 μm (d and f) or 8 μm (a–c, e, and g–n).
Table 2.
Selected genes modulated during the yeast to hyphae transition induced by FBS/37°C
| Gene | Ref. number | Fold | Function |
|---|---|---|---|
| Upregulated genes | |||
| Secreted and cell surface proteins | |||
| ECE1 | orf6.2886 | 23.1 | Unknown |
| SAP5 | orf6.4427 | 9.9 | GPI-anchored aspartic protease |
| HWP1 | orf6.4883 | 6.5 | Cell surface flocculin |
| SAP4 | orf6.3803 | 5.7 | GPI-anchored aspartic protease |
| SAP6 | orf6.3624 | 4.8 | GPI-anchored aspartic protease |
| SOD5b | orf6.7495 | 4.4 | Cu, Zn superoxide dismutase |
| RBT1 | orf6.2929 | 3.1 | Unknown |
| DDR48 | orf6.6854 | 2.9 | Flocculent specific protein |
| PHR1 | orf6.7524 | 2.2 | Cell surface glycoprotein |
| RBT5-like | orf6.6914 | 1.8 | Mycelial surface antigen precursor |
| Small GTPases and cytoskeletal modulators | |||
| YBL060Wa | orf6.6814 | 2.4 | Homology to sec7 domain of GEF |
| YKE2 | 4-2790_0001 | 1.9 | Non-native actin binding complex |
| RDI1a | orf6.9069 | 1.5 | Rho GTPase inhibitor |
| PFY1 | orf6.6300 | 1.5 | Profilin |
| Other functions | |||
| PTP3 | orf6.8958 | 3.2 | Protein tyrosine phosphatase |
| GRE2 | orf6.1740 | 2.0 | Putative reductase |
| SNZ1a | orf6.6669 | 2.0 | Snooze: stationary phase-induced gene family |
| HAL9a | orf6.8931 | 2.0 | Putative zinc finger transcription factor |
| CHA1 | orf6.6363 | 1.9 | l-serine/l-threonine dehydratase |
| SEC24a | orf6.2185 | 1.5 | ER to Golgi transport |
| Unknown functions | |||
| IHD1b | orf6.6198 | 2.8 | Unknown, putative transmembrane |
| YMR90 | orf6.8783 | 2.3 | Conserved hypothetical protein |
| RBT4 | orf6.537 | 2.2 | Similar to plant PR-1 class of pathogen related proteins |
| IHD2b | orf6.3925 | 2.0 | Unknown |
| Downregulated genes | |||
| DNA-binding proteins | |||
| GIS2 | orf6.4479 | 2.1 | Cysteine-rich zinc finger motifs |
| CBF1 | orf6.4385 | 1.7 | Basic helix-loop-helix protein, binds to centromeres |
| YDR174 | orf6.6542 | 1.7 | HMG-class DNA-binding |
| TYE7 | orf6.6049 | 1.6 | Basic helix-loop-helix protein |
| CUP9 | orf6.7646 | 1.6 | Homeodomain |
| Lipid metabolism | |||
| YER73 | orf6.6640 | 2.0 | Aldehyde dehydrogenase |
| YKR70 | orf6.1739 | 1.9 | Phosphatidyl synthase |
| DAK2 | orf6.1906 | 1.8 | Dihydroxyacetone kinase |
| SOU1 | orf6.6126 | 1.7 | Peroxisomal 2,4-dienoyl-CoA reductase |
| PLB2 | orf6.795 | 1.6 | Phospholipase |
| Cell surface proteins | |||
| FLO1a | orf6.3288 | 3.8 | Putative cell wall glycoprotein |
| CSP37 | orf6.2388 | 2.8 | Cell surface virulence factor |
| Other functions | |||
| CHT2 | orf6.2344 | 5.3 | Endochitinase |
| YHB1 | orf6.2156 | 2.1 | Flavohemoglobin |
| RHR2 | orf6.8673 | 1.8 | DL-glycerol-3-phosphatase |
| HSP12 | orf6.2761 | 1.8 | 12 kDa heat shock protein |
| YLR63 | orf6.4669 | 1.8 | Oligopeptide transporter |
| PKC1 | orf6.9136 | 1.7 | Protein kinase C |
| Unknown functions | |||
| RHD1b | orf6.4552 | 5.0 | Unknown |
| RHD2b | orf6.5566 | 2.7 | Unknown, putative transmembrane |
| RHD3b | orf6.8294 | 2.4 | Unknown |
| YER67 | orf6.1218 | 2.1 | Unknown |
Name of closest S. cerevisiae homolog.
New gene names.
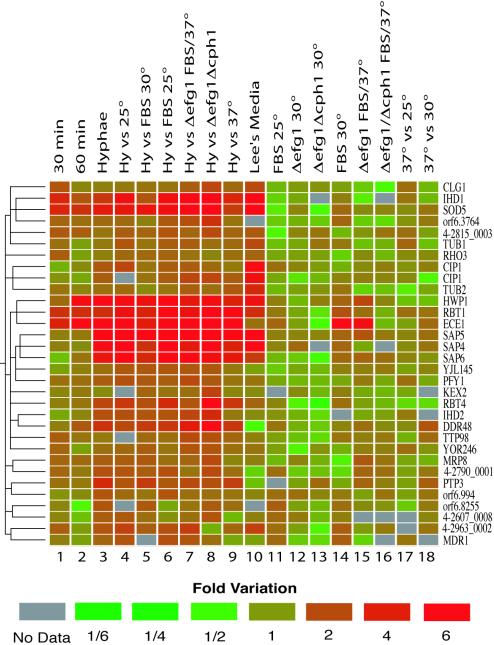
Subcluster A, illustrated in Figure 3, contains a large number of genes that were induced upon hyphal development. This section of the cluster is especially rich in genes that are unique to Candida (see Supplementary Data, Figure 1S). We identified several genes that had been previously recognized as hyphal-specific through independent analyses of differentially induced genes. ECE1 (Birse et al., 1993), SAP4,5,6 (Monod et al., 1994), RBT1 (Braun et al., 2000), and HWP1 (Sharkey et al., 1999) are induced fivefold or more in our microarray analysis. Other previously characterized hyphal-induced genes detected in this analysis include DDR48 (Lane et al., 2001a), PHR1 (Porta et al., 1999), and RBT4 (Braun et al., 2000). Results from some known hyphal-specific genes such as HYR1 (Bailey et al., 1996) and ALS3 (Hoyer et al., 1998) were omitted from the data analysis because of problems with PCR amplification, whereas others (PLD1, RFG1; Hube et al., 2001; Khalaf and Zitomer, 2001) were not detected in our initial search for ORFs in the version 4 assembly.
Figure 3.
Close-up representation of subcluster A (see Figure 1). This section of the two-dimensional clustering is especially enriched in genes that show a significant increase in expression in the hyphal phase (lane 3). Each of these genes is colored according to its change in expression. Downregulated genes are green, whereas upregulated genes are red. See Table 1 or Figure 1 for a detailed description of the experiments in the x-axis.
In addition to the expected genes, the transcription profiling revealed a number of genes whose expression had not previously been identified as being regulated by the yeast-to-hyphal transition. The upregulation of PFY1 (orf6.6300), a Candida homolog of Profilin, as well as the expression of a homolog of the budding yeast RDI1 inhibitor of Rho GTPases (orf6.6469), may reflect a need to modulate actin filament assembly during cell elongation (Pring et al., 1992; DiNubile and Huang, 1997; Pollard et al., 2000; Su et al., 2001). A homolog of the Saccharomyces cerevisiae YBL060W gene (orf6.6814) contains a Sec7 domain and is a putative guanidine nucleotide exchange factor (Sata et al., 1998). As in most other eukaryotes, the regulation of small GTPases is likely to play a role in Candida cell polarization. Other new genes include a previously undescribed superoxide dismutase that was named SOD5 as well as orf6.8958, which encodes a homolog of the S. cerevisiae Ptp3p tyrosine phosphatase (Wurgler-Murphy et al., 1997; Zhan et al., 1997). Increased expression of SEC24, an essential protein in budding yeast that is involved in vesicular transport may be necessary for the rearrangements of cell surface proteins (Pagano et al., 1999). Finally, two uncharacterized ORFs (orf6.6198 and orf6.3925) show significant increases in expression but do not share significant homologies with any other proteins. These were renamed IHD1 and IHD2 (Induced during Hyphae Development). The 392-aa peptide encoded by IHD1 is likely to be a trans-membrane protein because it contains hydrophobic domains at both its N- and C-terminal ends. The region next to the putative transmembrane domain is extremely rich in Ser/Gly residues. IHD1 was recently identified by Murad et al. (2001b) as one of the genes coregulated by the Nrg1p and Tup1p repressors.
A previous attempt at using filter arrays to identify genes that are repressed in hyphae relative to yeast cells had only detected two HSP12 homologues (Lane et al., 2001a). In this study, we have identified 46 ORFs that show a significant reduction in expression. Most of these genes did not cluster together in Figure 1 because they respond differently to individual environmental conditions. Generally, the extent of transcriptional repression was smaller than the levels of induction. In addition to HSP12, our analysis identified the previously reported hyphae-repressed gene CHT2 (McCreath et al., 1995). Csp37p, a cell surface protein whose absence leads to reduced virulence in a mouse model (Sentandreu et al., 1997), has its expression levels consistently reduced about threefold in hyphae compared with yeast cells. Among the most highly repressed genes are three ORFs (orf6.4552, orf6.5566, and orf6.8294) that have no sequence similarity to currently identified genes. These genes were renamed RHD1, RHD2, and RHD3 (Repressed during Hyphae Development). RHD3, like IDH2, is another putative membrane protein although it is rich in alanine and serine residues. Additional genes that are repressed upon induction of hyphal growth include Protein Kinase C (PKC1; orf6.9136; Paravicini et al., 1996), a homolog of the budding yeast FLO1 gene, which encodes a putative cell wall glycoprotein (Teunissen et al., 1993; Watari et al., 1994; Bidard et al., 1995) and RHR2, a DL-glycerol-3-phosphatase that controls glycerol levels (Pahlman et al., 2001). We also noted that five of the repressed transcripts encode enzymes that are directly or indirectly related to lipid metabolism. The list of repressed genes includes several putative transcription factors. These include homologues of the yeast zinc-finger protein Gis2 (Balciunas and Ronne, 1999), the High Mobility Group Protein Ydr174p, the Cup9p homeoprotein (Knight et al., 1994), and the bHLH proteins Tye7p (Nishi et al., 1995) and Cbf1p (Eck et al., 2001). In addition, our most recent arrays include NRG1 (Murad et al., 2001b), which is repressed threefold in the hyphal cells.
Time Course of Gene Induction
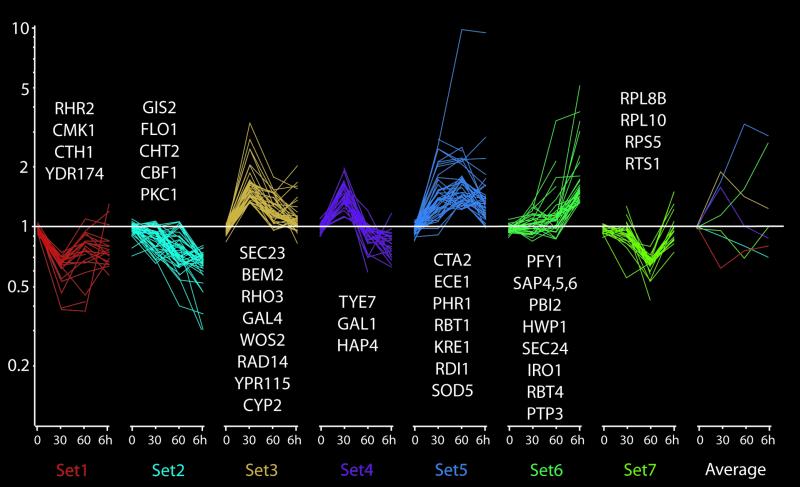
We investigated the timing of the change in gene expression profiles associated with the switch in growth conditions as well as the identity of any genes specifically expressed before germ tube outgrowth and therefore associated with initiation of hyphal growth. After yeast cells were grown at 30°C in YPD and switched to growth at 37°C in the presence of 10% serum, the global transcriptional profile was determined at 30 and 60 min. At 30 min, germ tubes are either absent or initiating, whereas at 60 min a significant number of yeast cells contain germ tubes (Figure 2c). We used K-means clustering to separate, into 7 distinct groups, 232 genes that show significant variation at the 30-min, 60-min, or 6-h time points. This clustering method separates genes according to the shape of their overall expression pattern and allows us to distinguish a variety of expression patterns during hyphal development (Figure 4). Many of the highly expressed genes at the 6-h time point are not strongly induced at the 30- and 60-min points; these include the SAPs, RBT4, HWP1, and PTP3 genes (set 6). In contrast, some genes, such as ECE1, RDI1, RBT1, and the new hyphal genes SOD5 and IHD1, are fully or almost fully induced by 60 min (set 5). Other genes show transient changes in expression. The transcripts of genes classified in sets 3 and 4 accumulate very rapidly but then decrease at the 60-min and 6-h points. These include a basic helix loop helix protein with homology to the S. cerevisiae transcription factor Tye7p and 4 chaperonins encoded by WOS2, RAD14, YPN115, and CYP2. Of note is the transient induction of orf6.7561, a homolog of S. cerevisiae BEM2, a Rho1-GAP protein involved in cell wall maintenance as well as the small GTPase Rho3p, a putative mediator of cell polarity (Wendland and Philippsen, 2001). YDR174 is another gene worthy of note because it shows a constant ∼40% reduction in all time points and encodes a transcription factor of the HMG class. Finally, many of the genes grouped in set 7 are transiently repressed at the 60-min time point encode proteins involved in translation.
Figure 4.
Changes in gene expression during a time course of the yeast-to-hyphae transition induced by FBS/37°C. The 232 genes that show a statistically significant modulation under the studied conditions were separated by K-means clustering according to their expression pattern at t = 30 min, 60 min, or 6 h. Each line represents one gene, and its change in expression, as defined by the y-axis, in the control arrays and the three times points. Lines are colored according to their K-means set. The rightmost graph shows the average change in gene expression for each of the seven K-means sets as defined by its color. Name of representative members of each sets are shown above or under each graph. For a complete list see the supplemental material.
Alternate Hyphal Induction Conditions
Several other conditions have been identified that induce the yeast-to-hyphal transition. One of these include growth in Lee's medium followed by a switch from 25 to 37°C. We found that hyphal induction under these conditions generated a slightly different pattern of gene expression than that found with the serum plus 37°C treatment (Figures 1a and 6a). The majority of the genes strongly induced by the serum regime are also induced in the Lee's medium hyphae. However, the increase in ECE1 transcripts is not as pronounced as in serum-treated cells because yeast cells grown in Lee's medium already express this gene to significant levels. In addition, other genes are induced under the serum regime that are not induced and are even repressed in Lee's medium induction; these include DDR48 and the PTP3 phosphatase. There are also significant differences in the reduction in transcript abundance of RHR2 and CHT2. Finally, hyphae induced in Lee's medium show reduced expression in a set of conserved genes whose products are involved in protein translation possibly as a consequence of the reduced nutrient levels in Lee's medium and the longer growth period (24 vs. 6 h) necessary for hyphae development in this medium. A direct comparison between hyphae induced in YPD and Lee's medium was not done because the changes necessary for adaptation to completely different medium (synthetic vs. complex) are likely to obscure those changes that might be responsible for the differences between hyphal structures.
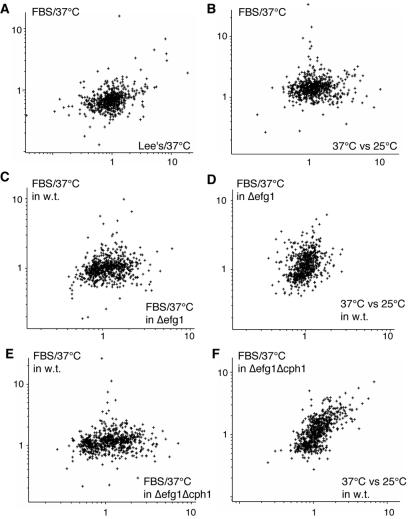
Figure 6.
Comparison of the expression ratios of 742 significantly modulated genes. (A) Scatter plot of changes during the yeast-to-hyphae transition induced by treatment with FBS/37°C compared with a shift to 37°C in Lee's medium. (B) Scatter plot of changes during the yeast-to-hyphae transition induced by treatment with FBS/37°C compared with a 25–37°C temperature shift in YPD. (C) Scatter plot of the gene expression pattern during the response of wild-type or Δefg1 cells to a treatment to FBS/37°C. (D) Comparison of the changes in gene expression during the response of an efg1 mutant to FBS/37°C compared with the changes induced by a 25°C to 37°C shift in wild-type cells. (E) Scatter plot of the gene expression pattern during the response of wild-type or efg1Δcph1 cells to a treatment to FBS/37°C. (F) Comparison of the changes in gene expression during the response of an Δefg1Δcph1 mutant to FBS/37°C compared with the changes induced by a 25°C to 37°C shift in wild-type cells.
Separating Signals from Serum or Increased Temperature
Because the standard hyphal induction regime involves changes in two environmental parameters (temperature shift and the addition of serum), we characterized the transcription profile of cells undergoing either the addition of serum at lower temperatures of 25 or 30°C or the effects of 25–37°C and 30–37°C temperature shifts in the absence of serum. Treatment with FBS or incubation at 37°C alone are usually sufficient to induce some cell elongation (Figure 2, h–j), whereas serum at 25°C has no significant effects on morphology and gene expression (Figures 1, lane 11, and 2i). As shown in Figure 1, the increase in growth temperature from 30 to 37°C has only minor effects on gene expression, whereas the 25–37°C shift causes more pronounced changes (lanes 17 and 18). Although the upregulation of a few genes during the yeast-to-hyphal transition can be attributed to the increased temperature (see below), experiment clustering, and scatter plot analysis (Figures 1, lanes 3 and 17, and 6b) demonstrates that the transcriptional program induced by addition of FBS at 37°C is significantly different from the one initiated by adaptation to growth at 37°C. Two notable exceptions to this are the downregulation of CHT2 and RHD2, both of which are repressed at 37°C in YPD medium. Growth temperature alone or the addition of serum at lower temperatures has very little effect on the induction of hyphal-specific genes with the notable exception of ECE1 and RBT1, which respond well to serum alone (Figure 3). Thus, these experiments allow us to separate changes in gene expression that are caused by environmental conditions from those specific to the yeast-to-hyphal switch. We also compared the expression profiles of FBS/37°C-induced hyphae with cells treated with partially inductive conditions. The profiles measured in these experiments clustered close to the standard FBS/37°C vs. YPD/30°C profile (Figure 1), and the only significant difference was in those genes that are especially sensitive to increased temperature (Figure 5). For example, genes encoding G protein subunits alpha (CAG1) and beta (Contig4–3039_0017) were up- and downregulated, respectively, during the switch to the hyphal form. These changes were maintained when hyphae were compared with cells treated with FBS at 30°C but were lost when hyphae were compared with cells incubated at 37°C. Finally, a 25 to 37°C switch also reproduced this change in Galpha/Gbeta ratio, suggesting that these signaling proteins could be involved in an environmental response. Interestingly, a group of ∼40 genes from subcluster B, illustrated in Figure 5, were repressed only at the 30- and 60-min time points (enriched in Figure 4, K-means sets 1 and 7) as well as with the addition of serum at low temperature. The expression of these genes then appears to increase during the later stages of hyphae development, potentially as a result of the long-term adaptation to growth at 37°C. This time-dependent response to two different environmental signals appears to be controlled by EFG1 and CPH1 as demonstrated below.
Figure 5.
Close-up representation of subcluster B (see Figure 1). This section of the two-dimensional clustering is especially enriched in genes that are inversely modulated by the addition of serum or an increase in growth temperature. Each of these genes is colored according to its change in expression. Downregulated genes are green, whereas upregulated genes are red. See Table 1 for a detailed description of the experiments in the x-axis.
Role of Transcription Factors
Several transcription factors have been implicated in the yeast-to-hyphal transition. Efg1p, a bHLH transcription factor predicted to be the target of a cAMP-dependent kinase signaling pathway (Stoldt et al., 1997; Bockmuhl and Ernst, 2001) has been shown to play a major role in the hyphal transition, because true hyphae fail to form in the absence of the gene, although pseudohyphal formation still occurs (Figure 2l). Deletion of a second transcription factor, CPH1, together with the EFG1 disruption, creates cells that are totally defective in serum-induced hyphal formation (Lo et al., 1997). To date, the consequences of deleting these transcription factors has only been studied on a few target genes.
We examined the transcriptional profiles of cells defective in EFG1 or EFG1/CPH1 under both yeast and hyphal formation conditions. A comparison of mutant and wild-type cells grown under yeast growth conditions showed that the absence of one or both of these transcription factors increases transcripts levels of 30 genes, whereas the expression of 44 genes appears to be reduced (see Supplementary Material). Many of these modulated genes encode proteins classified as transport modulators (HGT1, CTR1, a homolog of budding yeast HST7), whereas a significant number of the repressed genes encode ribosomal proteins or translation initiation factors. Some of the repressed genes (RHR2, HSP12, GLK1, SNO1, ECM4, and GRE2) have been shown, in S. cerevisiae, to be involved in stress response (Gasch et al., 2000). The apparent repression of IRO1 is probably due to the fact that the 3′ end of this gene is missing from the deletion strains used in these studies. In contrast, the comparison of the wild-type and mutant cells under hyphal induction conditions (FBS/37°C), where the mutants produced only yeast cells and pseudohyphae, revealed a very noticeable shift in the response to environmental cues. As seen in the experimental clustering of Figures 1, 3, and 5 and the scatter plots in Figure 6, c–f, most of the hyphal-modulated genes do not respond to FBS/37°C in the Δefg1 mutants, and none of them are activated in the double mutant. It might be assumed that the double knockout would fail to produce changes in gene expression patterns under hyphae-inducing conditions. Instead the transcriptional profiles suggest that Δefg1Δcph1 cells are unable to respond to the presence of serum and show instead a change in global gene expression patterns that mimics the response of wild-type cells that have adapted to growth at 37°C compared with cells grown at 25°C.
Finally, we compared the transcription profile of cells lacking both Cph1p and Efg1p to those lacking only Efg1p. Under the three environmental conditions tested, the transcription factor double mutant had a very similar profile to the single EFG1 disruption (Figure 1). Among hyphal-modulated genes, CPH1 appears to be necessary for the FBS/37°C-dependent modulation of the secreted protein Ece1p, the transcription factor Tye7p, the cell surface protein Hwp1p, the flavohemoglobin Yhb1p and an unknown protein encoded by the orf6.8909 gene. These were fully or significantly modulated by FBS/37°C in the Δefg1 strain but not in the double mutant. Others, like SAP5, were only mildly induced by FBS/37°C in the Δefg1 strain, and the deletion of the Cph1 alleles was necessary to completely abolish the response. There is little significant difference between both deletion strains when grown in YPD at 30°C, which suggests that CPH1 does not have an EFG1-independent role in yeast morphology.
DISCUSSION
C. albicans is an important opportunistic human pathogen and can cause deadly systemic infections of immunocompromised individuals such as HIV-infected patients, tissue transplant recipients, and patients undergoing cancer chemotherapy. This organism has several cellular forms but is distinguished primarily by a dimorphic shift from a yeast-like growth pattern to a hyphal growth pattern. This morphogenesis appears important for the virulence of the organism (Lo et al., 1997) and is regulated in part by transcription factors that are controlled by signaling pathways responding to a variety of extracellular conditions (Brown et al., 2000; Ernst, 2000; Whiteway, 2000; Liu 2001). Molecular genetics can provide powerful tools for the analysis of pathogens. However the diploid nature of C. albicans and the absence of a natural sexual cycle have made classical genetics impossible. The detailed information generated by genome sequencing programs provides the opportunity to apply the tools of the postgenomic era to better understand virulence in this fungal pathogen.
We have examined the transcriptional profiles of ∼6300 ORFs defined in C. albicans from the 4.0 release of the Stanford Candida genome sequencing project (Tzung et al., 2001; Scherer, 2002). Out of 6580 identified Candida ORFs, 4821 (73%) had homologues in the genomes of the fungi S. cerevisiae or Schizosaccharomyces pombe, providing a strong measure of confidence in the ORF designation strategy. Because we used an early assembly of the Candida genome, some genes were spotted in multiple locations on the array. The fact that copies of the same gene typically clustered next to each other (see CIP1 in Figure 3 and GLK1, ADH1, and HSP12 in Figure 5) demonstrates that the large number of replicates used in this study has produced highly consistent data.
We initially chose to study hyphal induction by treatment with serum and a growth temperature of 37°C because these conditions most closely mimic those encountered during a systemic blood infection. We identified a number of genes whose expression is modulated during the switch to hyphal growth independently of the response to serum or temperature alone. Many of these genes were found in a single expression cluster along with previously characterized hyphal and virulence genes such as those encoding secreted aspartyl proteases (SAPs 4 through 6). Some genes in this cluster have no known function and no obvious homologues in other organisms, whereas others encode proteins with predicted functions including a phosphatase, a superoxide dismutase homolog, and a Rho family GTPase inhibitor.
A common characteristic of signaling pathways is the need for their downregulation, and downregulating effectors are often induced by the signaling pathway they regulate (Burchett et al., 1998; Garrison et al., 1999). Induction of a PTPase and a Rho GTPase inhibitor therefore may implicate dual specificity kinases and Rho GTPases in the signaling pathways leading to hyphal induction. The induced superoxide dismutase is a member of a group of 3 Cu/Zn dismutases that are quite distinct from the CaSOD1 gene identified through its strong homology with the S. cerevisiae Cu/Zn protein (Hwang et al., 1999). Intriguingly, the induced gene is closely linked (2 kb) to a second family member that shows no change in transcript abundance in hyphal conditions. Thus there are 4 Cu/Zn superoxide dismutase proteins and 2 Mn proteins identified in C. albicans, and one of the Cu/Zn family members is strongly induced during the yeast-to-hyphal transition. Hyphal formation has been shown to be associated with increased generation of reactive oxygen species (Schmidt and Geschke, 1996).
The global transcriptional profiling approach has also permitted us to identify genes that were repressed in response to the hyphal switch. We observed a reduction in transcripts encoding protein kinase C, the endochitinase Cht2p, several DNA-binding proteins, and enzymes involved in lipid metabolism or glycerol biosynthesis. Deletions of the C. albicans PKC1 genes are necessary for survival in hypo-osmotic medium but had no effect on dimorphism (Ernst, 2000), whereas the Cht2p endochitinase may be involved in cell wall reorganization or in the separation of daughter cells.
A time course of the yeast-to-hyphae transition induced by serum and high temperature has demonstrated that a large number of transcripts change in their abundance. These changes occur before cell elongation becomes apparent. The transient upregulation of homologues of the small GTPases Rho3p and the Rho-GAP protein Bem2p is of interest because these proteins have already been shown to play a role in the determination of cell polarity in fungi (Kim et al., 1994; Cid et al., 1998). We have also identified a large cluster of genes (Figure 5) that are coordinately downregulated in the early stages of the yeast-to-hyphal transition, possibly as part of an Efg1p/Cph1p-mediated response to the presence of serum. The levels of these transcripts then increase in response to an unknown pathway that appears to be modulated by temperature. Examination of the global gene expression profiles of Δefg1Δcph1 cells show them to be incapable of responding to serum, an observation that would have been difficult to substantiate from the study of individual target genes. These results are consistent with a report showing that an activated mutant of the Candida Ras1 gene, which is proposed to regulate Efg1p, can bypass the requirement for serum in hyphal induction (Feng et al., 1999). In serum-dependent hyphal development, the role of the Cph1p transcription appears to be relatively minor, but still significant. Lane et al. (2001b) have shown that Cph1p plays a much more significant role in hyphal development when cells are grown in SS Medium.
In addition to Efg1 and Cph1, many other transcription factors have been shown to modulate the yeast-to-hyphae transition. These include Cph2p, Tec1p, Czf1p, Rim101p, and Tup1p, some of which exhibit changes in their transcript levels. It should be noted that the spot intensity for most of these genes was generally very weak, resulting in poor reproducibility. We have observed the Efg1-dependent repression of Cph2 and Tup1 transcription in hyphae. The gene for Tec1p, another transcription factor that was recently shown to be an effector of Efg1p and Cph2p (Schweizer et al., 2000; lane et al., 2001b), was only recently spotted on our microarray and thus lacks the high number of replicates necessary for statistical significance. We still observed a transient 2.5-fold increase in TEC1 transcripts 30 min after treatment with FBS/37°C (p = 0.06). Another recent addition to our microarrays is the NRG1 gene whose strong downregulation during hyphal development (Murad et al., 2001b) has been confirmed in our very latest experiments. Our results have further identified several more significantly modulated transcription factor genes whose expression patterns may point toward a role in morphogenesis.
In conclusion, this study has revealed a significant number of genes whose transcriptional profiles are similar to those of known virulence factors and markers of the yeast-to-hyphae transition. These results provide new insights into the mechanisms for the initiation and maintenance of filamentous growth. We have also confirmed that the main function of the Efg1p and Cph1p transcription factors is to transmit signals induced by the presence of serum. The molecular roles of all of these genes can now be analyzed further through disruption analysis.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the comments and assistance of past and present members of the Whiteway and Thomas laboratories. C.B. and A.P.B. were supported by grants from the National Science and Engineering Research Council of Canada. We thank the Stanford Genome Technology Center for publishing the Candida sequence data and the Ontario Cancer Institute for advice on the establishment of our microarray facility. This project was funded by the Genome Health Initiative of the National Research Council of Canada and the Canadian Institutes of Health Research grant MOP-42516 to MW. This is NRC Publication number 44834.
Footnotes
Online version of this article contains supplemental data. Online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0272. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0272.
REFERENCES
- Ashman RB. Candida albicans: pathogenesis, immunity and host defense. Res Immunol. 1998;149:281–288. doi: 10.1016/s0923-2494(98)80752-9. ; discussion 494–496. [DOI] [PubMed] [Google Scholar]
- Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Ronne H. Yeast genes GIS1–4: multicopy suppressors of the Gal- phenotype of snf1 mig1 srb8/10/11 cells. Mol Gen Genet. 1999;262:589–599. doi: 10.1007/s004380051121. [DOI] [PubMed] [Google Scholar]
- Bidard F, Bony M, Blondin B, Dequin S, Barre P. The Saccharomyces cerevisiae FLO1 flocculation gene encodes for a cell surface protein. Yeast. 1995;11:809–822. doi: 10.1002/yea.320110903. [DOI] [PubMed] [Google Scholar]
- Birse CE, Irwin MY, Fonzi WA, Sypherd PS. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157:1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Head WS, Wang MX, Johnson AD. Identification, and characterization of TUP1-regulated genes in Candida albicans. Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Brown AJ, et al. Gene regulation during morphogenesis in Candida albicans. Contrib Microbiol. 2000;5:112–25. doi: 10.1159/000060347. [DOI] [PubMed] [Google Scholar]
- Burchett SA, Volk ML, Bannon MJ, Granneman JG. Regulators of G protein signaling: rapid changes in mRNA abundance in response to amphetamine. J Neurochem. 1998;70:2216–2219. doi: 10.1046/j.1471-4159.1998.70052216.x. [DOI] [PubMed] [Google Scholar]
- Calinski T, Harabasz J. A dendrite method for cluster analysis. CommunStat. 1974;3:1–27. [Google Scholar]
- Cid VJ, Cenamor R, Sanchez M, Nombela C. A mutation in the Rho1-GAP-encoding gene BEM2 of Saccharomyces cerevisiae affects morphogenesis and cell wall functionality. Microbiology. 1998;144:25–36. doi: 10.1099/00221287-144-1-25. [DOI] [PubMed] [Google Scholar]
- Corner BE, Magee PT. Candida pathogenesis: unraveling the threads of infection. Curr Biol. 1997;7:R691–R694. doi: 10.1016/s0960-9822(06)00357-5. [DOI] [PubMed] [Google Scholar]
- Cowen L, Nantel A, Whiteway M, Thomas DY, Tessier DC, Kohn LM, Anderson JB. Population Genomics of drug resistance in Candida albicans. Proc Natl Acad Sci USA ( 2002;99:35719–35724. doi: 10.1073/pnas.102291099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer MD, Ilyina T, Ma XJ, Vandoninck S, Luyten WH, Vanden Bossche H. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob Agents Chemother. 2001;45:1660–1670. doi: 10.1128/AAC.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNubile MJ, Huang S. Capping of the barbed ends of actin filaments by a high-affinity profilin-actin complex. Cell Motil Cytoskelet. 1997;37:211–225. doi: 10.1002/(SICI)1097-0169(1997)37:3<211::AID-CM3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Eck R, Stoyan T, Kunkel W. The centromere-binding factor Cbf1p from Candida albicans complements the methionine auxotrophic phenotype of Saccharomyces cerevisiae. Yeast. 2001;18:1047–1052. doi: 10.1002/yea.757. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF. Regulation of dimorphism in Candida albicans. Contrib Microbiol. 2000;5:98–111. doi: 10.1159/000060348. [DOI] [PubMed] [Google Scholar]
- Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaasterland T, Sensen CW. MAGPIE: automated genome interpretation. Trends Genet. 1996;12:76–78. doi: 10.1016/0168-9525(96)81406-5. [DOI] [PubMed] [Google Scholar]
- Garrison TR, Zhang Y, Pausch M, Apanovitch D, Aebersold R, Dohlman HG. Feedback phosphorylation of an RGS protein by MAP kinase in yeast. J Biol Chem. 1999;274:36387–36391. doi: 10.1074/jbc.274.51.36387. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum A, Tsay E, Kirsch D. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Hube B, Hess D, Baker CA, Schaller M, Schafer W, Dolan JW. The role, and relevance of phospholipase D1 during growth, and dimorphism of Candida albicans. Microbiology. 2001;147:879–889. doi: 10.1099/00221287-147-4-879. [DOI] [PubMed] [Google Scholar]
- Hwang CS, Rhie G, Kim ST, Kim YR, Huh WK, Baek YU, Kang SO. Copper- and zinc-containing superoxide dismutase and its gene from Candida albicans. Biochim Biophys Acta. 1999;1427:245–255. doi: 10.1016/s0304-4165(99)00020-3. [DOI] [PubMed] [Google Scholar]
- Khalaf RA, Zitomer RS. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics. 2001;157:1503–1512. doi: 10.1093/genetics/157.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Francisco L, Chen GC, Marcotte E, Chan CS. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J Cell Biol. 1994;127:1381–1394. doi: 10.1083/jcb.127.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Tamai KT, Kosman DJ, Thiele DJ. Identification and analysis of a Saccharomyces cerevisiae copper homeostasis gene encoding a homeodomain protein. Mol Cell Biol. 1994;14:7792–7804. doi: 10.1128/mcb.14.12.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Lane S, Birse C, Zhou S, Matson R, Liu H. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J Biol Chem. 2001a;276:48988–48996. doi: 10.1074/jbc.M104484200. [DOI] [PubMed] [Google Scholar]
- Lane S, Zhou S, Pan T, Dai Q, Liu H. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol Cell Biol. 2001b;21:6418–6428. doi: 10.1128/MCB.21.19.6418-6428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Rega M, Watson R, Campbell C. An amino acid liquid medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Liu H. Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol. 2001;4:728–735. doi: 10.1016/s1369-5274(01)00275-2. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lortholary O, Dupont B. Antifungal prophylaxis during neutropenia and immunodeficiency. Clin Microbiol Rev. 1997;10:477–504. doi: 10.1128/cmr.10.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreath KJ, Specht CA, Robbins PW. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc Natl Acad Sci USA. 1995;92:2544–2548. doi: 10.1073/pnas.92.7.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod M, Togni G, Hube B, Sanglard D. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 1994;13:357–368. doi: 10.1111/j.1365-2958.1994.tb00429.x. [DOI] [PubMed] [Google Scholar]
- Murad AM, et al. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1, and CaNrg1. Mol Microbiol. 2001a;42:981–993. doi: 10.1046/j.1365-2958.2001.02713.x. [DOI] [PubMed] [Google Scholar]
- Murad AM, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001b;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Park CS, Pepper AE, Eichinger G, Innis MA, Holland MJ. The GCR1 requirement for yeast glycolytic gene expression is suppressed by dominant mutations in the SGC1 gene, which encodes a novel basic- helix-loop-helix protein. Mol Cell Biol. 1995;15:2646–2653. doi: 10.1128/mcb.15.5.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano A, Letourneur F, Garcia-Estefania D, Carpentier JL, Orci L, Paccaud JP. Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem. 1999;274:7833–7840. doi: 10.1074/jbc.274.12.7833. [DOI] [PubMed] [Google Scholar]
- Pahlman AK, Granath K, Ansell R, Hohmann S, Adler L. The yeast glycerol 3-phosphatases Gpp1p, and Gpp2p are required for glycerol biosynthesis, and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J Biol Chem. 2001;276:3555–3563. doi: 10.1074/jbc.M007164200. [DOI] [PubMed] [Google Scholar]
- Paravicini G, Mendoza A, Antonsson B, Cooper M, Losberger C, Payton MA. The Candida albicans PKC1 gene encodes a protein kinase C homolog necessary for cellular integrity but not dimorphism. Yeast. 1996;12:741–756. doi: 10.1002/(sici)1097-0061(19960630)12:8<741::aid-yea967>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Porta A, Ramon AM, Fonzi WA. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol. 1999;181:7516–7523. doi: 10.1128/jb.181.24.7516-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring M, Weber A, Bubb MR. Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry. 1992;31:1827–1836. doi: 10.1021/bi00121a035. [DOI] [PubMed] [Google Scholar]
- Rocha CRC, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth, and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata M, Donaldson JG, Moss J, Vaughan M. Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc Natl Acad Sci USA. 1998;95:4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S. Gene discovery, and comparative genomics. Progress and prospects. In: Calderone RA, editor. Candida and Candidiasis. Washington, DC: ASM Press; 2002. pp. 259–265. [Google Scholar]
- Schmidt A, Geschke U. Comparative virulence of Candida albicans strains in CFW1 mice and Sprague-Dawley rats. Mycoses. 1996;39:157–160. doi: 10.1111/j.1439-0507.1996.tb00119.x. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Rupp S, Taylor BN, Rollinghoff M, Schroppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development, and virulence in Candida albicans. Mol Microbiol. 2000;38:435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- Sentandreu M, Nieto A, Iborra A, Elorza MV, Ponton J, Fonzi WA, Sentandreu R. Cloning and characterization of CSP37, a novel gene encoding a putative membrane protein of Candida albicans. J Bacteriol. 1997;179:4654–4663. doi: 10.1128/jb.179.15.4654-4663.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey LL, McNemar MD, Saporito-Irwin SM, Sypherd PS, Fonzi WA. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999;181:5273–5279. doi: 10.1128/jb.181.17.5273-5279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Knoblauch R, Garabedian MJ. Rho GTPases as modulators of the estrogen receptor transcriptional response. J Biol Chem. 2001;276:3231–3237. doi: 10.1074/jbc.M005547200. [DOI] [PubMed] [Google Scholar]
- Teunissen AW, Holub E, van der Hucht J, van den Berg JA, Steensma HY. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast. 1993;9:423–427. doi: 10.1002/yea.320090413. [DOI] [PubMed] [Google Scholar]
- Tzung KW, et al. Genomic evidence for a complete sexual cycle in Candida albicans. Proc Natl Acad Sci USA. 2001;98:3249–3253. doi: 10.1073/pnas.061628798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari J, et al. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast. 1994;10:211–225. doi: 10.1002/yea.320100208. [DOI] [PubMed] [Google Scholar]
- Wendland J, Philippsen P. Cell polarity, and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics. 2001;157:601–610. doi: 10.1093/genetics/157.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M. Transcriptional control of cell type, and morphogenesis in Candida albicans. Curr Opin Microbiol. 2000;3:582–588. doi: 10.1016/s1369-5274(00)00144-2. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Deschenes RJ, Guan KL. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.