Abstract
CNG channels are cyclic nucleotide-gated Ca2+-permeable channels that are suggested to be involved in the activity-dependent alterations of synaptic strength that are thought to underlie information storage in the CNS. In this study, we isolated an endogenous RNA transcript antisense to CNGα1 mRNA. This transcript was capable of down-regulating the expression of sense CNGα1 in the Xenopus oocyte expression system. RT-PCR, Northern blot, and in situ hybridization analyses showed that the transcript was coexpressed with CNGα1 mRNA in many regions of human brain, notably in those regions that were involved in long-term potentiation and long-term depression, such as hippocampal CA1 and CA3, dentate gyrus, and cerebellar Purkinje layer. Comparison of expression patterns between adult and fetal cerebral cortex revealed that there were concurrent developmental changes in the expression levels of anti-CNG1 and CNGα1. Treatment of human glioma cell T98 with thyroid hormone T3 caused a significant increase in anti-CNG1 expression and a parallel decrease in sense CNGα1 expression. These data suggest that the suppression of CNGα1 expression by anti-CNG1 may play an important role in neuronal functions, especially in synaptic plasticity and cortical development. Endogenous antisense RNA-mediated regulation may represent a new mechanism through which the activity of ion channels can be regulated in the human CNS.
INTRODUCTION
Cyclic nucleotide-gated (CNG) channels are Ca2+-permeable nonselective cation channels that exist as heteromeric complexes consisting of α and β subunits. Four distinct α subunits and two β subunits have been identified. α1–3 may form functional channels on their own. α4 and two β subunits do not exhibit channel properties themselves but are able to modify the channel properties (Kaupp, 1995; Finn et al., 1996; Zagotta and Siegelbaum, 1996). CNG channels have fairly widespread tissue distribution, including sensory neurons, CNS, heart, kidney, blood vessels, and spleen. In the CNS, electrophysiological and molecular biological evidence has demonstrated that CNG channels are present in many regions of rat brain, including hippocampus, cerebral cortex, and cerebellum (Ahmad et al., 1994; Kingston et al., 1996; Bradley et al., 1997; Samanta Roy and Barnstable, 1999; Strijbos et al., 1999). Research has linked the nitric oxide (NO)–cGMP pathway to the CNG channel activity in neurons (Ahmad et al., 1994). CNG channels may act as one of the downstream effectors for the NO–cGMP pathway, modulating neurotransmitter release and causing the activity-dependent alterations of synaptic strength that are thought to underlie information storage, such as long-term potentiation (LTP) and long-term depression (LTD). In addition to their proposed role in synaptic plasticity, CNG channels may also play an important role in brain development. Evidence suggests that CNG channels may control axon guidance (Coburn and Bargmann, 1996) and influence cortical dendritic outgrowth in the development of the CNS (Samanta Roy and Barnstable, 1999).
An important feature of CNG channels is their Ca2+-permeability (Finn et al., 1996; Dzeja et al., 1999). CNG channels open in response to cyclic nucleotides and link cGMP/cAMP signaling to Ca2+ homeostasis. Activation of CNG channels would raise cytosolic Ca2+ levels, and this could trigger secondary pathways that contribute to short-term and long-term alterations in neural functions (Zufall et al., 1997). Substantial amounts of data are available regarding the modulation of CNG channel activity by cellular factors, including phosphorylation enzymes, Ca2+/calmodulin, and diacylglycerol (Hsu and Molday, 1993; Molokanova et al., 1997; Crary et al., 2000). However, the regulation of CNG channels at the gene transcription and/or protein translation levels is largely unknown. In this study, we report the isolation of a cDNA clone representing an endogenous antisense transcript against the mRNA of human CNGα1 channels. This endogenous antisense transcript is expressed in many regions of human brain, and it may down-regulate the sense CNGα1 channels by suppressing the amount of channel proteins.
MATERIALS AND METHODS
Library Screening
A cDNA fragment for CNGα1 (underlined by dots in Figure 1) was amplified by RT-PCR using total RNA isolated from the human epithelial cell line ECV304. The primers used for PCR were (+)TTGGTCCACAGGTAGTC and (−) TCATCATTATCCACTGGAA. The fragment was labeled by random primer extension with α-32P-labeled dCTP (Amersham), and it was then used to screen a commercial human cDNA library that was primed with oligo-dT and unidirectionally cloned in pCMV-SPORT1 (Life Technologies-BRL). The hybridizations were performed on Hybond nylon membranes in Rapid-Hyb buffer (Amersham) at 55°C overnight. The membranes were washed in 2× SSC/0.1% SDS at room temperature for 1 h, followed by 0.2× SSC/0.1% SDS at 42°C for 45 min as described previously (Yao et al., 1995). Positive clones were verified with Southern blot using the same CNGα1-specific probe. A 1.66-kilobase (kb) clone was isolated, and the nucleotide sequence was analyzed by Sanger sequencing with Sequenase (US Biochemicals).
Figure 1.
Nucleotide sequence of human anti-CNG1 RNA transcript. mRNA sequence is deduced from the DNA sequence of a cDNA clone isolated from adult human brain cDNA library. The probe used for library screening was derived from the cDNA fragment underlined by dots. Amino acid sequence of a putative translation product is illustrated. Bold, the 1311-bp overlap between the sense and antisense transcripts; double underlines, potential poly(A) signals; thick underlines, primer sites for RT-PCR of Figure 3A.
Reverse Transcription–Polymerase Chain Reaction
Total RNAs were isolated at autopsy from adult human brain tissue and human glioma cell lines by the acid guanidinium thiocyanate method (Chomczynski and Sacchi, 1987). One of the human brain total RNA samples was a DNase-treated sample purchased from Invitrogen. RNAs were transcribed into first-strand cDNAs using Superscript reverse transcriptase (Life Technologies-BRL). Anti-CNG1–specific primers [(+)GATGACGATATACATAACAAGG and (−)CTCAGCAGAATATTTTCTACAGCC] were used to amplify a 450-base pair (bp) PCR fragment. The primer sites are shown in Figure 1. PCR reactions of 100 μl contained 1 μl of the first-strand cDNA, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTP, 1.0 μM primers, and 2.5 U Taq DNA polymerase (Life Technologies-BRL). Thirty cycles (94°C for 1 min, 50°C for 1 min, 72°C for 1 min) were performed with a Robocycler (Stratagene). Negative controls applied the same samples and experimental procedures except that the step of reverse transcription was omitted. The amplified PCR products were sequenced by ABI 310 autosequencer (Perkin Elmer-Cetus). Human kidney and liver cDNA libraries were purchased from Life Technologies-BRL.
Hormone Effect
T98 cells were treated with 1 μM T3 (Calbiochem) or 100 ng/ml human growth hormone (Calbiochem) for 4 d in a culture medium consisting of 90% Ham's F-12 and 10% fetal calf serum (Life Technologies). Total RNAs were isolated thereafter for semiquantitative RT-PCR assays. The primers used for anti-CNG1 detection were (+)GGTATCAGTGACAGAACATCAA and (−)TACAGCCATAGGTTTATTAGTAT. The primers used for sense CNGα1 detection were (+)GAATTTGGCCGTTTGGCTAG and (−)CGTTGATGGCAATTTCTGCT. PCR reactions of 50 μl contained 1 μl of the first-strand cDNA, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTP, 1.0 μM primers, and 2.5 U Taq DNA polymerase (Life Technologies-BRL). Thirty-five cycles (94°C for 1 min, 55°C for 1 min, 72°C for 1 min) were performed with a Robocycler (Stratagene). For anti-CNG1, a single PCR product of 271 bp was amplified. For sense CNGα1, a single amplified product of 386 bp was detected. The authenticity of the amplified products was confirmed by ABI 310 autosequencer. Equal volumes of the PCR products were loaded onto a 1%-agarose gel and stained with ethidium bromide. The intensity of the bands was analyzed by FluorChem 8000 imaging system (Alpha Innotech). As a control for analysis, we used the expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a normalizing basis for comparison. The primers for GAPDH were (+)ACCACAGTCCATGCCATCAC and (−)TCCACCACCCTGTTGCTGTA.
Northern Blot
A 211-bp DNA fragment near the 3′ end of anti-CNG1 (from position 1308 to 1518 as in Figure 1, or P2 as in Figure 2) was subcloned into pCMVsport1 and then in vitro transcribed into a 32P-labeled riboprobe antisense to anti-CNG1 with the T7 MAXIscript transcription kit (Ambion). The molecular size of synthesized riboprobe was confirmed by gel electrophoresis. This strand-specific riboprobe was used to hybridize with a Human Brain Multiple Tissue mRNA Blot (ClonTech) at 60°C overnight with ExpressHyb. The blot was then washed twice in 2× SSC/0.5% SDS for 45 min, followed by 2× 20-min washes in 0.1× SSC/0.1% SDS at room temperature, and then exposed to x-ray film overnight. The Multiple Tissue mRNA Blot was reprobed with human β-actin gene (Clontech) to demonstrate that equal amounts of poly(A) RNA was loaded onto each lane.
Figure 2.
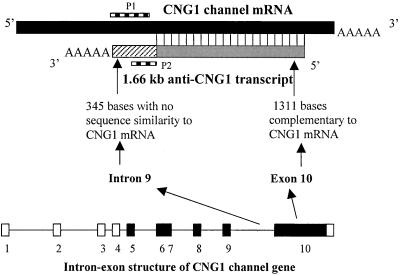
Gene structure of anti-CNG1. Top, schematic illustration of the nucleotide complementarity of anti-CNG1 to sense CNGα1. Black bar, sense CNGα1; gray bar, region of anti-CNG1 complementary to sense CNGα1; hatched bar, region of anti-CNG1 lacking complementarity to sense CNGα1. P1, the region in which the CNGα1-specific riboprobe for in situ hybridization (Figures 5 and 6) was derived; P2, the region in which anti-CNG1–specific riboprobe for Northern blot (Figure 3B) and in situ hybridization (Figures 5 and 6) was derived. Bottom, intron-exon structure of CNGα1 channel gene. Black bars, coding regions; white bars, 5′ and 3′ untranslated regions; lines, introns. Anti-CNG1 is transcribed from the region of intron 9 and exon 10 of CNGα1 channel gene but in reverse orientation.
Cloning of Sense CNGα1 Gene
Sense CNGα1 was amplified by RT-PCR using total RNA isolated from the human epithelial cell line ECV304. The PCR primers [(+)TCCATGAACAATATTATCAAT and (−)TCAAAAGGATCATGAGGCAT] were designed on the basis of the published nucleotide sequence of human CNGα1 mRNA (GenBank Accession Number NM 000087) (Dhallan et al., 1992). The amplified PCR product of 2112 bp was cloned into pPCR-ScriptAmp cloning vector (Stratagene). The amplified PCR products were sequenced by ABI 310 autosequencer (Perkin Elmer-Cetus). The DNA sequencing confirmed that the clone represented authentic human CNGα1.
Oocyte Expression
Anti-CNG1, CNGα1, and Kv1.5 were subcloned into pgh21 vector and then expressed in Xenopus oocytes by microinjecting in vitro transcribed cRNAs as previously described (Yao et al., 1995). For CNGα1, oocyte membrane was clamped at −100 mV. For anti-CNG1 suppression studies, 25 ng of anti-CNG1 cRNA was injected into oocytes 2 h before the injection of sense CNGα1 cRNA. For Kv1.5, the outward currents were elicited by two-microelectrode voltage clamp using 800-ms pulses of +80 mV from a holding potential of −80 mV. The experimental bath contained 88 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 2.5 mM NaH2CO3, and 5 mM HEPES, pH 7.4. Expressed currents were measured with OC-725 oocyte clamp 2 days after cRNA injection as described previously (Yao et al., 1995). Measured currents were analyzed using Pulse and Pulse-fit software (Heka Lambretch, Germany).
In Situ Hybridization
The same 211-bp DNA fragment as used in Northern blot (illustrated as P2 in Figure 2) was in vitro transcribed into DIG-labeled RNA probes with a DIG-labeling kit (Roche Biochemicals). The strand complementary to anti-CNG1 was used to detect anti-CNG1, and the sense strand was used as control. For the detection of sense CNGα1 transcript, a 259-bp DNA fragment illustrated in Figure 2 as P1 was subcloned into pPCR-ScriptAmp cloning vector (Stratagene) and then in vitro transcribed into DIG-labeled riboprobes. The strand complementary to CNGα1 was used to detect CNGα1 mRNA, and the sense strand was used as control. These probes were used to hybridize with the sections cut from the brain tissues embedded in paraffin. The brain tissues of human adult and 15-wk-old fetus were from autopsy cases with the consent of family members and the approval of the university clinical research ethics committee. Tissues were fixed overnight with 4% paraformaldehyde in phosphate-buffered saline (PBS). The postmortem delay was ∼7 h. The tissues were dehydrated through graded ethanol, cleared with xylene, and embedded in Parafilm, and 6-μm-thick sections were prepared. After dewaxing and hydration, the sections were washed briefly with diethylpyrocarbonate-treated water followed by PBS for 10 min. They were then digested with Proteinase K (10 μg/ml) at 37°C for 15 min. Hybridization was performed at 48°C in a hybridization buffer containing 4× SSC, 10% dextran sulfate, 1× Denhardt's solution, 5 mM EDTA, 0.1% CHAPS, 50% deionized formamide, 200 μl/ml herring sperm DNA, and 200 ng/ml DIG-labeled probe (Yew et al., 1999). The slides were then washed four times for 15 min each in 2× SSC/0.1% SDS and then twice for 15 min each in 0.2× SSC/0.1% SDS at 42°C. Colorimetric detections were performed using an anti-DIG antibody conjugated to alkaline phosphatase followed by incubation with NBT/BCIP color substrates using a digoxygenin-nucleic acid detection kit (Roche, Germany) as described previously (Yew et al., 1999). For anti-CNG1, positive signals appeared ∼5 min after incubation in NBT/BCIP color substrate solution. For the detection of sense CNGα1, ∼30 min was needed for the color development in NBT/BCIP color substrate solution.
RESULTS
A CNGα1-related cDNA clone was isolated by screening an adult human brain cDNA library with a DNA probe specific for CNGα1. Nucleotide sequence of the isolated cDNA clone was analyzed by Sanger's sequencing. Because the library was a commercial cDNA library that was primed with oligo-dT and unidirectionally cloned in pCMVsport1, we were able to determine the 5′-3′ orientation of the clone on the basis of the location of the 18-base oligo(A) (Figure 1). Comparison of nucleotide sequence of this clone with that of human CNGα1 mRNA (GenBank Accession Number NM 000087) revealed that a 1311-bp region at the 5′ portion of this clone was complementary to CNGα1 mRNA, whereas the rest of the clone (345 bp at the 3′ end) had no similarity to CNGα1 mRNA (Figure 2). Therefore, this transcript represented a natural antisense transcript complementary to CNGα1 mRNA. We named this transcript anti-CNG1. Comparison of the nucleotide sequence of anti-CNG1 with that of human chromosome 4 showed that the 345 bp at the 3′ end of the clone was actually transcribed from an immediate downstream region following the CNGα1-coding 1311-bp upstream region in chromosome 4 between 4p12 and the centromere (Figure 2). It should be noted that anti-CNG1 and CNGα1 were transcribed in the same locus but in reverse orientation. On the basis of the published intron-exon structure of human CNGα1 gene (Dhallan et al., 1992), the transcription of anti-CNG1 started from exon 10 of CNGα1 and extended into intron 9. The isolated clone might contain a genuine 3′ end, because oligo(A) was located at the end and multiple AAUAAA motifs, the signal for processing of mRNA at the 3′ end (Nevins, 1983), could be found near the A-rich region. It was not clear whether this transcript might encode any protein, but the longest possible open reading frame was only 243 bp long, which is equivalent to 81 amino acids. The putative amino acid sequence from this open reading frame is illustrated in Figure 1. Blast GenBank search with this putative protein showed no significant similarity to any known protein.
A suspicion could be raised that the isolated clone might represent a cloning artifact caused by a piece of cDNA inserted in the wrong direction. But this was unlikely, because a detailed sequence analysis of the clone showed that the 3′ end of the clones contained an intact oligo(dT) that was flanked by a NotI restriction site specially designed for unidirectional cloning, and this NotI restriction site was followed immediately by a pCMV-SPORT1 vector sequence. In addition, the 5′ end of the clone contained a complete EcoRI adaptor specially designed for unidirectional cloning, and this EcoRI site was followed immediately by a pCMV-SPORT1 vector sequence. The correct insertion pattern argued against false insertion. The clone was certainly not a result of the false fusion of cDNA for CNG1 with another cDNA encoding different protein, because the analyses of genomic DNA showed that the cDNA was continuously transcribed from a single locus (Figure 2).
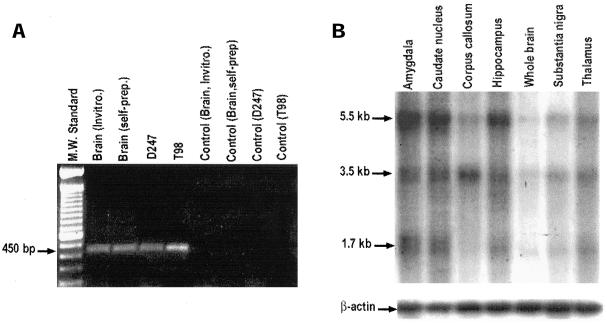
RT-PCR was used to examine the expression pattern of this endogenous anti-CNG1 transcript. PCR primers were carefully designed so that the targeted 450-bp amplification products extended across the boundary of intron 9 and exon 10 of CNGα1 (Figure 1). In this way, only anti-CNG1 but not CNGα1 could be amplified. RT-PCR experiments revealed the expression of anti-CNG1 in adult human brain and two human glioma cell lines, T98 and D247 (Figure 3A). No expression could be detected in the total RNA samples isolated from human adult stomach tissue and rat pheochromocytoma cell line PC12 (data not shown). DNA sequencing confirmed that the amplified PCR products represented the authentic anti-CNG1. The amplified anti-CNG1 did not originate from the residual genomic DNA contaminations in total RNA samples, because PCR reactions alone without reverse transcription did not produce any detectable product (Figure 3A). Furthermore, the same anti-CNG1 product of ∼450 bp could be amplified by RT-PCR from a commercially available DNase-treated total RNA sample (Figure 3A). In addition to brain, anti-CNG1 transcript was also expressed in kidney and liver, because the same 450-bp product could be amplified by PCR from commercially available human cDNA libraries generated from kidney and liver (data not shown).
Figure 3.
Detection of anti-CNG1 transcripts by RT-PCR and Northern blot. (A) RT-PCR detected the expression of anti-CNG1 in total RNAs prepared from brain tissue, glioma cell lines D247 and T98, and a commercial DNase-treated human brain total RNA (Invitrogen). Reverse transcription step was omitted in negative controls. (B) Multiple Tissue Northern blot analysis detected three anti-CNG1–related transcripts with molecular size of 5.5, 3.5, and 1.7 kb in many human brain regions. Each lane contained 2 μg poly(A) RNA. The probe for Northern blot was a strand-specific riboprobe derived from P2 region as labeled in Figure 2. The blot was reprobed with human β-actin gene to demonstrate that equal amounts of poly(A) RNA were loaded onto each lane.
Northern blot analysis was used to further examine the expression of anti-CNG1 in brain. A human Multiple Tissue mRNA Blot was hybridized with a 32P-labeled riboprobe specific for anti-CNG1. Three different transcripts with molecular sizes of 5.5, 3.5, and 1.7 kb were detected in poly(A) RNAs from different brain regions (Figure 3B). The 1.7-kb transcript agreed well with the size of anti-CNG1 we isolated, and this transcript was expressed in amygdala, caudate nucleus, hippocampus, and thalamus. Weak hybridization signals could also be observed in substantia nigra and whole brain tissue. Large transcripts of 5.5 and 3.5 kb were expressed more abundantly in amygdala, caudate nucleus, and hippocampus. These large transcripts might represent alternatively spliced forms of anti-CNG1. It should be noted that the 211-bp riboprobe used for hybridization was derived from a region close to the 3′ end of anti-CNG1 transcript (Figure 2). This probe was highly specific for anti-CNG1, because BLAST GenBank search with anti-CNG1 did not reveal significant similarity to any known gene. A point worth mentioning was that the riboprobe was strand-specific. It could recognize only the transcripts containing anti-CNG1 but not those containing the complementary strand.
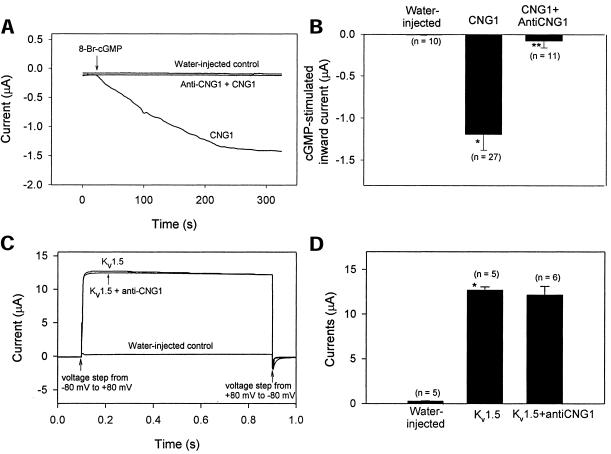
To examine whether the isolated anti-CNG1 was capable of down-regulating sense CNGα1, in vitro transcribed anti-CNG1 cRNA was microinjected into Xenopus oocytes to determine whether it could suppress the expression of sense CNGα1 currents. We used membrane-permeable 8-Br-cGMP to activate sense CNGα1 channels in the oocytes injected with CNGα1 cRNA. In Xenopus oocytes injected with 5 ng of CNGα1 cRNA, activation of CNGα1 channels by 8-Br-cGMP initiated inward currents (Figure 4, A and B). Conversely, 8-Br-cGMP had no effect in the control oocytes, which did not receive the injection of CNGα1 cRNA. Importantly, cGMP-activated inward currents in CNGα1-injected oocytes were abolished if the oocytes were preinjected with 25 ng anti-CNG1 cRNA 2 h before the injection of CNGα1 (Figure 4, A and B). This suppression was not caused by any nonspecific effect associated with the injection of anti-CNG1 transcript, because in control experiments, the expression of a voltage-gated potassium channel, Kv1.5, was not affected by anti-CNG1 transcript (Figure 4, C and D).
Figure 4.
The suppression of CNGα1-mediated inward currents by anti-CNG1 cRNA. (A) cGMP stimulated the Ca2+-dependent Cl− inward currents in Xenopus oocytes that were injected with 5 ng of CNGα1 cRNA. The currents were abolished by prior injection of 25 ng anti-CNG1 cRNA. Each trace is a representative experiment. Membrane was clamped at −100 mV. (B) Average amplitude of cGMP-activated Cl− currents from different experiments. (C) Oocytes injected with 0.5 ng of Kv1.5 cRNA expressed voltage-dependent outward currents. The Kv1.5 currents were not affected by prior injection of 25 ng anti-CNG1 cRNA. (D) Average amplitude of Kv1.5 currents from different experiments. *p<0.01 compared with the water-injected control; **p<0.01 compared with the CNGα1 injection alone.
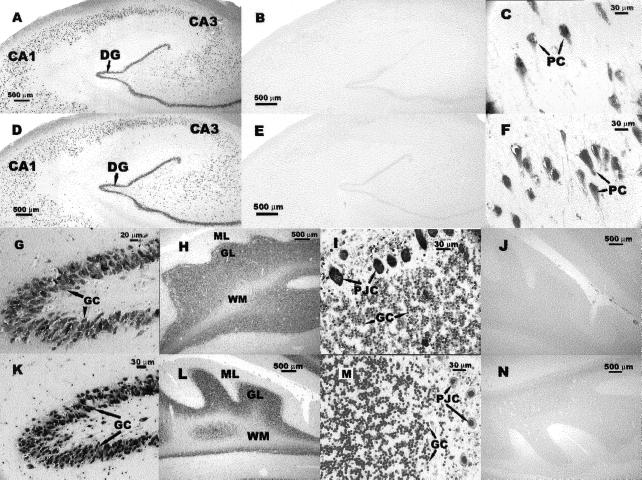
In situ hybridizations were performed to examine the expression of anti-CNG1 and sense CNGα1 mRNA in different human brain regions. DIG-labeled anti-CNG1 riboprobe for in situ hybridization contained the same anti-CNG1–specific 211-base nucleotides as that used in Northern blot. The 259-base riboprobe for CNGα1 was derived from a region of CNGα1 with no complementarity to anti-CNG1 (Figure 2). Our data showed that the expression of anti-CNG1 was widespread in adult human CNS. This expression could be observed in pyramidal neurons of hippocampal CA1 and CA3 of Ammon's horn (Figure 5, A and C), granule neurons in dentate gyrus (Figure 5, A and G), and Purkinje cells and granular cells in cerebellum (Figure 5, H and I). Similar expression patterns were observed for sense CNGα1 (Figure 5, D and F–M). Very little hybridization signal could be seen in the experiments with control riboprobes (Figure 5, B, E, J, and N). It was notable that virtually every visible hippocampal pyramidal, dentate granule, and cerebellar Purkinje neuron was labeled with both anti-CNG1 and sense CNGα1 probes. Two additional sets of DIG-labeled riboprobes, one covering the region from position 1–667 and the other covering the region from position 567 to 952 of anti-CNG1 (the positions were as labeled in Figure 1), were used for in situ hybridization. Because these two probes covered the regions in which both anti-CNG1 and sense CNGα1 were transcribed, as expected, we were able to detect the hybridization signals with the probes generated from both sense and antisense strands (data not shown).
Figure 5.
Detection of anti-CNG1 and sense CNGα1 transcripts in the hippocampus and cerebellum of adult human brain by in situ hybridizations. Anti-CNG1 expression was observed in hippocampus (A, C, G) and cerebellum (H, I). B and J are the control experiments for anti-CNG1 in hippocampus and cerebellum, respectively. Sense CNGα1 expression was detected in hippocampus (D, F, K) and cerebellum (L, M). E and N are the control experiments for CNGα1 in hippocampus and cerebellum, respectively. (C and F) High-magnification pictures for hippocampal CA1 region; G and K, high-magnification pictures for dentate gyrus; I and M, high-magnification pictures for cerebellar Purkinje/granular layer. DG, dentate gyrus; PC, pyramidal cells; GC, granule cells; WM, white matter; PJC, Purkinje cells.
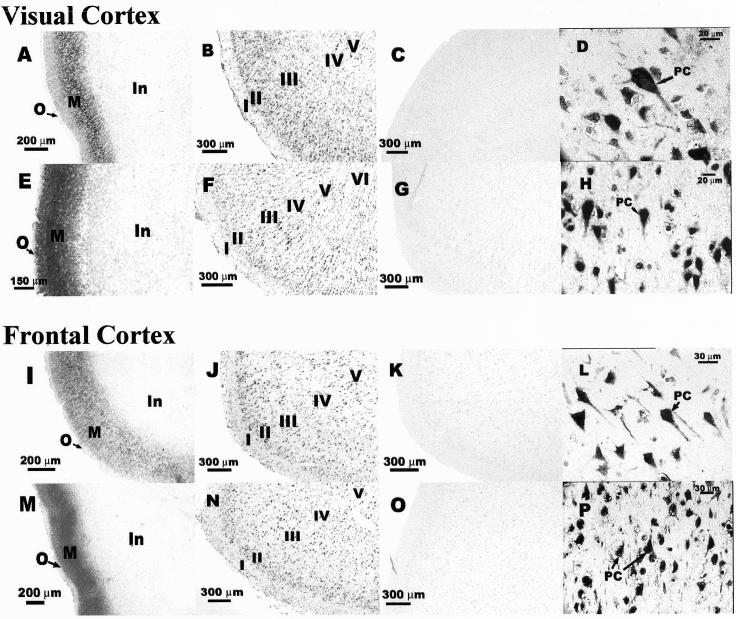
Because CNG channels were suggested to play a crucial role in cortical development (Samanta Roy and Barnstable, 1999), attempts were made to study the developmental changes of anti-CNG1 and sense CNGα1 in cerebral cortex. We found that in fetal cortex, the expression of anti-CNG1 was observed predominantly in a cortical layer adjacent to the outermost layer (Figure 6, A and I). Fetal cortex at 15 wk old was still not well differentiated and could not be divided into classic six-layered cortical structures. But on the basis of the anatomical location, these anti-CNG1–expressing neurons should correspond to the layer II and layer III cells of the adult cortex. Unlike fetal cortex, the expression of sense anti-CNG1 in adult cortex was more widespread and could be found in almost all cortical layers except layer I (Figure 6, B and J). High-magnification pictures showed that anti-CNG1 transcript was expressed in the soma as well as in the primary dendrite of pyramidal and granule neurons (Figure 6, D and L). Very little stain was observed in cortical layer I, which contained primarily dendrites and axons of cortical neurons. Similar expression patterns were observed for sense CNGα1. The expression was located predominantly in a narrow layer in fetal cortex (Figure 6, E and M), but it was widespread and existed in almost all cortical layers except layer I in adult cortex (Figure 6, F and N). High-magnification pictures showed that both soma and primary dendrite of cortical neurons were stained (Figure 6, H and P). No hybridization signal could be seen for the experiments with control riboprobes (Figure 6, C, G, K, and O).
Figure 6.
Developmental changes of anti-CNG1 and sense CNGα1 expression in human cerebral cortex. (A–D) Anti-CNG1 expression in visual cortex; (E–H) CNGα1 expression in visual cortex; (I–L) anti-CNG1 expression in frontal cortex; (M–P) CNGα1 expression in frontal cortex. A, E, I, and M show samples of fetal cerebral cortex; B, F, J, and N show samples of adult cerebral cortex. D, H, L, and P are high-magnification pictures taken from adult cortical layer III/II; C, G, K, and O show control experiments. O, outermost layer; M, middle layer; In, inner layer; I–VI, cortical layers I to VI; PC, pyramidal cells.
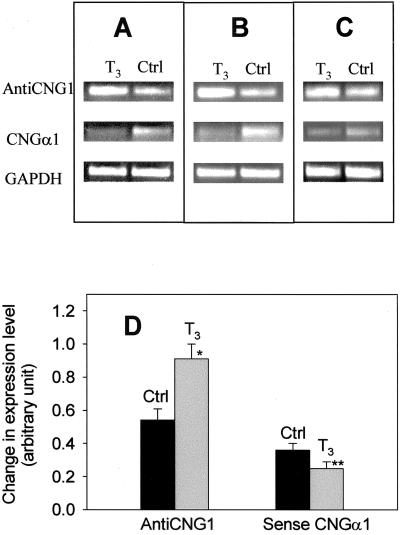
We also explored the possible endogenous factor(s) that could regulate the expression level of anti-CNG1. As shown in Figure 7, treatment of human glioma cell line T98 with thyroid hormone T3 (1 μM) caused a significant increase in the expression level of anti-CNG1. The same treatment reduced the expression of sense CNGα1. Thyroid hormone is a well-recognized agent involved in brain development and maturation. The deficiency of thyroid hormone during critical periods of development is associated with irreversible mental retardation and profound neurological defects, including deafness and movement disorders (Dussault and Ruel, 1987; Oppenheimer and Schwartz, 1997). The effect of T3 on the expression of anti-CNG1 and sense CNGα1 was consistent with the hypothesis that anti-CNG1 might play a critical role in down-regulating the expression of sense CNGα1 during brain development. We also tested the effect of human growth hormone. Human growth hormone at 100 ng/ml had no effect on the expression of either anti-CNG1 or sense CNGα1 in T98 cells (data not shown).
Figure 7.
The effect of thyroid hormone (T3) on the expression of anti-CNG1 and sense CNGα1 in human glioma cell T98 as detected by RT-PCR. (A–C) Three independent experiments. Top row, the expression of anti-CNG1; middle row, the expression of sense CNGα1; bottom row, the expression of GAPDH as control (Ctrl). T3 treatment increased the expression of anti-CNG1 but suppressed the expression of sense CNGα1. D, Relative expression. The intensity of the bands for anti-CNG1 and sense CNGα1 was divided by the intensity of the bands for the internal marker GAPDH (anti-CNG1/GAPDH or sense CNGα1/GAPDH). Values are means ± SEM (n = 8). *p<0.01 compared with control. **p<0.05 compared with control.
DISCUSSION
Endogenous natural antisense RNAs are widely distributed in the prokaryotic and eukaryotic world. Numerous natural antisense RNAs have also been reported in mammals, including rats, mice, cows, and human (for review, see Dolnick, 1997; Simons, 1988; Vanhee-Brossollet and Vaquero, 1998). These antisense RNAs are complementary to sense transcripts encoding proteins involved in extremely diverse biological functions: hormonal response, proliferation control, development, etc. In fact, it is believed that endogenous antisense regulation may be part of a general mechanism in the control of gene expression (Wutz et al., 1997; Simons, 1988; Vanhee-Brossollet and Vaquero, 1998; Kimelman and Kirschner, 1989; Thrash-Bingham and Tartof, 1999). Several best-characterized examples in eukaryotes include a native antisense RNA to basic fibroblast growth factor in Xenopus (Kimelman and Kirschner, 1989), an antisense RNA to Igf2r (the receptor for insulin-like growth factor type-2) in mice (Wutz et al., 1997), an antisense RNA to HIF1α (hypoxia inducible factor α) in humans (Thrash-Bingham and Tartof, 1999), and an antisense RNA to eb4-psv gene in Dictyostelium (Hildebrandt and Nellen, 1992). In this study, we isolated a cDNA clone representing an endogenous antisense transcript against the mRNA of human CNGα1 channels. The presence of anti-CNG1 transcript(s) was verified by RT-PCR, Northern blot hybridization, and in situ hybridization. The expression of the transcript(s) could be found in many regions of human brain (Figures 3 and 5) as well as in human glioma cell lines T98 and D247 (Figure 3A). Interestingly, the molecular size of the isolated anti-CNG1 cDNA clone matched that of one of the transcripts detected on the Northern blot (1.7 kb), suggesting that the isolated anti-CNG1 clone might represent a complete transcript. This study represents the first demonstration of an endogenous antisense RNA transcript against any ion channel.
A unique function of antisense transcripts is to regulate the expression of their sense counterparts. It has been recognized that antisense transcripts may hybridize with their complementary sense mRNAs to form RNA–RNA duplexes. These duplexes can be digested by the RNases that are specific for double-stranded RNA. Alternatively, because of the structural similarity between the sense and antisense transcripts, antisense transcripts may down-regulate the sense gene by depriving sense mRNAs from the proteins necessary for their functions (Vanhee-Brossollet and Vaquero, 1998). To test whether the isolated 1.66-kb anti-CNG1 transcript was capable of down-regulating the expression of sense CNGα1, we used the Xenopus oocyte expression system. Xenopus oocytes have endogenous Ca2+-dependent Cl− channels that are sensitive to the Ca2+ concentration beneath the plasma membrane. Therefore, this expression system can be used as an amplification system to detect Ca2+ influx (Petersen and Berridge, 1994). We chose to use this Ca2+-activated Cl− channel reporter system instead of conventional inside-out patch clamp for the detection of CNG channel expression, because the effect of antisense RNA should presumably decrease the overall density (or number) of CNGα1 channel protein. A decrease in the overall density of the channel protein could be better resolved in the whole-cell recording mode, which represented the overall expression of CNGα1, rather than in the patch recording mode, which would not reflect the overall channel density. We microinjected the in vitro transcribed cRNA for sense CNGα1 into Xenopus oocytes. The injected oocytes exhibited cGMP-activated inward currents (Figure 4, A and B). The inward currents were caused by the activation of Ca2+-permeable CNGα1 channels by 8-Br-cGMP. The opening of CNGα1 channels promoted Ca2+ influx, which in turn stimulated Ca2+-dependent Cl− channels. A critical piece of evidence that supported the functional role of anti-CNG1 was provided by anti-CNG1 preinjection study. The prior injection of oocytes with anti-CNG1 cRNA before the injection of sense CNGα1 effectively “knocked out” the cGMP-activated inward currents, indicating that the isolated anti-CNG1 transcript possessed the function of down-regulating CNGα1 channels.
CNG channels may play a general role in a number of activity-dependent modulatory and adaptive changes in synaptic strength, such as LTP and LTD (Kingston et al., 1996; Savchenko et al., 1997; Zufall et al., 1997). A growing body of evidence suggests that CNG channels are important downstream mediators for the effects of the diffusible messengers NO and carbon monoxide (CO) (Shiells and Falk, 1990; Ahmad et al., 1994; Leinders-Zufall et al., 1995), agents known to stimulate the activity of soluble guanylyl cyclase and then cGMP level. The resultant activation of CNG channels may subsequently increase the release of neurotransmitter(s) in presynaptic terminals through Ca2+ influx–mediated exocytosis (Zufall et al., 1997). This mechanism may be widely used in brain as a retrograde signaling pathway to modulate synaptic transmission (Reike and Schwartz, 1994; Savchenko et al., 1997), and it may represent an important feature of a number of forms of activity-dependent synaptic plasticity (Arancio et al., 1995; Zufall et al., 1997). In agreement with the above notion, our data showed that CNGα1 transcript was expressed in many brain regions that were known to be important for LTP and LTD, such as hippocampal CA1 and CA3, dentate gyrus, and the cerebellum Purkinje layer. If the function of anti-CNG1 is to regulate the expression of sense CNGα1, it is likely that they will be coexpressed in the same type of tissues or cells (Laabi et al., 1994; Knee et al., 1994; Thrash-Bingham and Tartof, 1999). Our experiments demonstrated that anti-CNG1 and CNGα1 transcripts were indeed coexpressed in many different brain regions. It was also noticed in our experiments that a long color development time for in situ hybridization was needed for the detection of sense CNGα1. This was in agreement with several previous in situ hybridization studies of CNGα1 in rat hippocampus and cerebral cortex (Kingston et al., 1996; Samanta Roy and Barnstable, 1999), and it suggested that the level of CNGα1 mRNA in neurons was low. It was possible that the suppressive effect of anti-CNG1 might have contributed to the low levels of sense CNGα1 mRNA.
CNG channels play a role in the development of CNS. In rat brain, the CNG channels are highly expressed in developing visual cortex during dendritic outgrowth, and the expression level changes in an age-dependent manner (Samanta Roy and Barnstable, 1999). In Caenorhabditis elegans, mutation of CNG channels has been shown to cause defects in axon outgrowth (Coburn and Bargmann, 1996). Because CNG channels are calcium-permeable channels and because calcium levels in growth cones of neurons are known to be important in regulating growth cone motility, it is conceivable that CNG channels, by influencing [Ca2+]i levels, may exert a great effect on neuronal growth in cortical development. In agreement with what was reported by Samanta Roy and Barnstable (1999) for rat visual cortex, we found that the expression of CNGα1 in human cerebral cortex was developmentally regulated. In fetal visual and frontal cortex, the expression was concentrated primarily on a narrow neuronal layer that corresponded to the layer II and layer III of adult cortex. In adult cortex, CNGα1 was universally expressed in almost all areas of cortical structure except layer I, which contained primarily dendrites and axons of cortical neurons. Interestingly, the expression pattern of anti-CNG1 was remarkably similar to that of CNGα1 (Figure 6). It was expressed predominantly in a narrow layer in fetal cortex, but it was universally expressed in all cortical layers except layer I in adult cortex. The expression of anti-CNG1 in the developing fetal cortex suggests that anti-CNG1 may down-regulate CNG channel level, thereby influencing neuronal growth during cortical development. Concurrent expression of sense CNGα1 and anti-CNG1 together with the parallel developmental changes in the expression levels of these two transcripts supports the notion that anti-CNG1–mediated regulation might be a general mechanism for the control of CNGα1 expression in the CNS. The regulatory role of T3 on the expression of anti-CNG1 and sense CNGα1 in human glioma cell line T98 further substantiates the argument that anti-CNG1 may play a critical role in down-regulating the expression of sense CNGα1 during brain development.
In conclusion, we have isolated a 1.66-kb endogenous transcript (anti-CNG1) that is antisense to CNGα1 mRNA. This transcript is coexpressed with sense CNGα1 mRNA in many different brain regions, noticeably in those regions involved in LTP and LTD, and there are parallel changes of anti-CNG1 and CNGα1 transcripts during brain development. It is likely that the suppression of CNGα1 expression by anti-CNG1 may play an important role in neuronal functions, especially in LTP/LTD and cortical development. Endogenous antisense RNA–mediated regulation may represent a new mechanism through which the activity of ion channels can be regulated in human CNS.
ACKNOWLEDGMENTS
We thank Dr. K.H. Lee for advice, F. Tang and M.W. Leung for technical support, and P.C. Leung for manuscript correction. This study was supported by Hong Kong Research Grant Council CUHK4259/99 M and the Chinese University Research Committee.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0127. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0127.
REFERENCES
- Ahmad I, Leinders-Zufall T, Kocsis JD, Shepherd GM, Zufall F, Barnstable CJ. Retinal ganglion cells express a cGMP-gated cation conductance activatable by nitric oxide donors. Neuron. 1994;12:155–165. doi: 10.1016/0896-6273(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Arancio O, Kandel ER, Hawkins RD. Activity-dependent long-term enhancement of transmitter release by presynaptic 3′,5′-cyclic GMP in cultured hippocampal neurons. Nature. 1995;376:74–80. doi: 10.1038/376074a0. [DOI] [PubMed] [Google Scholar]
- Bradley J, Zhang Y, Bakin R, Lester HA, Ronnett GV, Zinn K. Functional expression of the heteromeric “olfactory” cyclic nucleotide-gated channel in the hippocampus: a potential effector of synaptic plasticity in brain neurons. J Neurosci. 1997;17:1993–2005. doi: 10.1523/JNEUROSCI.17-06-01993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Crary JI, Dean DM, Nguitragool W, Furshan PT, Zimmerman AL. Mechanism of inhibition of cyclic nucleotide-gated ion channels by diacylglycerol. J Gen Physiol. 2000;116:755–768. doi: 10.1085/jgp.116.6.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhallan RS, Macke JP, Eddy RL, Shows TB, Reed RR, Yau KW, Nathans J. Human rod photoreceptor cGMP-gated channel: amino acid sequence, gene structure, and functional expression. J Neurosci. 1992;12:3248–3256. doi: 10.1523/JNEUROSCI.12-08-03248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnick BJ. Naturally occurring antisense RNA. Pharmacol Ther. 1997;75:179–184. doi: 10.1016/s0163-7258(97)00050-8. [DOI] [PubMed] [Google Scholar]
- Dussault JH, Ruel J. Thyroid hormones and brain development. Annu Rev Physiol. 1987;49:321–334. doi: 10.1146/annurev.ph.49.030187.001541. [DOI] [PubMed] [Google Scholar]
- Dzeja C, Hagen V, Kaupp UB, Frings S. Ca2+ permeation in cyclic nucleotide-gated channels. EMBO J. 1999;18:131–144. doi: 10.1093/emboj/18.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn JT, Grunward ME, Yau KW. Cyclic nucleotide-gated ion channels: an extended family with diverse functions. Annu Rev Physiol. 1996;58:395–426. doi: 10.1146/annurev.ph.58.030196.002143. [DOI] [PubMed] [Google Scholar]
- Hildebrandt M, Nellen W. Differential antisense transcription from the dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell. 1992;69:197–204. doi: 10.1016/0092-8674(92)90130-5. [DOI] [PubMed] [Google Scholar]
- Hsu YT, Molday RS. Modulation of the cGMP-gated channel of rod photoreceptor cells by calmodulin. Nature. 1993;361:76–79. doi: 10.1038/361076a0. [DOI] [PubMed] [Google Scholar]
- Kaupp UB. Family of cyclic nucleotide gated ion channels. Curr Opin Neurobiol. 1995;5:434–442. doi: 10.1016/0959-4388(95)80002-6. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner MW. An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell. 1989;59:687–696. doi: 10.1016/0092-8674(89)90015-9. [DOI] [PubMed] [Google Scholar]
- Kingston PA, Zufall F, Barnstable CJ. Rat hippocampal neurons express genes for both rod retinal and olfactory cyclic nucleotide-gated channels: novel targets for cAMP/cGMP function. Proc Natl Acad Sci USA. 1996;93:10440–10445. doi: 10.1073/pnas.93.19.10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee RS, Pitcher SE, Murphy PR. Basic fibroblast growth factor sense (bFGF) and antisense (gfg) RNA transcript are expressed in unfertilized human oocytes and in differentiated adult tissues. Biochem Biophys Res Commun. 1994;205:577–583. doi: 10.1006/bbrc.1994.2704. [DOI] [PubMed] [Google Scholar]
- Laabi Y, Gras MP, Brouet JC, Berger R, Larsen CJ, Tsapis A. The BCMA gene, preferentially expressed during B lymphoid maturation, is bidirectionally transcribed. Nucleic Acids Res. 1994;22:1147–1154. doi: 10.1093/nar/22.7.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T, Shepherd GM, Zufall F. Regulation of cyclic nucleotide-gated channels and membrane excitability in olfactory receptor cells by carbon monoxide. J Neurophysiol. 1995;74:1498–1508. doi: 10.1152/jn.1995.74.4.1498. [DOI] [PubMed] [Google Scholar]
- Molokanova E, Trivedi B, Savchenko A, Kramer RH. Modulation of rod photoreceptor cyclic nucleotide-gated channels by tyrosine phosphorylation. J Neurosci. 1997;17:9068–9076. doi: 10.1523/JNEUROSCI.17-23-09068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Oppenheimer JH, Schwartz HL. Molecular basis of thyroid hormone-dependent brain development. Endocr Rev. 1997;18:462–475. doi: 10.1210/edrv.18.4.0309. [DOI] [PubMed] [Google Scholar]
- Petersen CCH, Berridge MJ. The regulation of capacitative calcium entry by calcium and protein kinase C in Xenopus oocytes. J Biol Chem. 1994;269:32246–32253. [PubMed] [Google Scholar]
- Reike F, Schwartz EA. A cGMP-gated current can control exocytosis at cone synapses. Neuron. 1994;13:863–873. doi: 10.1016/0896-6273(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Samanta Roy DR, Barnstable CJ. Temporal and spatial pattern of expression of cyclic nucleotide-gated channels in developing rat visual cortex. Cereb Cortex. 1999;9:340–347. doi: 10.1093/cercor/9.4.340. [DOI] [PubMed] [Google Scholar]
- Savchenko A, Barnes S, Kramer RH. Cyclic-nucleotide-gated channels mediate synaptic feedback by nitric oxide. Nature. 1997;390:694–698. doi: 10.1038/37803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiells RA, Falk G. Glutamate receptors of rod bipolar cells are linked to a cyclic GMP cascade via a G-protein. Proc R Soc Lond B. 1990;242:91–94. doi: 10.1098/rspb.1990.0109. [DOI] [PubMed] [Google Scholar]
- Simons RW. Naturally occurring antisense RNA control: a brief review. Gene. 1988;72:35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- Strijbos PJL, Pratt GD, Khan S, Charles IG, Garthwaite J. Molecular characterization and in situ localization of a full-length cyclic nucleotide-gated channel in rat brain. Eur J Neurosci. 1999;11:4463–4467. doi: 10.1046/j.1460-9568.1999.00893.x. [DOI] [PubMed] [Google Scholar]
- Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- Vanhee-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- Yao X, Segal AS, Welling P, Zhang X, McNicholas CM, Engel D, Boulpaep EL, Desir GV. Primary structure and functional expression of a cGMP-gated potassium channel. Proc Natl Acad Sci USA. 1995;92:11711–11715. doi: 10.1073/pnas.92.25.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew DT, Wong HW, Li WP, Lai HWL, Yu WHA. Nitric oxide synthase neurons in different areas of normal aged and Alzheimer's brains. Neuroscience. 1999;89:675–686. doi: 10.1016/s0306-4522(98)00383-2. [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Siegelbaum SA. Structure and function of cyclic nucleotide-gated channels. Annu Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- Zufall F, Shepherd GM, Barnstable CJ. Cyclic nucleotide gated channels as regulators of CNS development and plasticity. Curr Opin Neurobiol. 1997;7:404–412. doi: 10.1016/s0959-4388(97)80070-0. [DOI] [PubMed] [Google Scholar]