Abstract
We used immunofluorescence to study the distribution and targeting of RNA polymerase (pol) III subunits and pol III transcription factors in the Xenopus laevis oocyte nucleus. Antibodies against several of these proteins stained Cajal bodies and ∼90 specific sites on the lampbrush chromosomes. Some of the chromosomal sites had been identified previously by in situ hybridization as the genes for 5S rRNA. The remaining sites presumably encode tRNAs and other pol III transcripts. Pol III sites were often resolvable as loops similar to the much more abundant pol II loops, but without a matrix detectable by phase contrast or differential interference contrast. This morphology is consistent with the transcription of short repeated sequences. Hemagglutinin-tagged transcripts encoding core subunits and transcription factors were injected into the oocyte cytoplasm, and the distribution of newly translated proteins inside the nucleus was monitored by immunostaining. Cajal bodies were preferentially targeted by these proteins, and in some cases the chromosomal sites were also weakly stained. The existence of pol III subunits and pol III transcription factors in Cajal bodies and their targeting to these organelles are consistent with a model of Cajal bodies as sites for preassembly of the nuclear transcription machinery.
INTRODUCTION
Cajal bodies (CBs) are nuclear organelles first described in vertebrate neurons by Ramón y Cajal nearly 100 years ago (Cajal, 1903) and since then demonstrated in a variety of organisms and cell types, including both animals and plants (reviewed in Gall, 2000), and almost certainly the yeast Saccharomyces cerevisiae (Verheggen et al., 2001, 2002). They contain a marker protein named coilin along with numerous factors involved in the transcription and processing of all major types of RNA. Many discussions of CB function emphasize their possible involvement in assembly, modification, or storage of RNA processing factors, especially small nucleolar ribonucleoprotein particles (snoRNPs), splicing small nuclear ribonucleoprotein particles (snRNPs), and the U7 snRNP. However, some evidence suggests that CBs play an even wider role in transcription. On the basis of the composition of CBs in the amphibian oocyte nucleus and the targeting of various transcription and processing factors to CBs, we suggested that CBs may be the primary site for assembly of the transcription machinery of the nucleus (Gall et al., 1999). We postulated that all three eukaryotic RNA polymerases associate in CBs with a variety of specific factors required for transcription and processing of RNA. According to this model, complexes that we call transcriptosomes would assemble in CBs and then move to the respective polymerase (pol) I, pol II, and pol III sites of transcription on the chromosomes.
Central to this model is the demonstration that all three eukaryotic RNA polymerases occur in CBs. In our original article we gave preliminary evidence for the three polymerases in CBs from Xenopus oocyte nuclei. Later, we presented detailed data showing that a phosphorylated form of pol II exists in these CBs (Morgan et al., 2000). On the basis of inhibitor experiments and the kinetics of targeting, we argued that pol II transits through the CBs when it first enters the nucleus.
Herein, we present more complete data for pol III. We show that CBs in Xenopus oocyte nuclei are stained by antibodies against subunits of the pol III core polymerase and pol III transcription factors. Several of these antibodies also stain a specific subset of transcription loops at ∼90 loci on the lampbrush chromosomes. We also show that epitope-tagged versions of pol III subunits and a pol III transcription factor are targeted to the CBs and to the same chromosome loci that are stained by the antibodies. We suggest that pol III and associated transcription factors transit through the CBs on their way to the chromosomal loci at which pol III-specific genes are transcribed.
MATERIALS AND METHODS
Oocytes, Oocyte Injections, and GV Spreads
Techniques for removal and storage of Xenopus laevis oocytes, injection of various probes into the germinal vesicle (GV) or cytoplasm, and preparation of cytological spreads from individual GVs were described in detail previously (Gall et al., 1991, 1999; Gall, 1998). Postfixation of GV spreads in 2% paraformaldehyde was limited to no >2 h.
Immunofluorescence Staining and Microscopy
GV spreads were rinsed in phosphate-buffered saline (PBS; 135 mM NaCl, 2.5 mM KCl, 4.3 mM Na2HPO4, 1.5 mM KH2PO4) and blocked with 10% horse serum for 5–15 min before incubation in primary antibody for 1 h at room temperature. Rabbit polyclonal sera were diluted 1:100–1:2500 with 10% horse serum. Monoclonal antibodies were used at 0.5–1.0 μg/ml when the concentration was known. Otherwise, culture supernatant was used undiluted or diluted up to 1:20 with 10% horse serum. Secondary antibodies were Alexa 488-conjugated or Alexa 594-conjugated goat anti-mouse IgG, goat anti-rat IgG, or goat anti-rabbit IgG (Molecular Probes, Eugene, OR). They were used for 1 h at room temperature. After immunostaining, GV spreads were rinsed with PBS, stained 5 min with 0.01 μg/ml 4′,6 diamidino-2-phenylindole, and mounted in 50% glycerol containing 1 mg/ml phenylenediamine to retard fading. Slides were stored in a freezer at −20°C when not being observed. Images were taken with a Micromax charge-coupled device camera (Princeton Instruments, Trenton, NJ) by using the IPLab (3.5.5) image acquisition and analysis program (Scanalytics, Fairfax, VA). Confocal laser scanning microscopy was carried out with the TCS NT system (Leica, Heidelberg, Germany).
Antibodies
All antibodies against core pol III subunits and pol III transcription factors were produced in rabbits by injection of bacterially expressed proteins. The polyclonal sera were affinity purified against their corresponding antigens (Wang and Roeder, 1995, 1997; Hsieh et al., 1999). No34 is a mouse monoclonal antibody (mAb) raised against proteins derived from the high-speed pellet of fractionated X. laevis oocyte nuclei (Hügle et al., 1985). It immunoprecipitates several pol I and pol III components from cell lysates of Xenopus XLKE-A6–cultured cells, but because it does not stain proteins on a Western blot, its specificity is not known. It may recognize one of the two subunits shared by pol I and pol III, namely, RPC40 or RPC16. Hemagglutinin (HA)-tagged proteins were detected with rat mAb 3F10 (Roche Applied Science, Indianapolis, IN), which gives exceptionally low backgrounds on Xenopus GV spreads. Myc-tagged RPC53 was detected with mouse mAb 9E10 (Evan et al., 1985).
Clones and Transcripts
Human pol III core subunits and pol III transcription factors were originally cloned in FLAG-tagged vectors for biochemical studies. In preliminary immunostaining experiments, we had difficulty detecting FLAG-tagged proteins, and for that reason we used either myc- or HA-tagged probes in this study. Details of the clones and mode of transcript synthesis are as follows.
RPC15 (RPB6).
Clone pTRF RPB6 consists of a FLAG-tagged full-length cDNA encoding RPB6 (Acker et al., 1994) in the pRSET vector (Invitrogen, San Diego, CA). A polymerase chain reaction (PCR) product was made with primers CM120 and CM121 and then transcribed with T3 RNA polymerase to give an HA-tagged transcript. CM120 contains a T3 promoter, the Kozak consensus sequence (Kozak, 1989), the HA sequence, and the FLAG sequence. CM121 is from the pRSET vector.
RPC39.
Clone pTRF RPC39 consists of a FLAG-tagged full-length cDNA encoding RPC39 (Wang and Roeder, 1997) in the pRSET vector. We first made a PCR product from the clone by using primers CM120 and CM165 and then transcribed it with T3 RNA polymerase to give an HA-tagged transcript. CM165 contains the 3′ end of RPC39.
RPC53.
Clone pTRF RPC53 consists of a FLAG-tagged full-length cDNA encoding RPC53 (Ittmann, 1994) in the pRSET vector. A PCR product was made from the open reading frame of pTRF RPC53 with primers CM110 and CM111. The open reading frame was ligated into pCRII (Invitrogen) and then subcloned into pBluescript modified to contain a nuclear localization sequence and six repeats of the c-myc tag (Roth et al., 1991; Wu et al., 1994). For transcription with T3 RNA polymerase the pBluescript clone was linearized with XbaI.
RPC62.
Clone pTRF RPC62 consists of a FLAG-tagged full-length cDNA encoding RPC62 (Wang and Roeder, 1997) in the pRSET vector. A PCR product was made with primers CM120 and CM121 as for RPC15 and then transcribed with T3 RNA polymerase to give an HA-tagged transcript.
TFIIIB90.
Clone pFLAG (S)-7 TFIIIB90 consists of a FLAG-tagged full-length cDNA encoding TFIIIB90 (Wang and Roeder, 1995) in the pGEM-7Z vector (Promega, Madison, WI). An HA tag was produced by annealing oligonucleotides CM170 and CM168 and inserting at the XbaI and NdeI sites upstream of and in frame with the TFIIIB90 coding region. HA-tagged transcripts were synthesized by linearizing with SalI and transcribing with T7 polymerase.
TFIIIC63.
Clone pFLAG (AS)-7 TFIIIC63 consists of a FLAG-tagged full-length cDNA encoding TFIIIC63 (Hsieh et al., 1999) in the pGEM-7Z vector. An HA tag was produced by annealing oligonucleotides CM173 and CM174 and inserting at the BglII and NdeI sites upstream of and in frame with the TFIIIC63 coding region. HA-tagged transcripts were synthesized by linearizing with SalI and transcribing with Sp6 polymerase.
Deoxyoligonucleotides are as follows: CM110, CCGGATCCCAT-GGTTGCAGGCAACATGTCGGAAGGAAAC; CM111, CCTCTAGACCGGTGTTTGTGATCCAAGAGGGATTC; CM120, GCAATTA-ACCCTCACTAAAGGGAACAAAAGGCCGCCACCATGGAGTACCCATATGATGTGCCAGATTACGCTGACTACAAAGACGAT-GAC; CM121, CTAGTTATTGCTCAGCGGTGGCAG; CM165, TTAAAATTCGAGCCACTCTGTCATG; CM168, TATGGGATCCAGCGTAATCTGGCACATCATATGGGTACTCCATGGTGGCT; CM170, CTAGAGCCACCATGGAGTACCCATATGATGTGCCAGATTA-CGCTGGATCCCA; CM173, GATCTGCCACCATGGAGTACCCATATGATGTGCCAGATTACGCTTCTAGACA; and CM174, TATGTCTAGAAGCGTAATCTGGCACATCATATGGGTACTCCATGGTGGCA.
In Vitro Transcripts
Capped sense-strand transcripts were synthesized with T3, T7, or Sp6 RNA polymerase (Stratagene, La Jolla, CA) from 1 μg of the linearized DNA. The RNA was analyzed for size on a 1% agarose, 1.3% formaldehyde gel. Aliquots were precipitated and resuspended at ∼1 μg/μl. Then 23 or 46 nl of RNA solution was injected into the cytoplasm of oocytes by using a Nanoject injection apparatus (Drummond Scientific, Broomall, PA). Injected oocytes were transferred to OR2 saline (Wallace et al., 1973) and stored at 18°C until used for GV spreads or Western analysis.
Western Blots
Isolated GVs were disrupted by pipetting and then centrifuged at 20,000 × g for 10 min to separate organelles from nucleoplasm. All samples were boiled with the appropriate amount of 2× gel buffer for 5–10 min and electrophoresed on a 10% polyacrylamide-SDS gel (Laemmli, 1970). Proteins were electroeluted from the gel onto polyvinylidene fluoride membranes (Immobilon P; Millipore, Bedford, MA). Membranes were blocked in 3% horse serum in PBS and incubated with the appropriate primary antibody. Detection of signal was by enhanced chemifluorescence (Amersham Biosciences, Piscataway, NJ) with the Storm 860 scanner (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Polymerase III in CBs and Lampbrush Chromosomes
Major features of the pol III transcription machinery are conserved in S. cerevisiae, Schizosaccharomyces pombe, and human (Paule and White, 2000; Huang and Maraia, 2001). The core enzyme consists of 16–17 subunits that share sequence homology among the three organisms. Functional similarity is demonstrated by the fact that several human genes encoding pol III subunits can substitute for the corresponding genes in S. cerevisiae. This evolutionary conservation permitted us to study pol III and pol III transcription factors in Xenopus by using the corresponding cloned human genes and antibodies against human proteins.
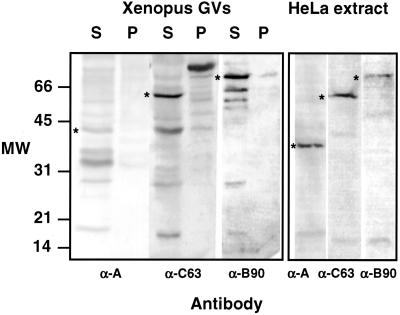
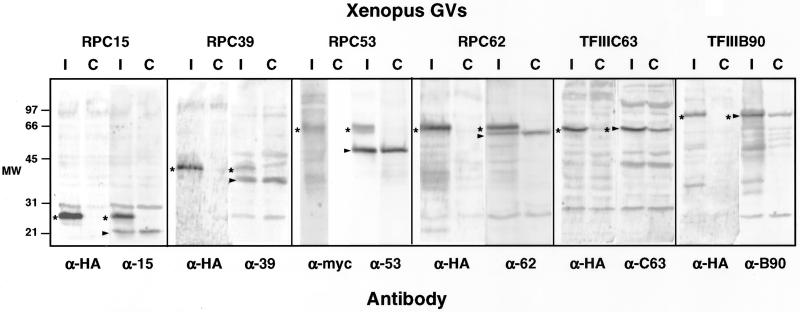
We examined three subunits that are unique to pol III (RPC39, RPC53, and RPC62), and one that is shared by pol I, pol II, and pol III (RPC15). In previous publications (Gall et al., 1999; Gall, 2001), we erroneously referred to RPC39 as RPC19. The protein in S. cerevisiae that corresponds to RPC15 is also known as RPB6 or ABC23. Antibodies raised against the four recombinant human proteins were used to probe Western blots of Xenopus GV proteins (Figure 1). In each case, a major band was detected at the position expected for the pol III subunit, suggesting that the antibodies correctly recognize the corresponding Xenopus proteins.
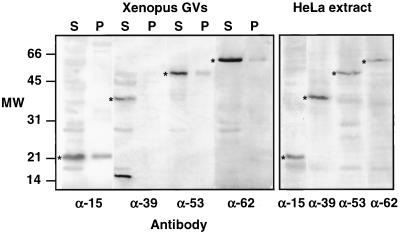
Figure 1.
Western blots of proteins from Xenopus GVs and HeLa nuclear extract probed with antibodies against human pol III core subunits RPC15, RPC39, RPC53, and RPC62. GVs were separated into supernatant (S) and pellet (P) fractions before solubilization and electrophoresis. In the HeLa extract each antibody recognizes a single major band of the expected size. Similar bands (*) in the Xenopus GVs presumably correspond to Xenopus pol III subunits. Anti-RPC39 recognizes an additional protein or breakdown product of ∼15 kDa in the GV. In the GVs most pol III is in the supernatant (nucleoplasm), not in the pellet (organelles). Because this is a montage of gels run at different times under different conditions, the molecular weight markers at the left should be considered approximate only.
These four antibodies were used to stain cytological preparations of GV contents spread on microscope slides. A single GV of Xenopus contains 18 lampbrush chromosomes, ∼1500 extrachromosomal nucleoli, 50–100 CBs, and a highly variable number of B-snurposomes (Figure 2). CBs were stained well by all four antibodies (Figure 3 and Table 1). CBs commonly have B-snurposomes attached to their surface or embedded in their interior. In all cases, staining was limited to the matrix of the CB, the attached or internal B-snurposomes being very weak or negative. Anti-RPC39, anti-RPC53, and anti-RPC62 stained nucleoli slightly above control levels, whereas anti-RPC15 gave significant nucleolar stain. Nucleolar stain with anti-RPC15 was anticipated, because RPC15 is a component of pol I, which is located in the fibrillar region of nucleoli.
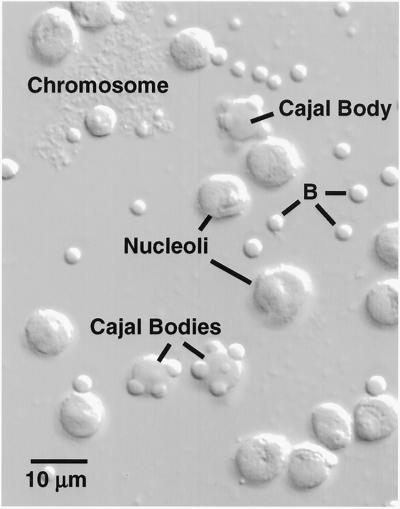
Figure 2.
A small portion of the contents of a single Xenopus GV centrifuged onto a microscope slide and viewed by DIC. The major nonchromosomal structures are nucleoli, Cajal bodies, and B-snurposomes (B). Cajal bodies often have one or more B-snurposomes embedded in them or attached to their surface.
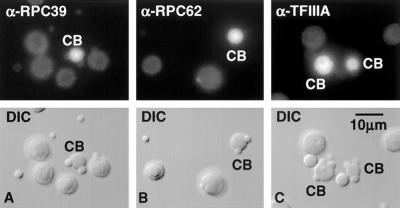
Figure 3.
CBs are stained by antibodies against subunits of the core pol III enzyme and pol III transcription factors. Shown herein is staining by α-RPC39, α-RPC62, and α-TFIIIA. Nucleoli and B-snurposomes are near the background level of staining.
Table 1.
Summary of staining and targeting experiments
| Protein | Antibody stain
|
Targeting
|
||
|---|---|---|---|---|
| Pol III loops | CBs | Pol III loops | CBs | |
| RPC39 | +++ | ++ | ++ | + |
| RPC53 | +++ | ++ | + | (−) |
| RPC62 | +++ | ++ | (−) | + |
| RPC15 (RPB6) | +++ | ++ | Weak pol II + III | +++ |
| Antigen for No34 | +++ | + | n.d. | n.d. |
| TFIIIA | (−) | +++ | n.d. | n.d. |
| TFIIIB90 | (−) | + | + | ++ |
| TFIIIC63 | +++ | + | (−) | + |
n.d., not done.
The two columns on the left summarize staining results with 8 antibodies against pol III core subunits and pol III transcription factors. All 5 antibodies against core subunits stain CBs and pol III loops. Antibodies against 3 transcription factors stain CBs but only one is positive on the loops (α-TFIIIC63). The two columns on the right summarize targeting of the corresponding proteins after cytoplasmic injection of in vitro trascripts from the cloned genes. Details of the injection experiments are discussed in the text.
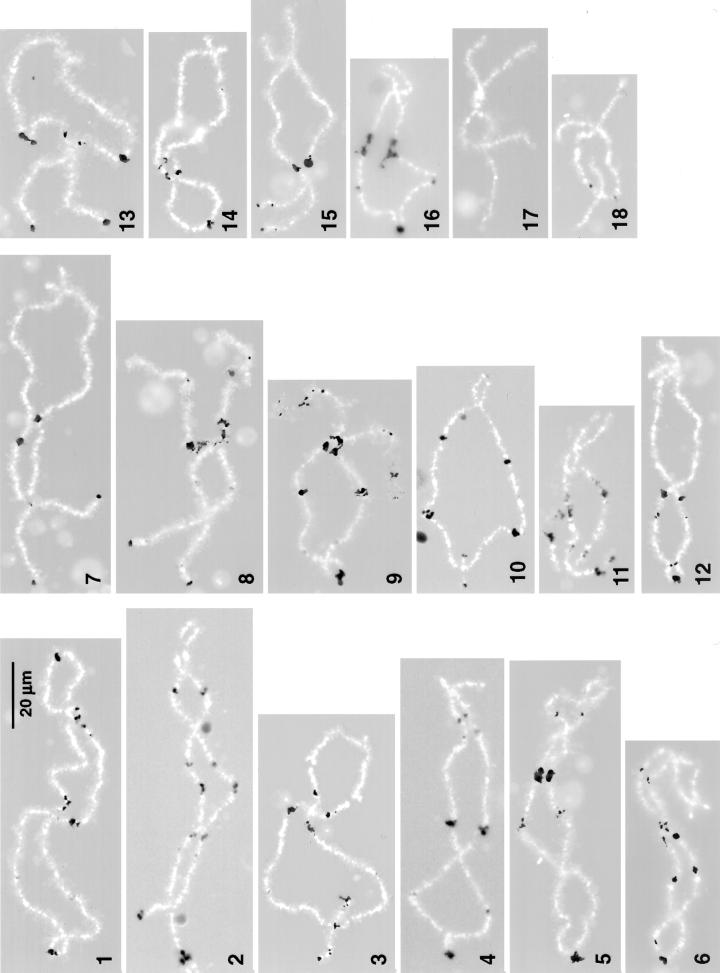
All four anti-pol III antibodies stained a specific set of loci on the Xenopus lampbrush chromosomes. Staining with anti-RPC15 and anti-RPC53 was especially striking in both intensity and specificity. Each gave an extraordinarily brilliant reaction at ∼90 loci distributed along 17 of the 18 chromosomes in the haploid set (Figure 4). Only chromosome 17, the next to shortest in the complement, completely lacked staining. The others each displayed 1–9 positive loci, which varied from minute spots to large irregular patches of stain. The staining patterns are unique for each chromosome, allowing for the first time unequivocal identification of all 18 Xenopus lampbrush chromosomes on the basis of a single stain.
Figure 4.
Pol III sites in the lampbrush chromosomes of Xenopus. This figure consists of a montage of chromosomes from several different nuclei stained with antibodies against RPC15 (chromosomes 2, 4, 5, 9–11, and 16–18), RPC53 (chromosomes 1, 3, 7, 8, and 12–15), or RPC62 (chromosome 6). Pol III loops are shown black against the white chromosome axis (4′,6 diamidino-2-phenylindole stain). Stain at the left end of all chromosomes except 17 and 18 corresponds to previously identified oocyte-type 5S gene loci. Each chromosome of the set can be recognized solely on the basis of its pattern of pol III loops. The 18 chromosomes are numbered according to their relative lengths and are displayed with their long arms to the left. The relative lengths shown herein differ somewhat from those published previously (Callan et al., 1987). Updated cytological maps of X. laevis lampbrush chromosomes can be obtained from us.
A fifth antibody, mAb No34, gave similar strong staining of chromosomal loci and nucleoli. No34 was raised against GV proteins of Xenopus. It immunoprecipitates subunits from both pol I and pol III of Xenopus, but because it does not recognize proteins on Western blots, its specificity has not yet been defined (Schmidt-Zachmann, personal communication). Nevertheless, because No34 is a mouse mAb, it could be used in double-label experiments with the four antibodies against human pol III subunits, which are rabbit polyclonal sera. In this way we verified that all five antibodies stained precisely the same regions on the chromosomes.
Sixteen chromosomes displayed a patch of stain very near the end of the longer arm. Previous in situ hybridization studies showed that the genes coding for Xenopus oocyte-type 5S rRNA are located at approximately these same sites (Pardue et al., 1973; Callan et al., 1987, 1988). Because 5S genes are transcribed by pol III, we assume that the stained regions near the chromosome ends correspond to the 5S genes. None of the other sites are at known gene loci, although this is not surprising, because only a handful of genes have been mapped on the Xenopus chromosomes.
Pol III Loops Are Insensitive to a Low Concentration of α-Amanitin
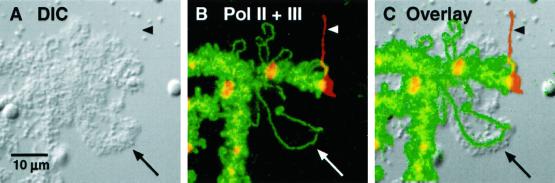
Whereas pol II transcription is inhibited by 0.5–1.0 μg/ml α-amanitin, pol III transcription is insensitive in this range, being inhibited only by concentrations some 50–100× higher (Roeder, 1976). We used this differential sensitivity to demonstrate that the loops on Xenopus chromosomes that are stained by pol III antibodies are uniquely unaffected by the inhibitor. We injected 0.9 ng of α-amanitin into the cytoplasm of stage IV oocytes, which have a diameter of ∼1.0 mm and a volume of ∼0.5 μl. On the assumption that the inhibitor distributes uniformly throughout the cell, the final concentration in the GV should be ∼2 μg/ml. GV spreads were prepared 30 min later and examined after immunostaining with anti-RPC15 for pol III and mAb H14 for pol II. As expected from previous studies (Bucci et al., 1971; Mancino et al., 1971), the morphology of the lampbrush chromosomes was dramatically affected. The chromosomes became much shorter than normal, and no loops were visible by phase contrast or differential interference contrast (DIC) microscopy. These changes are familiar consequences of any agent that inhibits pol II transcription, the assumption being that lampbrush loops retain their extended state only when coated with nascent transcripts (Callan, 1986). Remarkably, immunostaining with anti-RPC15 revealed the otherwise invisible set of 90 pol III loci on the shrunken chromosomes (Figure 5). The fact that these loops do not collapse in the presence of α-amanitin is compatible with the notion that they are, in fact, transcribed by pol III.
Figure 5.
Pol III loops are resistant to α-amanitin. Oocytes were injected with 0.9 ng of α-amanitin, an amount that inhibits pol II but not pol III transcription. Spread GV contents were double stained with mAb H14 against the phosphorylated C-terminal domain of RPB1, which stains pol II loop axes (red), and α-RPC15, which stains pol III loop axes (green). Amanitin causes a striking overall shortening of the chromosome (B). After amanitin treatment pol II loops contract and lose most of their matrix (compare control pol II loops in Figure 7). At the same time, pol III loops remain extended (A) and presumably continue to transcribe.
Pol III Antibodies Stain Axes of Lampbrush Loops
Although the regions stained by pol III antibodies usually appear as somewhat fuzzy or amorphous patches, loops of stain often extend from the patches. These loops may be quite short, but some are >20–30 μm in total length, rivaling the longest pol II loops found on Xenopus lampbrush chromosomes (Figure 6). In most cases, the stain appears as an exceedingly thin line of nearly uniform intensity around the loop. This line is ∼0.4 μm in width, suggesting that it might be the diffraction-limited image of a still finer structure. An obvious possibility is that the line corresponds to the DNA axis of the loop coated with pol III complexes made visible by the antibody staining.
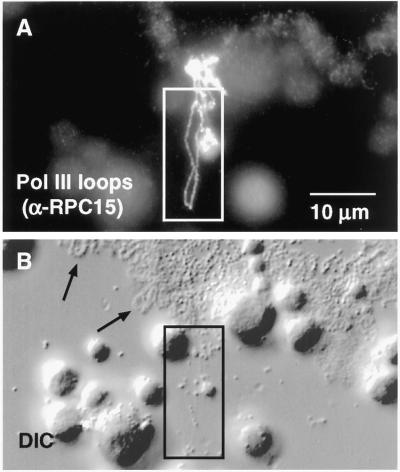
Figure 6.
Pol III sites on the chromosomes consist of irregular patches of stain from which distinct loops may extend laterally. (A) After staining with α-RPC15, loops appear as a more-or-less uniform line ∼0.4 μm in diameter, suggesting that they are diffraction-limited images of polymerase attached to the underlying DNA loop axis. (B) Pol III loops (box) are barely detectable by DIC microscopy, even after digital image processing, whereas pol II loops (arrows) are readily visible.
Pol III Loops Lack a Detectable Matrix
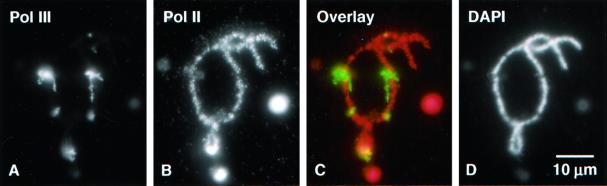
Lampbrush loops represent transcriptionally active regions of the chromosome. Typical pol II loops consist of a DNA axis surrounded by a ribonucleoprotein matrix of nascent RNA chains associated with heterogeneous nuclear ribonucleoproteins and various processing factors. The matrix on a given loop is in the form of one or more thin-to-thick regions, which correspond to single transcription units with short nascent transcripts at one end and longer ones at the other (Miller and Hamkalo, 1972; Angelier and Lacroix, 1975; Scheer et al., 1976; Gall et al., 1983; Callan, 1986). These features are most easily demonstrable in the giant loops of urodele lampbrush chromosomes, but they can be seen clearly in some of the larger loops of Xenopus. The axis of a loop can be visualized by staining with an antibody against phosphorylated RPB1, such as mAb H5 or mAb H14 (Figure 7B, arrow); the matrix can be detected simultaneously by antibody staining or by direct visualization with phase contrast or DIC microscopy (Figure 7A, arrow).
Figure 7.
Left end of chromosome 12 showing loop axes stained with mAb H14 against the phosphorylated C-terminal domain of RPB1, which stains pol II loop axes (green), and α-RPC53, which stains pol III loop axes (red). Pol II loops (arrows) consist largely of nascent transcripts with associated proteins that form a bulky thin-to-thick matrix (arrow in A) covering the axis (arrow in B). The matrix is visible by phase contrast or DIC microscopy. On the other hand, pol III loops have essentially no matrix. Their axis is visible when stained (ar-rowhead in B) but the loop is basically undetectable by DIC (arrowhead in A). The majority of loops are transcribed by pol II, whereas pol III is limited to ∼90 sites scattered on the 18 chromosomes.
In sharp contrast to the majority of loops on the lampbrush chromosomes, pol III loops are essentially undetectable by phase contrast or DIC microscopy (Figures 6 and 7, arrowhead). If a pol III loop is first identified by antibody staining, one can take a phase contrast or DIC image of the region, process it digitally, and make out the faint outline of the loop (Figure 6B). Under ordinary, nonfluorescent viewing conditions, however, one cannot see the pol III loops. The morphology of pol III loops is consistent with what is known about the organization and transcription of pol III genes. In general these genes are serially repeated and produce short noncoding RNA transcripts, including tRNAs, 5S rRNA, 7SK RNA, and U6 snRNA. Individual transcription units are at most a few hundred bases in length, and only nascent transcripts at the end of a transcription unit would be this long. The amount of protein associated with such nascent transcripts would be correspondingly small, so that one would not expect pol III transcription units to be visible by phase contrast or DIC microscopy. It is, in fact, somewhat surprising that the polymerase itself is so readily detectable by fluorescent antibody staining.
Chromosomes that were double stained with either H5 or H14 for pol II, and one of the pol III antibodies displayed completely nonoverlapping patterns for the two stains (Figure 7). We conclude, therefore, that lampbrush chromosome loops transcribe either pol II or pol III genes, but not both simultaneously.
Pol III Transcription Factors in CBs and Lampbrush Chromosomes
Pol III transcription factors have been less highly conserved during evolution than the pol III core enzyme subunits. Nevertheless, we have been able to use antibodies against human transcription factors to gain useful information about the Xenopus oocyte system. We examined antibodies raised against human recombinant proteins TFIIIA, TFIIIB90, and TFIIIC63. On Western blots of Xenopus GV proteins these antibodies cross-reacted with more than one protein, whereas on blots of HeLa cell extract, they were nearly specific for single proteins (Figure 8). When used to stain spread preparations of GV contents, the three antibodies gave significantly different results. All three stained CBs. Anti-TFIIIA was the most specific, with negligible staining of B-snurposomes and nucleoli (Figure 3). Despite the intense staining of CBs, anti-TFIIIA did not give a detectable reaction at the chromosomal pol III loci. Anti-TFIIIB90 also failed to stain the pol III loops. In contrast, anti-TFIIIC63 gave strong and specific staining at all 90 pol III chromosomal loci.
Figure 8.
Western blots of proteins from Xenopus GVs and HeLa nuclear extract probed with antibodies against human pol III transcription factors TFIIIA, TFIIIB90, and TFIIIC63. GVs were separated into supernatant (S) and pellet (P) fractions before solubilization and electrophoresis. In the HeLa extract each antibody recognizes a single major band of the expected size. Similar bands (*) in the Xenopus GVs probably correspond to Xenopus transcription factors, although there are additional bands that may be breakdown products or unrelated cross-reacting proteins. In the GVs transcription factors are more abundant in the supernatant (nucleoplasm) than in the pellet (organelles). Because this is a montage of gels run at different times under different conditions, the molecular weight markers at the left should be considered approximate only.
Targeting of pol III Subunits and pol III Transcription Factors
To examine the trafficking of pol III and its transcription factors within the GV, we injected myc- or HA-tagged transcripts of four pol III subunits (RPC15, RPC39, RPC53, and RPC62) and two transcription factors (TFIIIB90 and TFIIIC63) into the oocyte cytoplasm. In each case, we verified by Western blotting that the transcript was appropriately translated and a tagged protein of the predicted mobility was imported into the GV (Figure 9 and Table 1). At the same time we examined the distribution of the tagged protein products in GV spreads after immunostaining with antibodies against the myc or HA tag.
Figure 9.
Western blots to show epitope-tagged proteins in the GV. In each case transcripts of HA- or myc-tagged gene clones were synthesized in vitro. The tagged transcripts were injected into the cytoplasm of oocytes, where they were translated by the endogenous oocyte machinery and imported into the GV. Each lane contains proteins from 10 to 15 GVs isolated ∼24 h after injection. Duplicate samples from control (C) and injected (I) oocytes were probed with an antibody against the human protein or the epitope tag. In each case a tagged human protein of the appropriate size was made and imported into the GV. The endogenous Xenopus proteins are marked with an arrowhead. The tagged human proteins are marked with a star.
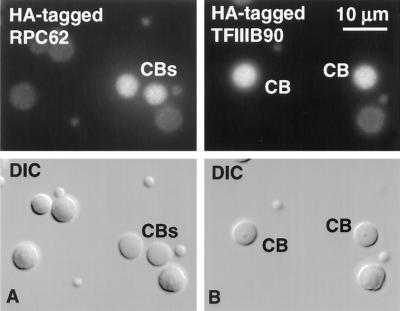
Five of the six tagged proteins were detectable by immunostaining in the CBs (Figure 10 and Table 1). As reported in a previous publication (Morgan et al., 2000), RPC15 gave unusually strong labeling, perhaps because all three RNA polymerases occur in CBs, and RPC15 is a component of each. Among the specific subunits of pol III core enzyme, RPC39 and RPC62 gave weak-to-moderate labeling of CBs, whereas RPC53 was not detectable. Full-length HA-tagged RPC39 and RPC62 were both readily demonstrable in the GV by Western blotting, but in several experiments myc-tagged RPC53 gave only a small amount of full-length product along with shorter myc-tagged molecules that represent either breakdown products or incompletely translated fragments. Of the two HA-tagged transcription factors that we examined by immunostaining, TFIIIB90 gave moderate staining of CBs, whereas TFIIIC63 was weaker but clearly positive (Figure 10 and Table 1). From the results with pol III core enzyme subunits and transcription factors, we feel confident that parts of the pol III machinery are not only present in CBs but are targeted there relatively quickly after translation in the cytoplasm.
Figure 10.
Targeting of pol III components to CBs. HA-tagged transcripts of TFIIIB90 and RPC62 were injected into the cytoplasm of Xenopus oocytes. Newly translated proteins entered the GV and concentrated in CBs. Images taken ∼48 h after injection.
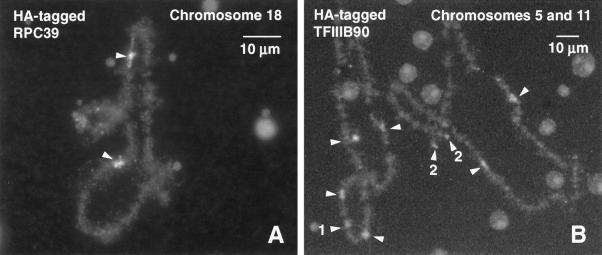
HA-tagged proteins were also detectable at the pol III loci on the lampbrush chromosomes (Figure 11 and Table 1), but there was no simple correlation between chromosomal and CB targeting. Surprisingly, myc-tagged RPC53 could be seen at the chromosomal loci, despite the deficiency of full-length product by Western blotting, whereas the opposite was true for RPC62, which was well translated but failed to appear on the chromosomes. RPC39 was detectable at the pol III loci, and RPC15 was weakly demonstrable on pol III loops and throughout the rest of the chromosome. RPC15 was also targeted to the fibrillar centers of the multiple oocyte nucleoli. The distribution of tagged RPC15 was unique among all the probes we examined, presumably reflecting the fact that it is a subunit of all three RNA polymerases. Of the two pol III transcription factors that we examined, HA-tagged TFIIIB90 was detected at the pol III loci on the chromosomes (Figure 11), whereas TFIIIC63 was not. Altogether, therefore, four of the six tested proteins were targeted to the chromosomal sites of pol III transcription.
Figure 11.
Targeting of pol III components to the chromosomes. HA-tagged transcripts were injected into the cytoplasm of Xenopus oocytes. Newly translated proteins entered the GV and concentrated preferentially in pol III loops on the chromosomes, where they were detected with an antibody against the HA tag. Images taken 48 h after injection. (A) Chromosome 18 from an oocyte injected with HA-tagged RPC39. Arrowheads point to the single pol III site on the homologous chromosomes. (B) Chromosomes 5 and 11 from an oocyte injected with HA-tagged TFIIIB90. Arrowheads point to the most prominent pol III sites. Site 1 is a terminal fusion at the left end of the two homologous chromosomes 11. Sites 2 are the separate left ends of the two homologous chromosomes 5. Sites 1 and 2 are known loci of oocyte-type 5S rRNA genes.
DISCUSSION
Assembly of Transcription Machinery
We recently proposed that CBs might function as sites for assembly of the transcription machinery of the nucleus (Gall et al., 1999). The basic hypothesis was that pol I, pol II, and pol III core enzymes associate in CBs with their respective transcription and processing factors to form large complexes, for which we suggested the name transcriptosomes. These complexes were postulated to travel to the appropriate sites for transcription, namely, nucleoli in the case of pol I transcriptosomes, or chromosomal sites in the case of pol II and pol III transcriptosomes. The distinguishing feature of this model is preassembly of the transcription machinery in CBs rather than assembly at promoter sites on the chromatin. The model requires, at a minimum, that all three RNA polymerases be present in CBs along with associated transcription and processing factors. Furthermore, these components should target to CBs and transit through them on their way to sites of transcription.
In previous studies, we presented evidence that pol II subunits and some pol II-associated factors are present in oocyte CBs and are targeted there when translated from transcripts injected into the cytoplasm. Furthermore, experiments with the kinase inhibitor 5,6-dichloro-β-d-ribofuranosylbenzimidazole suggested that the largest subunit of pol II may transit through oocyte CBs (Gall et al., 1999; Morgan et al., 2000; Gall, 2001).
Evidence concerning pol III and its associated transcription factors was originally limited to immunostaining data that demonstrated the presence of TFIIIA in oocyte CBs along with three subunits of the core enzyme (RPC39, RPC53, and RPC62) (Gall et al., 1999). Herein, we show that antibodies against two more human pol III transcription factors stain oocyte CBs (TFIIIB90 and TFIIIC63). Two additional antibodies against the core enzyme have been studied. One of these, mAb No34, was derived from a mouse that had been immunized with Xenopus GV proteins and was selected because it stained nucleoli. It does not recognize any band on a Western blot of GV proteins, but it does immunoprecipitate subunits from both pol I and pol III of Xenopus (Schmidt-Zachmann, personal communication). It may, therefore, recognize one of the two subunits shared by pol I and pol III, namely, RPC40 and RPC16 (AC40 and AC19 in S. cerevisiae). It stains oocyte CBs, the fibrillar portion of nucleoli, and the pol III loops on the chromosomes. It is possible, therefore, that mAb No34 stains both pol I and pol III in CBs. The other antibody we have examined is a serum against human RPC15, whose homologue in S. cerevisiae is referred to as RPB6 or ABC23. This antibody stains in a pattern similar to that of mAb No34, namely, CBs, the fibrillar portion of nucleoli, and the pol III loops. This pattern suggests that RPC15 is detected when part of the pol I and pol III enzymes, as expected, but for some reason not when part of the pol II enzyme, which is present in the majority of lampbrush loops. We can conclude that RPC15 occurs in oocyte CBs, but we cannot use its presence there as definitive proof of pol III. In summary, we have shown that oocyte CBs are stained by five antibodies that recognize subunits of the core pol III enzyme and three antibodies that recognize pol III transcription factors (Table 1).
Our model of CB function predicts that pol III subunits and pol III transcription factors are not stored for long periods in CBs nor are they permanent structural features of CBs. Instead, we suggest that these proteins are targeted to CBs, where they assemble into larger complexes before exiting on their way to the chromosomes. We have looked at targeting of four pol III subunits (RPC15, RPC39, RPC53, and RPC62) and two transcription factors (TFIIIB90 and TFIIIC63) after injection of tagged transcripts into the cytoplasm. Five of these were detectable in CBs within a few hours after the injection. Only RPC53 was negative in CBs, possibly because the full-length human protein was unstable in the Xenopus GV. The strength of signal in CBs was not simply correlated with the extent of translation: RPC15 was robustly translated and gave the strongest signal in CBs, whereas TFIIIC63 was equally well translated but gave a relatively weak signal in CBs (Table 1).
In summary, we have shown that pol III subunits and pol III transcription factors not only occur in oocyte CBs but also some, at least, are targeted there after translation in the cytoplasm. It is difficult to know exactly how to interpret the lack of a simple correlation between the efficiency of translation of a given probe and the intensity of its signal in the CBs. All proteins used herein were human in origin and some may interact better than others with Xenopus proteins. Furthermore, in these injection experiments the amount of newly translated human protein often equals or exceeds the amount of endogenous Xenopus protein. Under such circumstances the tagged human protein may overwhelm the pool of potential interacting partners. Our data suggest an important relationship between CBs and the pol III transcription machinery, but other techniques will be needed to determine what happens to these various proteins after they enter the CB and whether they do, in fact, assemble into larger complexes for delivery to the chromosomes.
Pol III Transcription Loops on Lampbrush Chromosomes
We have identified ∼90 loci on the lampbrush chromosomes of X. laevis where components of the core pol III enzyme and some associated transcription factors occur. Several lines of evidence argue strongly that these are sites at which transcription of pol III genes takes place:
- 1.
Five antibodies against core pol III subunits and one against a pol III transcription factor stain precisely the same set of ∼90 chromosome loci. Among the many antibodies that have been tested on Xenopus lampbrush chromosomes, this staining pattern has never been seen before.
- 2.
The same loci are targeted by epitope-tagged versions of two core pol III subunits and one pol III transcription factor.
- 3.
When oocytes are subjected to a low concentration of α-amanitin, the majority of chromosome loops, which are transcribed by pol II, collapse and lose their nascent transcripts. Under the same conditions, loops stained by pol III antibodies are unaffected (Figure 5), consistent with the known resistance of pol III to α-amanitin (Roeder, 1976).
- 4.
The oocyte-type 5S genes, which are transcribed by pol III, are found near the telomeres on the longer arm of 16 of the 18 Xenopus chromosomes (Pardue et al., 1973; Callan et al., 1988). Loops that stain with pol III antibodies occur near these same telomeres (Figure 4). We assume that 5S genes are located in the stained loops, although simultaneous in situ hybridization and immunofluorescence staining must be performed before this conclusion can be certain.
- 5.
Loops that stain with pol III antibodies do not stain with pol II antibodies.
- 6.
Loops that stain with pol III antibodies have little or no detectable RNP matrix and hence are essentially invisible by phase contrast or DIC microscopy (Figures 6 and 7).
These cytological data can be combined with information on the molecular organization of pol III genes to give a model of pol III transcription in oocyte chromosomes. The majority of genes known to be transcribed by pol III are both clustered and reiterated in the Xenopus genome. These include the genes coding for 5S rRNA (Brown et al., 1971; Brown and Sugimoto, 1973), tRNA (Clarkson et al., 1973a,b; Muller et al., 1987), and U6 small nuclear RNA (Krol et al., 1987). The coding regions are all short and separated from each other by noncoding sequences. Because transcription is limited to the coding regions, the nascent transcripts on the chromosome loops must be short and separated from each other by spacer regions. Scheer (1982) described loops of this sort in “Miller spreads” of GV contents from the salamander Pleurodeles and suggested that they might represent active 5S or tRNA genes. Because they contain almost no RNP matrix, such loops do not have a thin-to-thick matrix, as found in pol II transcription units, and they should be essentially invisible by conventional phase contrast or DIC microscopy. The loops that stain with pol III antibodies fit this description. They can be seen only when stained with a fluorescent probe (Figures 6 and 7) and even in this case they appear as a diffraction-limited line that presumably corresponds only to the DNA axis of the loop coated with pol III and associated transcription factors.
X. laevis has ∼24,000 copies of the oocyte-type 5S genes per haploid genome, at a single site on each of 16 chromosomes. Because the lampbrush stage occurs during prophase after the chromosomes have replicated, there will be an average of 3000 copies on each homologue. One 5S gene plus spacer is 0.9 kilobases (kb) or 0.3 μm in length, giving a total of ∼900 μm of 5S DNA on average at each chromosome locus. Because pol III loops tend to be compacted into unresolvable clumps, it is difficult to determine the precise morphological organization at the 5S loci. It is unlikely that the total loop material at one locus is 900 μm in length, suggesting either that the DNA is not fully extended or only a fraction of the genes are active at one time. In any case, there are more than enough gene copies to account for the length of transcribing material detected at the oocyte-type 5S loci. The chromosomal location of somatic-type 5S genes has been described previously (Callan et al., 1988), but we have not yet carried out detailed mapping studies to determine whether they correspond to any of the pol III loci identified by immunofluorescence.
Similar but less certain calculations apply to the tRNA genes of Xenopus, which have a total redundancy of 7800–9600 copies/haploid genome (Clarkson et al., 1973a,b). A cloned tRNA cluster studied by Clarkson's group (Clarkson and Kurer, 1976; Muller et al., 1987) contains two tRNA1met genes and single genes for six other tRNA species irregularly spaced on both strands of a 3.18-kb piece of DNA (∼0.4 kb or 0.13 μm/gene). This cluster is tandemly repeated ∼150 times in the haploid genome for a total of 1200 genes. It hybridizes to a single locus near the telomere on the long arm of one of the shorter acrocentric chromosomes (Fostel et al., 1984). Thus, a single chromosomal site may account for 1200 of the 7800–9600 tRNA gene copies. The chromosomal location of two other similar tRNA clones has also been determined by in situ hybridization (Narayanswami et al., 1995). One of them is at or very near the same locus as the 3.18-kb clone and the other is on another chromosome. The in situ hybridization was performed on chromosome spreads that were labeled with gold particles for electron microscopy. The gold particles appeared as clusters over the labeled site or as extraordinarily long loops extending out from the body of chromosome. These patterns are similar to those we see herein by immunostaining.
How the remaining ∼7500 tRNA genes are distributed among the 74 non-5S loci is not known. If they are evenly distributed and each occupies ∼0.4 kb of DNA, the total estimated length of DNA at each (replicated) site will be ∼81 kb or ∼27 μm. Although this is considerably less than the estimate for oocyte-type 5S genes, the total length of DNA is enough to account for the transcribing material detected by immunofluorescence.
There are ∼500 U6 snRNA genes in large tandem arrays in the genome of X. (Silurana) tropicalis (Krol et al., 1987). If the number of genes per site is similar to that for the oocyte-type 5S and tRNA genes, these might all be located at a single chromosomal locus. Information about U6 genes in X. laevis is not available, but their organization is probably similar to that for S. tropicalis. The Xenopus genome may also contain homologues of the mammalian middle-repetitive Alu sequences, which are transcribed by pol III.
In summary, by several independent criteria we have identified ∼90 sites on the lampbrush chromosomes of X. laevis at which pol III transcripts are being made in stage IV–VI oocytes. Whether these 90 loci represent all pol III sites in the genome is not certain, because there could be additional sites that are active only in somatic tissues or at other stages in oogenesis. The pol III sites are especially useful for mapping studies, because a single antibody produces a unique staining pattern on each of the 18 chromosomes (Figure 4). Before the identification of these loci, it was necessary to use a battery of probes to label all the chromosomes, and even then a few chromosomes were difficult to identify individually.
ACKNOWLEDGMENTS
We thank M. Schmidt-Zachmann for mAb No34 and unpublished data concerning this antibody. This work was supported by research grants GM-33397 from the National Institute of General Medical Sciences (J.G.G.) and CA-42567 from the National Cancer Institute (R.G.R.). J.G.G. is American Cancer Society Professor of Developmental Genetics.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0281. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0281.
REFERENCES
- Acker J, Wintzerith M, Vigneron M, Kedinger C. A 14.4 kDa acidic subunit of human RNA polymerase II with a putative leucine-zipper. DNA Seq. 1994;4:329–331. doi: 10.3109/10425179409020860. [DOI] [PubMed] [Google Scholar]
- Angelier N, Lacroix J-C. Complexes de transcription d'origines nucléolaire et chromosomique d'ovocytes de Pleurodeles waltlii et P. poireti (Amphibiens, Urodèles) Chromosoma. 1975;51:323–335. doi: 10.1007/BF00326319. [DOI] [PubMed] [Google Scholar]
- Brown DD, Sugimoto K. 5S DNAs of Xenopus laevis and Xenopus mulleri: evolution of a gene family. J Mol Biol. 1973;78:397–415. doi: 10.1016/0022-2836(73)90464-6. [DOI] [PubMed] [Google Scholar]
- Brown DD, Wensink PC, Jordan E. Purification and some characteristics of 5S DNA from Xenopus laevis. Proc Natl Acad Sci USA. 1971;68:3175–3179. doi: 10.1073/pnas.68.12.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci S, Nardi I, Mancino G, Fiume L. Incorporation of tritiated uridine in nuclei of Triturus oocytes treated with α-amanitin. Exp Cell Res. 1971;69:462–465. doi: 10.1016/0014-4827(71)90255-2. [DOI] [PubMed] [Google Scholar]
- Cajal SR. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Callan HG. In: Lampbrush Chromosomes. Solioz M, editor. Vol. 36. Berlin: Springer Verlag; 1986. [Google Scholar]
- Callan HG, Gall JG, Berg CA. The lampbrush chromosomes of Xenopus laevis: preparation, identification, and distribution of 5S DNA sequences. Chromosoma. 1987;95:236–250. doi: 10.1007/BF00294780. [DOI] [PubMed] [Google Scholar]
- Callan HG, Gall JG, Murphy C. The distribution of oocyte 5S, somatic 5S and 18S + 28S rDNA sequences in the lampbrush chromosomes of Xenopus laevis. Chromosoma. 1988;97:43–54. [Google Scholar]
- Clarkson SG, Birnstiel ML, Purdom IF. Clustering of transfer RNA genes of Xenopus laevis. J Mol Biol. 1973a;79:411–429. doi: 10.1016/0022-2836(73)90014-4. [DOI] [PubMed] [Google Scholar]
- Clarkson SG, Birnstiel ML, Serra V. Reiterated transfer RNA genes of Xenopus laevis. J Mol Biol. 1973b;79:391–410. doi: 10.1016/0022-2836(73)90013-2. [DOI] [PubMed] [Google Scholar]
- Clarkson SG, Kurer V. Isolation and some properties of DNA coding for tRNA1met from Xenopus laevis. Cell. 1976;8:183–195. doi: 10.1016/0092-8674(76)90002-7. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fostel J, Narayanswami S, Hamkalo B, Clarkson SG, Pardue ML. Chromosomal location of a major tRNA gene cluster of Xenopus laevis. Chromosoma. 1984;90:254–260. doi: 10.1007/BF00287032. [DOI] [PubMed] [Google Scholar]
- Gall JG. Spread preparation of Xenopus germinal vesicle contents. In: Spector D, Goldman R, Leinwand L, editors. Cells: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1998. pp. 52.51–52.54. [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall JG. A role for Cajal bodies in assembly of the nuclear transcription machinery. FEBS Lett. 2001;498:164–167. doi: 10.1016/s0014-5793(01)02461-9. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Diaz MO, Stephenson EC, Mahon KA. The transcription unit of lampbrush chromosomes. In: Subtelny S, Kafatos F, editors. Gene Structure and Regulation in Development. New York, NY: Alan R. Liss; 1983. pp. 137–146. [Google Scholar]
- Gall JG, Callan HG, Wu Z, Murphy C. Lampbrush chromosomes. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical Uses in Cell and Molecular Biology. San Diego, CA: Academic Press; 1991. pp. 149–166. [Google Scholar]
- Hsieh Y-J, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügle B, Scheer U, Franke WW. Ribocharin: a nuclear Mr 40,000 protein specific to precursor particles of the large ribosomal subunit. Cell. 1985;41:615–627. doi: 10.1016/s0092-8674(85)80034-9. [DOI] [PubMed] [Google Scholar]
- Ittmann MM. Cell cycle control of the BN51 cell cycle gene which encodes a subunit of RNA polymerase III. Cell Growth Differ. 1994;5:783–788. [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A, Carbon P, Ebel J-P, Appel B. Xenopus tropicalis U6 snRNA genes transcribed by Pol III contain the upstream promoter elements used by Pol II dependent U-snRNA genes. Nucleic Acids Res. 1987;15:2463–2478. doi: 10.1093/nar/15.6.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mancino G, Nardi I, Corvaja N, Fiume L, Marinozzi V. The effects of α-amanitin on Triturus lampbrush chromosomes. Exp Cell Res. 1971;64:237–239. doi: 10.1016/0014-4827(71)90218-7. [DOI] [PubMed] [Google Scholar]
- Miller OL, Hamkalo BA. Visualization of RNA synthesis on chromosomes. Int Rev Cytol. 1972;33:1–25. doi: 10.1016/s0074-7696(08)61446-1. [DOI] [PubMed] [Google Scholar]
- Morgan GT, Doyle O, Murphy C, Gall JG. RNA polymerase II in Cajal bodies of amphibian oocytes. J Struct Biol. 2000;129:258–268. doi: 10.1006/jsbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- Muller F, Clarkson SG, Galas DJ. Sequence of a 3.18 kb tandem repeat of Xenopus laevis DNA containing 8 tRNA genes. Nucleic Acids Res. 1987;15:7191. doi: 10.1093/nar/15.17.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanswami S, Doering JL, Fokta FJ, Rosenthal DS, Nguyen T-N, Hamkalo BA. Chromosomal locations of major tRNA gene clusters of Xenopus laevis. Chromosoma. 1995;104:68–74. doi: 10.1007/BF00352227. [DOI] [PubMed] [Google Scholar]
- Pardue ML, Brown DD, Birnstiel ML. Location of the genes for 5S ribosomal RNA in Xenopus laevis. Chromosoma. 1973;42:191–203. doi: 10.1007/BF00320940. [DOI] [PubMed] [Google Scholar]
- Paule MR, White RJ. Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Eukaryotic nuclear RNA polymerases. In: Losick R, Chamberlin M, editors. RNA Polymerase. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1976. pp. 285–329. [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U. A novel type of chromatin organization in lampbrush chromosomes of Pleurodeles waltlii: visualization of clusters of tandemly repeated very short transcriptional units. Biol Cell. 1982;44:213–220. [Google Scholar]
- Scheer U, Franke WW, Trendelenburg MF, Spring H. Classification of loops of lampbrush chromosomes according to the arrangement of transcriptional complexes. J Cell Sci. 1976;22:503–520. doi: 10.1242/jcs.22.3.503. [DOI] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine DLJ, Samarsky D, Mouaikel J, Blanchard J-M, Bordonné R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Mouaikel J, Thiry M, Blanchard J-M, Tollervey D, Bordonné R, Lafontaine DLJ, Bertrand E. Box C/D snoRNA trafficking involves snoRNP proteins, nucleolar factors, and a novel nuclear domain. EMBO J. 2001;20:5480–5490. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes: III. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell. 1994;5:1119–1127. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]