Abstract
A prerequisite for proteins to interact in a cell is that they are present in the same intracellular compartment. Although it is generally accepted that proteasomes occur in both, the cytoplasm and the nucleus, research has been focusing on cytoplasmic protein breakdown and antigen processing, respectively. Thus, little is known on the functional organization of the proteasome in the nucleus. Here we report that within the nucleus 20S and 26S proteasomes occur throughout the nucleoplasm and partially colocalize with splicing factor–containing speckles. Because proteasomes are absent from the nucleolus, a recruitment system was used to analyze the molecular fate of nucleolar protein fibrillarin: Subtoxic concentrations of mercuric chloride (HgCl2) induce subcellular redistribution of fibrillarin and substantial colocalization (33%) with nucleoplasmic proteasomes in different cell lines and in primary cells isolated from mercury-treated mice. Accumulation of fibrillarin and fibrillarin-ubiquitin conjugates in lactacystin-treated cells suggests that proteasome-dependent processing of this autoantigen occurs upon mercury induction. The latter observation might constitute the cell biological basis of autoimmune responses that specifically target fibrillarin in mercury-mouse models and scleroderma.
INTRODUCTION
The bulk of nonlysosomal proteolysis is carried out by the ATP-powered 26S proteasome, which is involved in the regulation of major cellular processes such as progression of the cell cycle, transcription, flux of substrates through metabolic pathways, elimination of abnormal proteins, and antigen processing (Hershko and Ciechanover, 1998; Kloetzel, 2001). In most cultured mammalian cells 80–90% of the protein breakdown occurs by the proteasome pathway (Lee and Goldberg, 1998). The 26S proteasome is composed of two distinct subcomplexes: the central 20S proteasome, in which proteins are degraded, and two flanking 19S complexes, which provide substrate specificity and regulation. The 20S proteasome forms the core subunit harboring multiple catalytic centers located within the hollow cavity of a cylinder (Finley, 2002). This topology sequesters the catalytic sites from potential substrates (Voges et al., 1999). Most of the substrates of the eukaryotic 26S proteasome must be marked by ubiquitination in order to be destroyed. This involves the covalent attachment of multiple ubiquitin molecules to the target protein (Ciechanover, 1998). However, exceptions to the rule such as ornithine decarboxylase (ODC), which is degraded by the 26S proteasome without ubiquitinylation are known, and more are currently under investigation (Murakami et al., 1992; Verma and Deshaies, 2000).
Proteasomes generate oligopeptides, most of which are further degraded by distinct endopeptidases and aminopeptidases into amino acids. However, a fraction of these peptides escape complete destruction and are subjected to antigen presentation. The peptides are transported to the endoplasmic reticulum via the transporter associated with antigen presentation (TAP; Neefjes et al., 1993), loaded onto major histocompatibility (MHC) class I molecules, and delivered to the cell surface for the continual surveillance by CD8+ T cells of the immune system (Rock and Goldberg, 1999). Thus, the spectrum of MHC class I–presented peptides accurately reflects which proteins are expressed within the cell.
Because antigen processing constitutes the prerequisite for every antigen-driven immune response, it seems noteworthy that the ubiquitin system degrades both intracellular “self” proteins and foreign “nonself” proteins such as viral proteins in a nondiscriminatory manner. Peptides from both populations are usually presented to CD8+ T cells, but T cells derived from self proteins normally do not elicit an immune response because they are selected for tolerance to self during their development in the thymus (reviewed in Sprent and Kishimoto, 2001). However, it is easy to imagine that aberations in the processing of endogenous proteins may lead to presentation of new self peptides that are incorrectly recognized as nonself and may well serve as the pathogenic basis for autoimmune diseases (Schwartz and Ciechanover, 1999).
Systemic rheumatic autoimmune diseases are common human diseases with an estimated prevalence of 2% in the United States (Jacobson et al., 1997) and in Europe. Autoantibodies that recognize intracellular nucleoprotein complexes such as nucleosomes, spliceosomal components, and nucleolus-associated proteins (reviewed in Tan, 1989; Hemmerich and von Mikecz, 2000) represent a hallmark of systemic rheumatic diseases. The factors triggering the formation of autoreactive B cells against nuclear proteins are still unknown, but genetic, hormonal, and environmental components are involved. A valuable model to study molecular mechanisms of systemic autoimmune responses is mercury-induced autoimmunity: Chronic administration of mercuric chloride (HgCl2) to susceptible H-2s mice generates a specific immune response against the nucleolar protein fibrillarin (Hultman et al., 1989; Reuter et al., 1989), which is also a target of antinuclear autoantibodies (ANA) produced by a subset of patients with systemic sclerosis (Arnett et al., 1996).
Fibrillarin is a 34-kDa protein that derives its name from its localization to both the fibrillar center (FC) and dense fibrillar component (DFC) of the nucleolus (Ochs et al., 1985). The nucleolus constitutes a highly dynamic substructure of the nucleus that forms around ribosome production (Scheer and Hock, 1999; Lewis and Tollervey, 2000; Carmo-Fonseca et al., 2001). Fibrillarin is associated with U3 and other small nucleolar RNAs (snoRNAs) required for rRNA processing (Smith and Steitz, 1997). As a component of all small nucleolar ribonucleoprotein particles (snoRNPs), fibrillarin seems to be involved in nearly all major posttranscriptional activities in ribosome synthesis, the first steps of rRNA processing, pre-rRNA modification, and ribosome assembly (Tollervey et al., 1993). Specific inhibition of nucleolar transcription by (1) microinjection of anti-RNA polymerase I antibodies into nuclei (Benavente et al., 1988) or (2) treatment of cells with subtoxic concentrations of HgCl2 (Chen and von Mikecz, 2000) induce redistribution of fibrillarin from the nucleolus to nucleoplasmic aggregates.
It is clear from studies using immunogold electron microscopy (Rivett, 1998), immunochemical procedures (Hügle et al., 1983), and observation of green fluorescent protein (GFP)-tagged proteasome subunits in living cells (Reits et al., 1997) that proteasomes are present in the cytoplasm and in the nucleoplasm of many different cell types but not within nucleoli. In the present study we describe how the nucleolar autoantigen fibrillarin may be subjected to proteasome-dependent proteolysis, nevertheless.
MATERIALS AND METHODS
Cell Culture
The following monolayer cell lines were used: human HEp-2 (HeLa derivative), NIH-3T3 fibroblasts, and mouse ET (thymic epithelial), all obtained from the American Type Culture Collection (ATCC, Rockville, MD) were grown in RPMI/10% FCS or DMEM/10% FCS to subconfluence. Where indicated, HgCl2 was added to the culture medium (5–20 μM, 4 h). Cells were detached by trypsinization, and viability was assessed by trypan blue exclusion.
Mice/Preparation of Murine Splenic Cells
B10.S female mice (6–8 weeks old) were obtained from Jackson Laboratories (Bar Harbor, ME) and housed in a pathogen-free facility. After 8 weeks of subcutaneous injection of 0.5 mg HgCl2/kg body weight or saline, given 3 d per week, spleens were removed from the mice and single-cell suspensions were prepared in PBS. Living cells isolated by gradient centrifugation on Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden) were depleted of T cells by MiniMax (Miltenyi Biotec, Auburn, CA) and seeded as monolayer on glass coverslips coated with 0.01% poly-l-lysine (Sigma, St. Louis, MO). After overnight culture in supplemented RPMI medium, indirect immunofluorescence was performed.
Immunofluorescence and Microscopy
Cells were seeded onto coverslips, grown to subconfluence, treated with 20 μM HgCl2 and 1 μM lactacystin where indicated, fixed with methanol/acetone or 3.7% formadehyde/0.25% Triton X-100, and incubated for 1 h at room temperature with rabbit polyclonal sera to 20S proteasomes from human placenta and to 26S proteasomes purified from rat muscle, both antibodies kindly provided by B. Dahlmann (Dahlmann et al., 1985; Stauber et al., 1987), and mouse monoclonal antibodies against the alpha 4-subunit XAPC7 from 20S proteasomes (Affinity, Exeter, UK). For double labeling immunofluorescence polyclonal rabbit sera were mixed with the following: mouse mAb to SC35 (Fu and Maniatis, 1990), mouse mAb 72B9 to fibrillarin (Reimer et al., 1987), mouse mAb to B23 (Finch et al., 1993), mouse mAb to nucleolin (Ginisty et al., 1998), and hamster antibodies to CD11c (N418) kindly provided by G. Kraal (Metlay et al., 1990). Bound antibodies were detected with fluorescein isothiocyanate (FITC) or rhodamine-conjugated anti-rabbit, anti-mouse, or anti-hamster IgG antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Secondary antibodies were diluted 1:100 in PBS and incubated with coverslips for 45 min at room temperature. After each antibody incubation, coverslips or slides were washed three times with PBS. DNA was counterstained by including 1 μg/ml 4′6-diamidino-2-phenylinole (DAPI, Sigma) in the last washing step. Images were obtained by laser confocal microscopy. Optical sections of double-stained samples were scanned with a laser scanning microscope from Olympus (Fluoview 2.0, IX70 inverted microscope; Lake Success, NY). A dual wavelength channel was used to excite FITC and rhodamine at 488 and 568 nm, respectively. Fluorescent signals of both fluorochromes were recorded simultaneously at one scan. Cy5 was excited at 647 nm. Controls established the specificity of fluorochrome-conjugated antibodies for their respective Igs, and that signals in green, red, and far red channels were derived from the respective fluorochrome. No cross talk was observed. For in situ accumulation studies confocal scans of lactacystin-treated and control cells were recorded with identical settings. Quantitative analysis of fluorescence intensity was determined using the Metamorph image analysis software package (Universal Imaging Corp., West Chester, PA). To measure fluorescence intensity within subnuclear compartments (nucleoli, No; nucleoplasm, Nu) regions of interest (ROIs) were positioned manually based on corresponding differential interference contrast (DIC) images. The total area of the nucleoplasm was obtained by subtracting the total area of nucleoli within the nucleus. For average intensity measurements of nucleoplasmic regions, the average fluorescence intensity of the nucleoli were subtracted in an area-corrected manner. Images were background-corrected by reference regions outside the cells but within the field of view, which corresponded to identical-sized ROIs within the nucleus. In double-labeling experiments, signals were defined as colocalizing in the range of Hue: 31–54, Intensity: 0–255, and Saturation: 106–251 (HIS color model, Metamorph software). For each experiment, the area-corrected intensity of 130 subnuclear compartments was determined. Digitalized image information was visualized using Adobe Photoshop (San Jose, CA). For visualization of colocalization in double-labeling experiments separate channels were converted to grayscale images, and colocalizing foci were determined by identification of pixels with high-intensity signals in both channels.
Immunoprecipitation
Immunoprecipitations were performed with HEp-2 whole cell lysates as described (von Mikecz et al., 2000) with the following modifications: 106 cells were resuspended in immunoprecipitation assay (RIPA) buffer (0.1% SDS; 0.5% Triton X-100; 1% sodium deoxycholate; 0.15 M NaCl; 0.01 M Tris-HCl, pH 7.4; 1 mM EDTA; 1 mM AEBSF; Pefabloc; Boehringer Mannheim, Indianapolis, IN). DNA was fragmented by shearing, and the resulting lysate cleared by centrifugation. The lysate was incubated overnight with polyclonal rabbit antiubiquitin antibodies (Sigma). Antibodies were diluted 1:50. Protein/antibody complexes were precipitated with protein A sepharose 6MB (Amersham Pharmacia Biotech, Piscataway, NJ), resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, and analyzed by SDS-PAGE and immunoblotting.
Immunoblotting
Proteins separated by SDS-PAGE were transferred to Hybond N (Amersham, Arlington Heights, IL) and reacted with either of the following: human autoimmune serum to fibrillarin (ANA-N, Sigma) diluted 1:100, rabbit anti–HIS-tagged fibrillarin diluted 1:200, mouse mAb to the 20S proteasome alpha 4-subunit XAPC7 (Affinity) diluted 1:10, mouse mAb to ornithine decarboxylase (Sigma) diluted 1:100, or human autoimmune serum to DNA topoisomerase I diluted 1:100 in PBS containing 0.1% Tween 20 with 5% nonfat dried milk. Bound antibodies were detected with horseradish peroxidase–conjugated anti-human, anti-rabbit, or anti-mouse IgG antibodies (Jackson ImmunoResearch Laboratories) at a dilution of 1:10.000 and the ECL system (Amersham) according to the manufacturer's instructions.
Proteasome Inhibition Studies
Measurement of substrate accumulation by inhibition of proteasome activity was performed as described previously (Rao et al., 1999). Briefly, HEp-2 cells were preincubated with increasing concentrations of lactacystin for 24 h before cell lysis, and accumulation of fibrillarin was determined by immunoblotting. HgCl2 at 20 μM was added to the cell culture for 4 h where indicated.
Plasmid Construction of HIS-tagged Fibrillarin
Plasmid pBS-mFib, containing mouse fibrillarin cDNA (Turley et al., 1993) was used to PCR-amplify a DNA fragment containing the open reading frame of fibrillain flanked by restriction enzyme sequences NcoI (5′) and BamHI (3′). This DNA was ligated into the NcoI and BamHI-prepared expression vector pEA12 (Rippmann et al., 1998). The resulting fusion protein contains a myc-tag and a 6xHis-tag at the C terminus. Fusion protein was expressed in Escherichia coli strain BL21(DE3) and purified by affinity chromatography on nickel-agarose columns as described before (von Mikecz et al., 1999).
RESULTS
Subnuclear Localization of the 20S Proteasome in Epithelial Cells
Although proteasomal function is generally thought to be confined to the cytoplasm, it has been well established from studies using immunogold electron microscopy, immunocytochemical procedures, and single-cell observation of living cells that proteasomes are present in the cytoplasm and the nucleus (Reits et al., 1997; Rivett, 1998). Moreover, immunofluorescence labeling showed nuclear localization of 20S proteasomes, the 19S regulatory complex (Peters et al., 1994), and the 11S (PA28) regulatory subunit (Lallemand-Breitenbach et al., 2001), suggesting that 20S, 26S proteasomes and immunoproteasomes are present in the nucleus as well as in the cytoplasm.
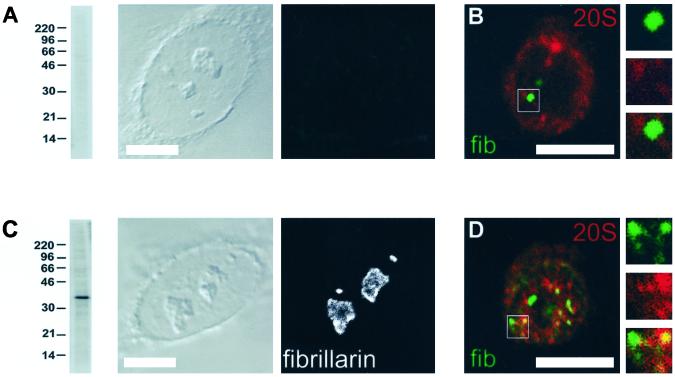
To determine the subnuclear distribution of proteasomes in more detail, we performed immunolabeling with antibodies raised against different biochemical preparations of 20S and 26S proteasomes from human placenta, rat muscle, or rat liver (Dahlmann et al., 1985) and with mouse monoclonal antibodies against alpha 4-subunits of 20S proteasomes. The results are shown in Figure 1. In HEp-2 cells anti-20S proteasome (Figure 1, B, D, and I) and anti-26S proteasome antibodies (Figure 1F) decorated 20–40 reticulated aggregates of variable size and shape in the nucleoplasm, reminiscent of splicing speckles. Such speckles represent subnuclear structures containing spliceosomal components and nuclear matrix proteins (reviewed in Misteli and Spector, 1998). Indeed, double-labeling experiments of antibodies against 20S proteasomes and splicing factor SC35 revealed partial colocalization of the proteins in splicing speckles (Figure 1I, yellow color, inset). Quantification of fluorescence intensities showed a significant overlap (34 ± 12.3%; mean ± SD) between SC35 and proteasomes within the nucleoplasm (Figure 1J). Confirmingly, a similar pattern of colocalization could be observed between proteasomes and spliceosomal components SmB/B′, U1–70k, or U1A/U2B′ (Rockel and von Mikecz, unpublished observation). As seen in Figure 1, anti-20S proteasome antibodies additionally label punctate and speckled structures throughout the nucleoplasm, which do not correspond to splicing speckles (Figure 1, H and I, green color). Nucleoli were devoid of staining in all experiments, suggesting that 20S proteasomes are not present in the nucleolus. Note, that the images in Figure 1 represent confocal optical sections of the nucleus. Therefore, labeling of proteasomes within the cytoplasm appeared weak or could hardly be observed at all (Figure 1, B and D). Localization of 20S proteasomes in speckles, and additional nucleoplasmic sites excluding nucleoli was confirmed (1) using five different rabbit antibodies raised against proteasomal preparations from rat muscle and rat liver (our unpublished results), (2) with mouse monoclonal antibodies to 20S proteasomes, (3) in cell lines such as thymic epithelial ET cells, and NIH-3T3 fibroblasts (our unpublished results), and (4) in primary cells (Figure 4), suggesting that this subnuclear distribution describes a general localization pattern of proteasomes in mammalian cells. The specificity of antiproteasomal antibodies was corroborated by immunoblotting on total cell extracts from HEp-2 cells (Figure 1G). All antibodies recognized bands between 20 and 30 kDa, corresponding to the molecular weight of proteasomal subunits. Mouse monoclonal antibodies against the alpha 4-subunit of 20S proteasomes label one distinct band at 28 kDa. The specificity of the antibodies for proteasomes was confirmed by immunocompetition experiments using purified proteasomes as blocking reagents in immunoblots (our unpublished results).
Figure 1.
Subnuclear localization of 20S and 26S proteasomes. Indirect immunofluorescence of HEp-2 cells with rabbit antibodies against 20S proteasomes purified from human placenta (B), and 26S proteasomes from rat muscle (F) revealed a speckled staining pattern within the nucleoplasm. Such a staining pattern was reproduced with mouse monoclonal antibodies to the alpha 4 subunit of 20S proteasomes (D). Colocalization of 20S proteasomes (H, green color) and splicing factor SC35 (H, red color) occurred in nucleoplasmic speckles (I, yellow color, inset). Note that cytoplasmic proteasome staining is weak due to confocal sectioning. (A, C, E, and H, gray color) represent corresponding differential interference contrast (DIC) images. Bars, 5 μm. (J) Fluorescence intensities of double-labeling experiments (I) were quantified with Metamorph analysis software according to MATERIALS AND METHODS. 20S proteasomes as well as SC35 are localized within the nucleoplasm (Nu) and excluded from the nucleolus (No). 34 ± 12.3% (mean ± SD) of SC35 colocalizes with 20S proteasomes within splicing speckles (*colocalization between SC35 and 20S proteasomes). (G) Characterization of the antibodies against 20S and 26S proteasomes by immunoblotting shows multiple bands between 20 and 30 kDa, representing proteasomal subunits. Molecular weight is given in kDa.
Figure 4.
Colocalization of fibrillarin and 20S proteasomes in splenic cells from mercury-treated mice. B10.S mice were treated with saline (A and B) or subLCs of HgCl2 (C and D) for 8 weeks. (A) Sera from saline-treated control mice did neither react in immunoblotting nor in indirect immunofluorescence of HEp-2 cells. (B) Splenic cells were isolated from these mice and subjected to double labeling and confocal microscopy: Fibrillarin (green color) is confined to the nucleolus, and thus no colocalization (merge, enlargements) could be observed with proteasomes (red). (C) Sera from mercury-treated mice reacted with a 34-kDa band corresponding to the molecular weight of fibrillarin by immunoblotting of HEp-2 cell lysates and showed an exclusive, clumpy staining of the nucleolus in indirect immunofluorescence of HEp-2 cells. (D) Double-labeling and confocal microscopy of splenic cells from HgCl2-treated mice revealed a redistribution of fibrillarin from the nucleolus to the nucleoplasm (green color) and colocalization with proteasomes (red color) in distinct nucleoplasmic aggregates (yellow color, merge, enlargements). Bars, 5 μm. Molecular weight is given in kDa. fib, fibrillarin.
HgCl2-induced Recruitment of Fibrillarin to Nucleoplasmic 20S Proteasomes
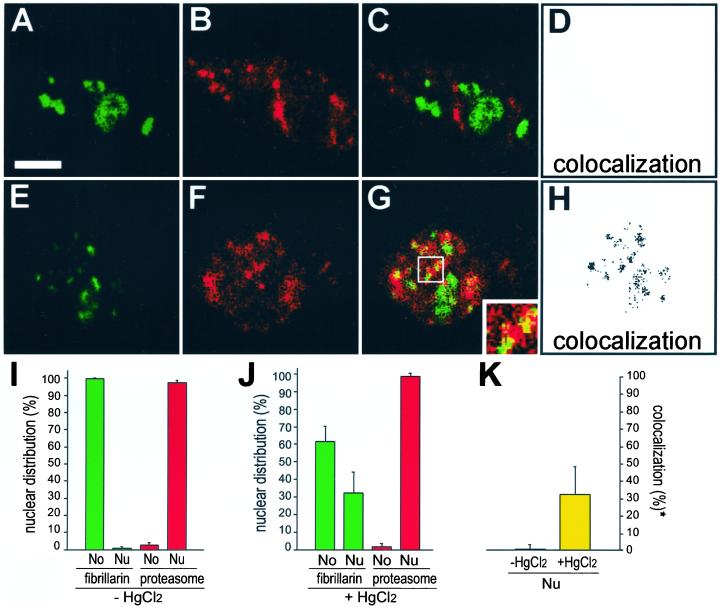
We reported elsewhere that HgCl2 induces inhibition of RNA polymerase I–dependent transcription of rRNA and a redistribution of fibrillarin from the nucleolus to the nucleoplasm (Chen and von Mikecz, 2000). To determine the molecular fate of nucleoplasmic fibrillarin dual-label immunofluorescence of interphasic HEp-2 cells was performed with antibodies against 20S proteasomes, because the latter are abundant in the nucleus and participate in antigen processing. In untreated control cells fibrillarin is exclusively confined to nucleoli (Figure 2, A and I, green color) and antiproteasome antibodies decorated the nucleoplasm in a speckled pattern excluding nucleoli (Figure 2, B and I, red color). The merged image shows no overlap between localization of fibrillarin with 20S proteasomes (Figure 2, C and D), suggesting that under normal conditions the two proteins are segregated from each other. However, when HEp-2 cells were treated with HgCl2 for 4 h, fibrillarin redistributed from the nucleolus to nucleoplasmic sites (Figure 2E, green color) where it colocalized with 20S proteasomes in nucleoplasmic aggregates (Figure 2G, yellow color). Colocalization occurred partially, e.g., cells also displayed nuclear regions where fibrillarin and the 20S proteasome did not overlap. Quantification of fluorescence signals revealed 33 ± 16.7% (mean ± SD) of colocalization between nucleoplasmic fibrillarin and 20S proteasomes (Figure 2K), suggesting that approximately one third of redistributed fibrillarin shares the same subcellular region with proteasomes. Because endogenous proteins are observed in our experiments, the extent of colocalization between nucleoplasmic fibrillarin and proteasomes is regarded as substantial. Moreover, the regions of exclusive fibrillarin localization may represent residual nucleoli and the fact that not all fibrillarin is recruited to the proteasomes simultaneously. To examine colocalization in more detail, separate channels were converted to grayscale images, and colocalizing foci were determined using Adobe Photoshop software, which identifies pixels with high-intensity signals in both channels. The visualization of such an analysis is shown in Figure 2H: In mercury-treated cells fibrillarin and proteasomes colocalize in distinct aggregates throughout the nucleoplasm. Overlap of fibrillarin and 20S proteasomes induced by mercuric chloride does not represent a special feature of HEp-2 cells, because it could as well be detected in mouse ET cells and NIH-3T3 fibroblasts (our unpublished results).
Figure 2.
Mercury-induced colocalization of fibrillarin with proteasomes in nucleoplasmic sites. Double-labeling of untreated HEp-2 cells (first panel) with (A) antifibrillarin (green color) and (B) anti-20S proteasome antibodies (red color). The merged image shows no colocalization (C). In HgCl2-treated HEp-2 cells (second panel) fibrillarin (E, green color) redistributes from the nucleolus to the nucleoplasm, and colocalizes with 20S proteasomes (F, red color) in nucleoplasmic aggregates (G, yellow color, inset). To visualize colocalization in more detail red and green channels were converted separately to greyscale images. Overlapping foci were determined by identification of pixels in which both red and green signals are within 30% of the maximum for that channel. Information of the pixel analysis is output in a superimposed channel showing no overlap in untreated HEp-2 cells (D) but colocalization of fibrillarin and proteasomes in distinct aggregations of fluorescent foci throughout the nucleoplasm in mercury-treated cells (H). Bar, 5 μm. Quantification of fluorescent data by means of Metamorph corroborated exclusive confinement of fibrillarin within nucleoli (No), and 20S proteasomes within the nucleoplasm (Nu) in untreated HEp-2 cells (I). However, upon mercury-treatment fibrillarin redistributes from the nucleolus (No) to the nucleoplasm (Nu), whereas 20S proteasomes maintain their subnuclear localization (J). Thus, no overlap could be observed in untreated cells, whereas 33 ± 16.7% (mean ± SD) of redistributed fibrillarin colocalizes with proteasomes within the nucleoplasm of HgCl2-treated cells (K). *Colocalization between nucleoplasmic fibrillarin and 20S proteasomes.
B23 and Nucleolin (C23) Do Not Colocalize with Proteasomes after HgCl2 Treatment
It has been well established by previous studies that the nucleolus is a dynamic structure that forms dependent on activity of rRNA transcription. Major nucleolar proteins such as B23, nucleolin, UBF, RNA polymerase I, and fibrillarin redistribute after inhibition of nucleolar transcription (Scheer and Benavente, 1990; Jordan and Carmo-Fonseca, 1998; Chen and von Mikecz, 2000). Thus, we wanted to know whether mercury-induced colocalization of fibrillarin and 20S proteasomes in nucleoplasmic aggregates is fibrillarin specific or does also occur with other prominent nucleolar proteins.
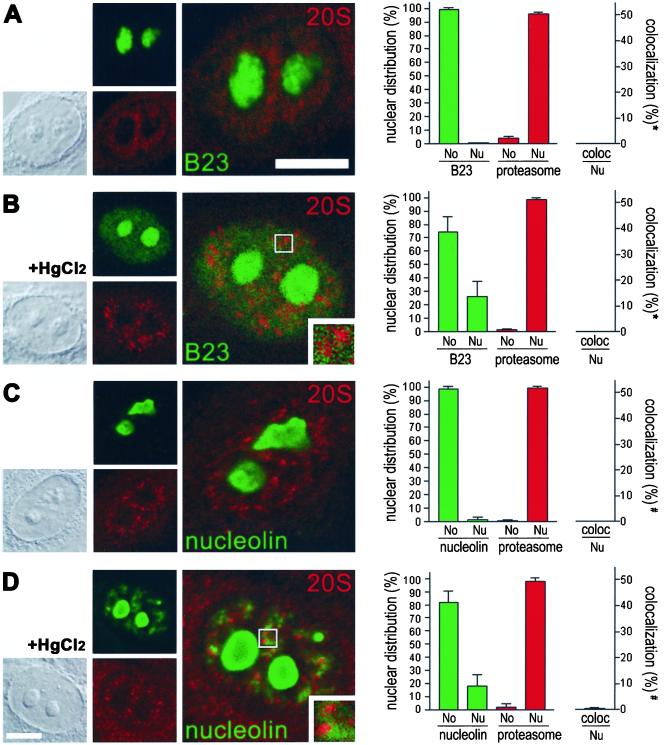
Double labeling of untreated HEp-2 cells revealed that B23 is confined to the nucleolus (Figure 3A, green color) and thus does not colocalize with nucleoplasmic proteasomes in interphasic cells (Figure 3A, merge, and graph). On HgCl2 exposure a subfraction of B23 redistributes from the nucleolus to the nucleoplasm where it localizes in numerous homogeneously distributed foci (Figure 3B, green color). However, no colocalization with nucleoplasmic proteasomes could be detected (Figure 3B, merge, inset, and graph). Similar results were obtained for nucleolin. In untreated cells antibodies to nucleolin exclusively decorated the nucleolus, whereas the nucleoplasm and cytoplasm was not stained (Figure 3C, green color). Thus, the subcellular distribution of nucleolin did not overlap with proteasomal localization (Figure 3C, merge, and graph). Upon HgCl2 treatment, a subfraction of nucleolin is distributed in numerous nucleoplasmic aggregates, similar to the aggregates formed by nucleoplasmic fibrillarin (Figure 3D, green color); however, no colocalization with proteasomes could be detected (Figure 3D, merge, inset, and graph). Rather, nucleoplasmic nucleolin aggregates and proteasomes seem to be juxtaposed in all cells observed. Taken together, the results suggest that major nucleolar proteins B23 and nucleolin redistribute from the nucleolus to the nucleoplasm because of mercury-induced inhibition of rRNA transcription, as does fibrillarin. However, because only fibrillarin colocalizes with proteasomes in nucleoplasmic aggregates, B23, nucleolin, and fibrillarin seem to be recruited to different nucleoplasmic sites after HgCl2 exposure, corroborating previous results (Chen and von Mikecz, 2000). Moreover, the results confirm that colocalization of fibrillarin and proteasomes is neither fortuitous nor due to cross-linking of proteins by HgCl2, because the latter should also lead to colocalization of proteasomes with B23 and nucleolin. Note that the morphology of neither the nucleus nor the nucleolus changed during treatment of cells with low concentrations of HgCl2 (5–20 μm), as shown by corresponding differential interference contrast (DIC, Figure 3, first column).
Figure 3.
Mercury-induced colocalization of proteasomes with fibrillarin is specific and does not occur with major nucleolar proteins B23 and nucleolin as revealed by double labeling immunofluorescence and confocal microscopy. (A) B23 (green color) is exclusively confined to the nucleolus in untreated control cells, whereas 20S proteasomes (red color) are localized within the nucleoplasm. Thus B23 does not overlap with nucleoplasmic proteasomes (merge, and graph). (B) On mercury-treatment a subfraction of B23 appears in numerous homogeneously distributed foci within the nucleoplasm (green color). However, no colocalization with proteasomes (red color) can be detected (merge, inset, and graph). (C) Nucleolin (green color) is exclusively localized to the nucleolus in untreated HEp-2 cells and does not colocalize with proteasomes (merge, and graph). (D) In contrast, a subfraction of nucleolin (green color) appeared in aggregates distributed throughout the nucleoplasm in HgCl2-treated cells. However, no colocalization between nucleoplasmic nucleolin aggregates and proteasomes (red color) could be observed (merge, inset, and graph). The first column shows corresponding DICs. Bars, 5 μM. Quantification of the fluorescent data by Metamorph (graphs, right column) confirmed that nucleolar proteins B23 and nucleolin alter their subnuclear distribution but do not colocalize with 20S proteasomes either in untreated or in mercury-treated cells. In contrast, 20S proteasomes remain confined to the nucleoplasm. No, nucleolus; Nu, nucleoplasm. *Colocalization of B23 with 20S proteasomes in the nucleoplasm; #colocalization of nucleolin and 20S proteasomes in the nucleoplasm.
In Vivo Colocalization of Fibrillarin and Nucleoplasmic Proteasomes in Splenic Cells from Mercury-treated Mice
Chronic subcutaneous administration of HgCl2 to susceptible H-2s mice generates a specific autoimmune response against fibrillarin (Hultmann et al., 1989; Reuter et al., 1989). We reported recently that the production of antifibrillarin antibodies is accompanied by redistribution of fibrillarin from the nucleolus to nucleoplasmic aggregates in splenic cells isolated from mercury-treated mice (Chen and von Mikecz, 2000). To confirm our results on subcellular localization of fibrillarin and proteasomes, in vivo susceptible B10.S mice were treated with either saline or HgCl2 for 8 weeks. Splenic cells were removed and subjected to double immunofluorescence (Figure 4) with monoclonal antibodies to fibrillarin (green) and rabbit antibodies to 20S proteasomes (red). In splenic cells from saline-treated control mice, fibrillarin is confined to the nucleolus and thus is segregated from nucleoplasmic proteasomes (Figure 4B, merge and enlargements). As revealed by immunoblotting and immunofluorescence of murine sera, there is no development of autoimmunity, e.g., production of autoantibodies, in saline-treated mice (Figure 4A). In contrast, mercury-treated mice develop antibodies against fibrillarin. A representative mouse serum shows (1) by immunoblotting of a HEp-2 cell lysate a 34-kDa band that corresponds to the molecular weight of fibrillarin, (2) by indirect immunofluorescence of HEp-2 cells an exclusive, clumpy staining of the nucleolus that is indicative of fibrillarin (Figure 4C) and positive reaction with recombinant fibrillarin in immunoblots (our unpublished results). Double labeling of splenic cells from HgCl2-treated mice revealed that fibrillarin is redistributed from the nucleolus to the nucleoplasm in splenic cells (Figure 4D, green color), where it partially colocalizes with proteasomes in distinct sites (Figure 4D, merge and enlargements. The number of colocalizing nucleoplasmic aggregates in one cell is less in murine splenic cells compared with cell lines (Figure 2), which may be attributable to better accessibility and penetration of mercuric chloride in cell culture. By means of triple labeling, the dendritic nature of murine splenic cells showing colocalization of fibrillarin and proteasomes was identified. Splenic cells displayed in addition to colocalization of fibrillarin and proteasomes a rim-like surface staining of CD11c, which could be further confirmed by (1) positive staining for dendritic cell-restricted surface antigen DEC-205 and (2) negative staining for B cell marker B220 (our unpublished results). Overlap of fibrillarin and proteasomes could be observed in 80% of dendritic cells isolated from mercury-treated mice.
Taken together, a mercury-induced recruitment of fibrillarin to nucleoplasmic proteasomes occurs in coincidence with the production of antifibrillarin autoantibodies in vivo and is not seen in saline-treated control mice that do not develop autoimmunity. The results suggest that altered processing of fibrillarin by proteasomes might play a role in the generation of mercury-induced autoimmunity.
Fibrillarin Is Accumulated in Lactacystin- and HgCl2-treated Cells
The turnover of proteasome substrates can be studied by its specific inhibition (reviewed in Lee and Goldberg, 1998). Lactacystin acts by inhibiting proteasome function as a pseudosubstrate that becomes covalently linked to the hydroxyl groups on the active site threonine of the β subunits (Fenteany et al., 1995). In consequence chymotryptic- and tryptic-like activities are inactivated, and substrates accumulate that are usually metabolized by the ubiquitin-proteasome pathway .
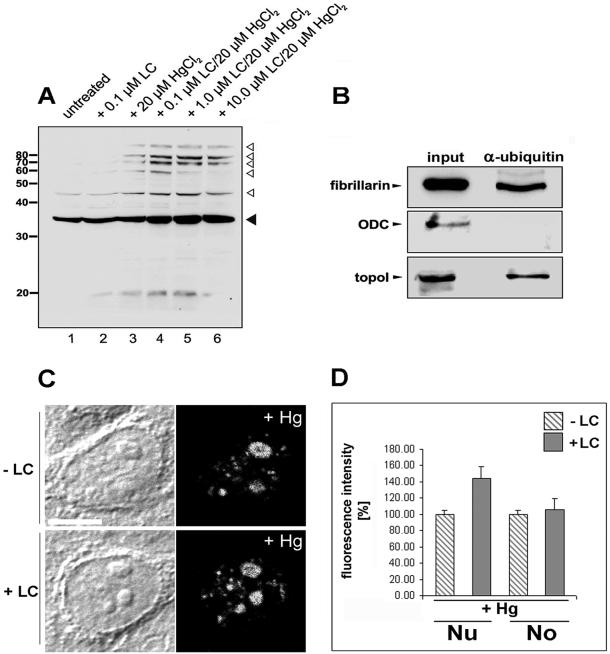
Accordingly, we tested by immunoblotting if fibrillarin constitutes a substrate for proteasomal processing by incubation of cells with increasing concentrations of lactacystin. In protein lysates of untreated HEp-2 cells, fibrillarin is detected as a 34-kDa band (Figure 5A, lane 1, filled arrowhead). In addition, a weaker band appeared at 43 kDa. Because ubiquitin has a molecular weight of 8.5 kDa, we analyzed if the additional, slower migrating band may represent ubiquitinylated fibrillarin. To this end HEp-2 cells were treated with proteasome inhibitor lactacystin in the presence or absence of HgCl2, and protein lysates from equal cell numbers were subjected to immunodetection of fibrillarin. Although lactacystin alone showed no significant change in the amount of fibrillarin (lane 2), HgCl2 treatment induced the appearance of slower migrating fibrillarin bands (lane 3). Addition of lactacystin leads to an accumulation of the fibrillarin band at 34 kDa (filled arrowhead) along with a prominent ladder of protein bands that were specifically recognized by the antifibrillarin antibodies (lanes 4–6, open arrowheads). Except for the highest molecular weight band, this protein ladder appeared as multiples of ∼9 kDa, suggesting that these bands represent fibrillarin, which is conjugated to multiubiquitin chains of different length. Coimmunoprecipitation studies corroborated that a subfraction of fibrillarin may be complexed to the ubiquitinylation machinery in untreated HEp-2 cells: antiubiquitin antibodies precipitate both full-length fibrillarin and a weaker slower migrating band at ∼43 kDa (Figure 5B), suggesting that fibrillarin is associated with proteins of the ubiquitinylation apparatus. In contrast, ornithine decarboxylase (ODC), which is processed by 26S proteasomes without being ubiquitinylated (Murakami et al., 1992), could not be coprecipitated with antibodies against ubiquitin (Figure 5B). Desai et al. (1997) reported previously that camptothecin induces ubiquitinylation of DNA topoisomerase I and its proteasome-dependent processing . We used the protein as a positive control and detected topoisomerase I in immunoprecipitates obtained with antiubiquitin antibodies (Figure 5B). The immunoprecipitation results were confirmed in untreated and mercury-treated HEp-2 cells using three different antiubiquitin antibodies raised in rabbits or mice. Neither fibrillarin nor topoisomerase I was precipitable with the respective presera (our unpublished results).
Figure 5.
Proteasome-dependent processing of fibrillarin induced by mercury. (A) HEp-2 cells were treated with increasing concentrations of proteasome inhibitor lactacystin for 24 h and HgCl2 where indicated and separated on SDS-PAGE, followed by immunoblotting. Antifibrillarin antibodies detected a 34-kDa band corresponding to the molecular weight of fibrillarin in untreated control cells (lane 1, filled arrowhead), and cells were treated with 0.1 μm lactacystin (lane 2). Simultaneous addition of increasing concentrations of lactacystin and HgCl2 lead to accumulation of the 34-kDa band and to the appearance of additional slower migrating bands that represent the molecular weight of fibrillarin (34 kDa) plus multiples of ∼8.5 kDa (lanes 3–6, open arrowheads). (B) Immunoblotting of untreated HEp-2 cells (first column) and coimmunoprecipitates obtained with antiubiquitin antibodies (second column). Fibrillarin (upper panel) and topoisomerase I (lower panel) were coprecipitated with antiubiquitin antibodies. Ornithine decarboxylase (ODC), which is degraded by 26S proteasomes without being ubiquitinylated, served as a negative control and was not precipitated by antiubiquitin (middle panel). (C) For in situ accumulation studies HEp-2 cells were treated with 20 μM HgCl2 and 1 μM lactacystin where indicated. Mercury-treatment induces redistribution of fibrillarin from the nucleolus to the nucleoplasm (second column) and accumulation of fibrillarin in the nucleoplasm of cells, which were additionally treated with lactacystin (lower panel). Fluorescence images from lactacystin-treated and control cells were recorded with identical settings of the confocal microscope. (D) Quantification of fluorescence intensities with Metamorph (see MATERIALS AND METHODS) revealed an accumulation of nucleoplasmic fibrillarin from 100 to 144 ± 14.8% (mean ± SD) in lactacystin-treated cells, whereas concentration of fibrillarin within the nucleolus remained constant at 105 ± 14.1% (mean ± SD, filled bars). Hg, mercuric chloride; Lc, lactacystin; Nu, nucleoplasm; No, nucleolus.
HgCl2-induced Accumulation of Fibrillarin Occurs within Nuclei
To determine the subcellular localization of proteasome-dependent fibrillarin processing, HEp-2 cells were treated with 20 μM mercury and 1 μM lactacystin and subjected to indirect immunofluorescence, and intensity of fluorescent signals was quantitated. In HgCl2-treated cells fibrillarin redistributed from the nucleolus to the nucleoplasmic aggregates (Figure 5C, second column) as observed before (Figure 2; Chen and von Mikecz, 2000). On inhibition of proteasomal proteolysis by lactacystin, fibrillarin accumulated in the nucleoplasm (Figure 5C, lower panel). Quantification of immunofluorescence intensities in subnuclear compartments revealed that lactacystin induces an increase of nucleoplasmic fibrillarin of 44 ± 14.79% (mean ± SD), whereas nucleolar staining appeared to be unchanged (Figure 5D, filled bars). The results suggest that inhibition of proteasome-dependent proteolysis induces accumulation of fibrillarin within the nucleoplasm of mercury-treated cells.
DISCUSSION
Research on proteasomes is mainly focused on processing within the cytoplasm. About 30% of substrates are derived from newly synthesized, faulty proteins that would never attain native structure and thus are subjected to ubiquitination and proteasome-dependent degradation (Schubert et al., 2000). A substantial fraction of the resulting peptides is then translocated via the TAP transporter into the endoplasmatic reticulum for binding to MHC class I molecules and for subsequent presentation to the immune system (Reits et al., 2000). However, proteasomes also play an important role in the turnover of flawless proteins that have simply reached the end of their half lives. Thus, the following question arises: how are nuclear proteins processed when they have come to the end of their working lives and/or if they cannot exert their function because of changes in nuclear structure?
A prerequisite for two proteins to interact in a cell is that they are present in the same intracellular region. In the present study we provide evidence that mercury-induced redistribution of fibrillarin leads to colocalization with nucleoplasmic proteasomes and proteasome-dependent processing of fibrillarin within nuclei. These results confirm recent reports describing As2O3-induced degradation of nuclear protein PML (Lallemand-Breitenbach et al., 2001) and intracellular localization of proteasomal proteolysis of a viral antigen (Anton et al., 1999). Both studies localize proteasome-dependent processing to nucleoplasmic aggregates, thus defining the nucleus as an intracellular site of proteasomal degradation. It has been well established over the last two decades that proteasomes occur in the cytoplasm and the nucleoplasm but not in nucleoli (Hügle et al., 1983; Stauber et al., 1987; Amsterdam et al., 1993; Reits et al., 1997; Rivett, 1998). By means of double labeling and confocal analysis, we were able to refine subnuclear localization of 20S as well as 26S proteasomes in nucleoplasmic speckles where they partially colocalize with the splicing factor SC35 (Figure 1). Additionally, proteasomes were distributed throughout the nucleoplasm in punctate and speckled aggregates but not within nucleoli, which is in perfect agreement with the literature. Thus it seems highly unlikely that nucleoli represent intracellular sites for proteasomal protein degradation. However, inhibition of nucleolar transcription by mercury induces redistribution of nucleolar protein fibrillarin (Chen and von Mikecz, 2000), resulting in (1) colocalization with nucleoplasmic proteasomes (Figure 2) and (2) proteasome-dependent processing of fibrillarin (Figure 5).
The latter observations do not only describe one possible turn over mechanism of a nucleolar protein but may as well provide new insights into the generation of systemic autoimmune responses, because redistribution of fibrillarin and colocalization with proteasomes could be observed in splenic cells from mercury-treated mice (Figure 4). Such mice also developed a specific autoimmunity against fibrillarin. The results suggest that fibrillarin, which is normally segregated from proteasomes within the nucleolus is recruited to proteasome-dependent degradation by mercury. This event may represent altered antigen processing, which in turn may lead to presentation of cryptic determinants to the immune system. The following findings corroborate our hypothesis: (1) Susceptibility for mercury-induced autoimmunity seems to be under the control of MHC genes (Hultman et al., 1992), suggesting that antigen processing and presentation may be involved in the generation of the autoimmune response. (2) Because treatment of IFN-γ knockout mice with subtoxic concentrations of HgCl2 does not provoke any autoimmune response, it was concluded that the prototypic autoimmunity induced by mercury is dependent on IFN-γ (Kono et al., 1998), the same cytokine that modulates the subunit composition and proteolytic activity of immunoproteasomes (Boes et al., 1994). (3) Like fibrillarin, major nucleolar proteins B23 and nucleolin redistributed to the nucleoplasm after HgCl2 treatment, but colocalization with proteasomes could not be observed (Figure 3). The specificity of subcellular colocalization of fibrillarin with proteasomes might reflect the specificity of mercury-induced autoimmunity, which is exclusively directed against fibrillarin in mice.
Apart from our results on altered degradation of fibrillarin there are recent studies that report similar recruitment of nuclear proteins to proteasomal processing when nuclear structure and function is disturbed. Cells infected by herpes simplex virus type 1 in the G2 phase of the cell cycle become stalled in mitosis. This block correlates with the viral immediate-early protein ICP0-induced, proteasome-dependent degradation of centromere proteins CENP-C (Everett et al., 1999), and CENP-A (Lomonte et al., 2001). The antitumor drug camptothecin inhibits the rejoining step of superhelical DNA relaxation, thereby trapping topoisomerase I in covalent linkage with DNA and preventing normal DNA replication. Desai et al. (1997) showed that the half-life of topoisomerase I dropped from 10–16 h down to 1–2 h and that conjugates of topoisomerase I and ubiquitin emerged upon camptothecin treatment . Because MG-132 and lactacystin prevented camptothecin-induced destruction, it is concluded by the authors that camptothecin stimulates proteasome-dependent processing of topoisomerase I. Interestingly, both examples describe how alteration of nuclear function leads to recruitment of nuclear autoantigens to proteasomal processing, because centromere proteins as well as topoisomerase I constitute frequent targets of the autoimmune response in scleroderma (Tan, 1989). The exposure to a number of environmental substances has been associated with scleroderma (Galperin and Gershwin, 1998); thus future studies should focus on further elucidation of proteasome-dependent processing of nuclear antigens and its role in the generation of systemic autoimmune responses.
We consider investigation on proteasomal proteolysis within nuclei an important research topic, because it may provide novel insights into (1) the turn over of nuclear proteins, (2) the regulation of nuclear processes such as transcription and splicing, and (3) the generation of systemic autoimmune responses against nuclear proteins.
ACKNOWLEDGMENTS
We extend our sincere thanks to those individuals who kindly provided us with reagents and made this study possible: Burkhardt Dahlmann, Herve Ginisty, George Kraal, Eng M. Tan, and Ben Valdez donated antibodies; John Aris donated human cDNA of fibrillarin; Mike Pollard donated murine cDNA of fibrillarin; Dieter Moosmayer and Jörg Rippmann provided us with plasmid pEA12-H398. This work was supported by Deutsche Forschungsgemeinschaft through SFB 503, and Deutsche Stiftung für Sklerodermie (DSS). M.C. was supported by Hochschulsonderprogramm III, and German Ministry of Education and Science through Rheumatology Competence Network.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–05–0083. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–05–0083.
REFERENCES
- Amsterdam A, Pitzer F, Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc Natl Acad Sci USA. 1993;90:99–103. doi: 10.1073/pnas.90.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton LC, et al. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett FC, Reveille JD, Goldstein R, Pollard KM, Leaird K, Smith EA, Leroy EC, Fritzler MJ. Autoantibodies to fibrillarin in systemic sclerosis (scleroderma). An immunogenetic, serologic, and clinical analysis. Arthritis Rheum. 1996;39:1151–1160. doi: 10.1002/art.1780390712. [DOI] [PubMed] [Google Scholar]
- Benavente R, Reimer G, Rose KM, Hügle-Dörr B, Scheer U. Nucleolar changes after microinjection of antibodies to RNA polymerase I into the nucleus of mammalian cells. Chromosoma. 1988;97:115–123. doi: 10.1007/BF00327368. [DOI] [PubMed] [Google Scholar]
- Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski UH, Kloetzel PM. Interferon γ stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2001;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Chen M, von Mikecz A. Specific inhibition of rRNA transcription and dynamic re-location of fibrillarin induced by mercury. Exp Cell Res. 2000;259:225–238. doi: 10.1006/excr.2000.4923. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann B, Kuehn L, Rutschmann M, Reinauer H. Purification and characterization of a multicatalytic high-molecular-mass proteinase from rat skeletal muscle. Biochem J. 1985;228:161–170. doi: 10.1042/bj2280161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SD, Liu LF, Vazquez-Abad D, D'Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- Everett RD, Earnshaw WC, Finlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Finch RA, Revankar GR, Chan PK. Nucleolar localization of nucleophosmin/B23 requires GTP. J Biol Chem. 1993;268:5823–5827. [PubMed] [Google Scholar]
- Finley D. Ubiquitin chained and crosslinked. Nat Cell Biol. 2002;4:E121–E123. doi: 10.1038/ncb0502-e121. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Galperin C, Gershwin ME. Systemic sclerosis (scleroderma) In: Rose NR, Mackay IR, editors. The Autoimmune Diseases. San Diego: Academic Press; 1998. pp. 119–126. [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, von Mikecz A. Antinuclear antibodies (ANA): fluorescent highlights on structure and function in the nucleus. Int Arch Allergy Immunol. 2000;123:16–27. doi: 10.1159/000024420. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hügle B, Kleinschmidt JA, Franke WW. The 22S cylinder particles of Xenopus laevis. II. Immunological characterization and localization of their proteins in tissues and cultured cells. Eur J Cell Biol. 1983;32:157–163. [PubMed] [Google Scholar]
- Hultman P, Eneström S, Pollard KM, Tan EM. Anti-fibrillarin antibodies in mercury-treated mice. Clin Exp Immunol. 1989;78:470–477. [PMC free article] [PubMed] [Google Scholar]
- Hultman P, Bell LJ, Enestrom S, Pollard KM. Murine susceptibility to mercury. I. Autoantibody profiles and systemic immune deposits in inbred, congenic, and intra-H-2 recombinant strains. Clin Immunol Immunopathol. 1992;65:98–109. doi: 10.1016/0090-1229(92)90212-7. [DOI] [PubMed] [Google Scholar]
- Jacobson DL, Gange SJ, Rose NR, Graham NMH. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol. 1997;84:223–243. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- Jordan P, Carmo-Fonseca M. Cisplatin inhibits synthesis of ribosomal RNA in vivo. Nucleic Acids Res. 1998;26:2831–2836. doi: 10.1093/nar/26.12.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetzel P. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–188. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- Kono DH, Balomenos D, Pearson DL, Park MS, Hildebrandt B, Hultman P, Pollard KM. The prototypic Th2 autoimmunity induced by mercury is dependent on IFN-γ and not Th1/Th2 imbalance. J Immunol. 1998;161:234–240. [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML/retinoic acid receptor α degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Lomonte P, Sullivan K, Everett RD. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J Biol Chem. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- Metlay IP, Witmer Pack MD, Agger R, Crowley MT, Lawless D, Steinman RM. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J Exp Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ishihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Neefjes JJ, Momburg F, Hämmerling GJ. Selective and ATP-dependent translocation of peptides by the MHC-encoded transporter. Science. 1993;261:769–771. doi: 10.1126/science.8342042. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Peters JM, Franke WW, Kleinschmidt JA. Distinct 19S and 20S subcomplexes of the 20S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- Rao S, Porter DC, Chen X, Herliczek T, Lowe M, Keyomarsi K. Lovastatin-mediated G1 arrest is through inhibition of the proteasome, independent of hydroxymethyl glutaryl-CoA reductase. Proc Natl Acad Sci USA. 1999;96:7797–7802. doi: 10.1073/pnas.96.14.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Busch H, Tan EM. Monoclonal antibody from a (New Zealand black x New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Reits EAJ, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits EA, Vos JC, Gromme M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- Reuter R, Tessars G, Vohr H-W, Gleichmann E, Lührmann R. Mercuric chloride induces autoantibodies against U3 small nuclear ribonucleoprotein in susceptible mice. Proc Natl Acad Sci USA. 1989;86:237–241. doi: 10.1073/pnas.86.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippmann JF, Klein M, Hoischen C, Brocks B, Rettig WJ, Gumpert J, Pfizenmaier K, Mattes R, Moosmayer D. Procaryotic expression of single-chain variable-fragment (scFv) antibodies: secretion in L-form cells of Proteus mirabilis leads to active product and overcomes the limitations of periplasmic expression in Escherichia coli. Appl Environ Microbiol. 1998;64:4862–4869. doi: 10.1128/aem.64.12.4862-4869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivett AJ. Intracellular distribution of proteasomes. Curr Opin Immunol. 1998;10:110–114. doi: 10.1016/s0952-7915(98)80040-x. [DOI] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Scheer U, Benavente R. Functional and dynamic aspects of the mammalian nucleus. BioEssays. 1990;12:14–21. doi: 10.1002/bies.950120104. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz J. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Sprent J, Kishimoto H. The thymus and central tolerance. Philos Trans R Soc Lond B Biol Sci. 2001;356:609–616. doi: 10.1098/rstb.2001.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber WT, Fritz VK, Maltin CA, Dahlmann B. Localization of a multicatalytic, high-molecular mass proteinase in the nuclei of muscle cells. Histochem J. 1987;19:594–597. doi: 10.1007/BF01687368. [DOI] [PubMed] [Google Scholar]
- Tan ET. Antinuclear antibodies: diagnostic markers for autoimmune disease and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-RNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Turley SJ, Tan EM, Pollard KM. Molecular cloning and sequence analysis of U3 snoRNA-associated mouse fibrillarin. Biochim Biophys Acta. 1993;1216:119–122. doi: 10.1016/0167-4781(93)90046-g. [DOI] [PubMed] [Google Scholar]
- Verma R, Deshaies RJ. A proteasome howdunit: the case of the missing signal. Cell. 2000;101:341–344. doi: 10.1016/s0092-8674(00)80843-0. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- von Mikecz A, Neu E, Krawinkel U, Hemmerich P. Human ribosomal protein L7 carries two nucleic acid-binding domains with distinct specificities. Biochem Biophys Res Commun. 1999;258:530–536. doi: 10.1006/bbrc.1999.0682. [DOI] [PubMed] [Google Scholar]
- von Mikecz A, Zhang S, Montminy M, Tan EM, Hemmerich P. CBP/p300 and RNA-polymerase II colocalize in transcriptionally active domains in the nucleus. J Cell Biol. 2000;150:265–274. doi: 10.1083/jcb.150.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]