Abstract
Life cycle differentiation of African trypanosomes entails developmental regulation of mitochondrial activity. This requires regulation of the nuclear genome and the kinetoplast, the trypanosome's unusual mitochondrial genome. To investigate the potential cross talk between the nuclear and mitochondrial genome during the events of differentiation, we have 1) disrupted expression of a nuclear-encoded component of the cytochrome oxidase (COX) complex; and 2) generated dyskinetoplastid cells, which lack a mitochondrial genome. Using RNA interference (RNAi) and by disrupting the nuclear COX VI gene, we demonstrate independent regulation of COX component mRNAs encoded in the nucleus and kinetoplast. However, two independent approaches (acriflavine treatment and RNA interference ablation of mitochondrial topoisomerase II) failed to establish clonal lines of dyskinetoplastid bloodstream forms. Nevertheless, dyskinetoplastid forms generated in vivo could undergo two life cycle differentiation events: transition from bloodstream slender to stumpy forms and the initiation of transformation to procyclic forms. However, they subsequently arrested at a specific point in this developmental program before cell cycle reentry. These results provide strong evidence for a requirement for kinetoplast DNA in the bloodstream and for a kinetoplast-dependent control point during differentiation to procyclic forms.
INTRODUCTION
African trypanosomes are unicellular blood-borne parasites transmitted by tsetse flies. In these organisms, the mitochondrial genome is highly unusual, comprising a mass of catenated DNA called the kinetoplast. Each cell has one kinetoplast such that replication and segregation of this organelle must be carefully controlled during the cell cycle (Klingbeil et al., 2001). The kinetoplast is composed of ∼50 maxicircles (of ∼23 kilobases [kb]) and several thousand 1-kb minicircles. Although the maxicircles contain the genes for mitochondrially encoded proteins, some transcripts require addition or deletion of uridine residues to encode functional proteins (Stuart et al., 1997). This unusual process, called RNA editing, is templated by minicircle-encoded guide RNAs.
The developmental regulation of mitochondrial activity is a central component of the trypanosome life cycle (Priest and Hajduk, 1994). This reflects their metabolic requirements in the bloodstream or in the tsetse. In mammalian hosts, ATP is generated from blood glucose by glycolysis (Tielens and Van Hellemond, 1998). As cell numbers increase during a parasitaemia, bloodstream parasites transform from proliferative “slender” to nonproliferative “stumpy” forms (Brown et al., 1973; Matthews, 1999). These activate the expression of some mitochondrial components (Bienen et al., 1991, 1993), although full development of cytochrome-dependent respiration does not occur until transmission to the tsetse. This metabolic adaptation is accompanied by changes in cell morphology, patterns of gene expression and surface antigen presentation, culminating in a fully differentiated procyclic form that inhabits the tsetse midgut.
The developmental regulation of mitochondrial activity requires coordination between the nucleus and kinetoplast (Schneider, 2001). This is exemplified by the cytochrome oxidase (COX) complex, which comprises ∼10 nuclear-encoded subunits and three components encoded in the kinetoplast (COX I, II, and III). RNA editing is restricted to COX II and III; a stage-regulated frame shift is generated in COX II RNA to allow translation in procyclic forms (Feagin and Stuart, 1988), whereas 50% of the COX III mRNA is generated by uridine addition/deletion. Of the nuclear-encoded components only one, COX VI, has been studied in detail (Matthews and Gull, 1998). As with the mitochondrially encoded components, the abundance of COX VI mRNA is stage regulated, being more abundant in procyclic forms. Similarly, the protein is procyclic stage specific (Tasker et al., 2001).
Coordination between the nucleus and mitochondrial genome has been studied in several organisms. Predictably, the nucleus can regulate the mitochondrial genome because 90% of mitochondrial proteins are nuclear encoded, including components of the transcriptional and translational machinery. However, yeast cells with defects in the mitochondrial genome (rho mutants) can also show altered transcription of some nuclear genes (Parikh et al., 1987), implicating cross talk between the mitochondrial and nuclear genome (Poyton and McEwen, 1996). Mitochondrial mutants of African trypanosomes can also be derived. For example, exposure to DNA intercalating dyes (acriflavine and ethidium bromide) generates dyskinetoplastid (DK) bloodstream forms that survive and multiply long term (Stuart, 1971; Hajduk, 1976). Similarly, ablation of mitochondrial topoisomerase II (Topo II) via RNA interference (RNAi) in procyclic forms produces dyskinetoplasty (Wang and Englund, 2001). These cells do not grow, however, presumably due to the requirement for respiratory activity at this life cycle stage. The consequences of these perturbations for the regulation of nuclear gene expression and other cellular processes are unknown.
Herein, we have investigated the coordinated regulation between the nuclear and mitochondrial genome during the trypanosome life cycle. Specifically, we have disrupted one nuclear-encoded component of COX to determine the consequences for the regulation of its kinetoplast-encoded subunits. Moreover, the developmental program that generates procyclic forms has been investigated in bloodstream parasites lacking a mitochondrial genome. These experiments demonstrate an absence of coordinated regulation between nuclear- and mitochondrial-encoded subunit mRNAs of the COX complex during differentiation. We also show that the kinetoplast is not required for transition from slender to stumpy bloodstream forms or for early events of differentiation to procyclic forms. However, our experiments provide strong evidence that the kinetoplast is required for the viability of bloodstream forms and for progression through a novel control point operating during differentiation to procyclic forms.
MATERIALS AND METHODS
Trypanosomes and In Vitro Cell Growth
Bloodstream-form trypanosomes used were either T. brucei rhodesiense EATRO 2340, or T. brucei brucei s427. For acriflavine treatment in vitro, culture-adapted monomorphic forms of T. brucei rhodesiense EATRO 2340 GUP2965 (Tasker et al., 2000) were used. For acriflavine treatment in vivo, we used a pleomorphic line of T. brucei rhodesiense EATRO 2340 GUP2962 (Matthews and Gull, 1994). For RNA interference experiments, a line of T. brucei s427 was used that expressed the tetracycline repressor protein and T7 RNA polymerase (T. brucei s427 ‘single-marker T7 RNAP/TETR’ line; SMB, a kind gift of Professor George Cross, Rockefeller University, New York, NY; Wirtz et al., 1999). The dyskinetoplastid line D2 is a derivative of T. brucei strain 164 and was a kind gift from Professor Kenneth Stuart (University of Washington, Seattle, WA; Stuart, 1971, 1983).
Bloodstream-form trypanosomes were grown in vivo either in BALB/c mice or Sprague-Dawley rats. For growth in mice, ∼1 × 106 parasites were inoculated i.p., and parasites were harvested when at a level of 5 × 108 parasites ml−1 (∼3 d postinfection for monomorphic and ∼5 d postinfection for pleomorphic lines). At this parasitaemia, pleomorphic lines were predominantly (>80%) stumpy in morphology. For infection of rats, ∼5 × 107 pleomorphic trypanosomes were inoculated i.p., resulting in a predominantly stumpy population after 5 d. For acriflavine treatment, mice were injected i.p. with 2.5 mg kg−1 acriflavine 4 d postinfection, usually resulting in >50% dyskinetoplastid cells 24 h later. In some cases, these mice were immunocompromised with cyclophosphamide (Tasker et al., 2001).
Bloodstream parasites were cultured in vitro in HMI-9 medium containing the appropriate drugs for selection of particular genetic manipulations. The SMB line was cultured routinely in G418 to maintain expression of the T7 polymerase and tetracycline repressor. For transfection, parasites were grown in vitro to mid-log phase (2 × 106 cell ml−1) and transfected as described previously (Tasker et al., 2000, 2001) by using three pulses at 1.7 kV in a BTX electroporator (Kramel Biotech, Cramlington, Northumberland, United Kingdom). Drug concentrations used for selection were neomycin G418 (2.5 μg ml−1), hygromycin (2 μg ml−1), and phleomycin (0.5 μg ml−1).
Monomorphic and pleomorphic bloodstream trypanosomes were differentiated to procyclic forms by diluting cells to 2 × 106 ml−1 with HMI-9 or SDM-79, respectively, and then adding 6 mM cis-aconitate and incubating cells at 27°C. Differentiation was monitored for 24–120 h by immunofluorescence for the expression of EP procyclin (Richardson et al., 1988; Roditi and Clayton, 1999) and for kinetoplast repositioning (Matthews et al., 1995). This was assayed by measuring the kinetoplast-posterior dimension in 100–200 cells by using Scion Image 1.62 (Scion, Frederick, MD).
Construct Preparation
For insertion of the COX VI gene into pZJM (Wang et al., 2000), the gene was amplified using primers based on the first and last 20 nucleotides of the open reading frame incorporating a 5′ HindIII site or 3′ XhoI site, respectively (5′COXVIZJM, 3′COXVIZJM; Table 1). For COX VI gene disruption, gene replacement and insertional inactivation constructs were generated in which sequences flanking the COX VI gene, or within the coding region were inserted either side of a neomycin or a hygromycin resistance cassette. The NEO insertion cassette comprised an EP1 promoter, TET repressor gene (flanked by aldolase gene untranslated regions) and neomycin resistance gene (flanked by actin gene untranslated regions). The HYG insertion cassette comprised the EP1 promoter and hygromycin resistance gene (flanked by actin gene untranslated regions). The flanking sequences were amplified with the oligonucleotides COX6-HYG-SAC, COX6 HYG NOT (Figure 3A, fragment A); COX6-HYG-XHO, COX6-HYG KPN (Figure 3A, fragment B); COX6-NEO-KPN, COX6-NEO-XHO (Figure 3A, fragment C); or COX6-NEO NOT, COX6-NEO-SAC (Figure 3A, fragment D). Each oligonucleotide sequence is shown in Table 1. Correct insertion of each cassette was verified by amplification of genomic DNA by using oligonucleotides flanking the deleted or disrupted allele in combination with primers specific for the hygromycin or neomycin resistance gene (our unpublished data). Precise details of the assembly of these knockout vectors can be obtained from us. The Topo II stem-loop cassette (Wang and Englund, 2001) was a generous gift from Zefeng Wang and Professor Paul Englund (Johns Hopkins University, Baltimore, MD).
Table 1.
Oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| 5′COXVIZJM | 5′CCCAAGCTTATGCCCTTTGTCGATCAC 3′ |
| 3′COXVIZJM | 5′CCGCTCGAGCTACTCATCATATACGGG 3′ |
| COX6 NEO KPN | 5′GGTACCGAGATTCGTCCTTCACAC 3′ |
| COX6 NEO XHO | 5′CTCGAGCTCGAAGACGCTAAGGAC 3′ |
| COX6 NEO NOT | 5′GCGGCCGCGCACCGATTCTCCATGAC 3′ |
| COX6 NEO SAC: | 5′GAGCTCCCAATTCCACAAACACTTC 3′ |
| COX6 HYG SAC | 5′GAGCTCGTAATCATGCCCTTTGTC 3′ |
| COX6 HYG NOT | 5′GCGGCCGCGAATGTTCCTCTCCCCAG 3′ |
| COX6 HYG XHO | 5′CTCGAGGCAAGAAGATGCGGTGGT 3′ |
| COX6 HYG KPN | 5′GGTACCGCAGAAATCCGAAGCATC 3′ |
Primer sequences used to ablate or disrupt COX VI.
Figure 3.
(A) Gene disruption for COX VI in culture adapted, monomorphic forms of T .b. rhodesiense EATRO 795. A nested insertion strategy was used to disrupt each allele of COX VI, to generate the cell line, ΔCOXVI::NEO/∧COXVI::HYG. The first allele was deleted by homologous integration of a neomycin resistance construct; the second allele of COX VI is disrupted by insertional inactivation. (B) Northern blot of RNAs isolated from the COX VI null mutant during differentiation to the procyclic form or in wild-type procyclic forms. A weakly hybridizing band for COX VI is retained, reflecting hybridization to a readthrough RNA after integration of the second drug-resistance cassette into the COX VI gene. COX I, II, and III expression are not affected by COX VI gene ablation. (C) EP procyclin expression of the cell line in which the COX VI gene is disrupted, or wild-type cells. At each time point, cells were assayed for EP procyclin by immunofluorescence. Although differentiation was efficient for each population, being monomorphic the differentiation of the wild-type and the COX VI null mutant was asynchronous with respect to that for stumpy populations.
Northern Blotting
RNA was prepared from cultured parasites according to Tasker et al. (2001). Approximately 3 μg of total RNA was resolved on 1% agarose gels containing formaldehyde. Blotted gels were hybridized overnight at 55 or 65°C in 6× SSC, 50% formaldehyde, 1% SDS, and 0.1% sarkosyl with digoxygenin-labeled riboprobes prepared according to the manufacturer's protocol (Roche Applied Science, Indianapolis, IN). Posthybridization, blots were processed as described previously (Tasker et al., 2000). Signal detection was carried out by enhanced chemiluminescence with CDP-star (Roche Applied Science).
Immunofluorescence
Immunofluorescence was carried out according to Tasker et al. (2000). Briefly, 1 ml of cell culture was harvested and the trypanosomes concentrated at 1000 × g for 1 min. The cells were resuspended in 30 μl of cell culture medium and then spread onto microscope slides to generate air-dried smears. These were fixed in methanol at −20°C for at least 30 min. Thereafter, slides were rehydrated in phosphate-buffered saline (PBS) for 5 min and then incubated for 30 min with EP-specific antibody (Richardson et al., 1988; Cedar Lane Laboratories, Hornby, ONT, Canada) diluted 1:500 in PBS or YL1/2 (Abcam, Cambridge, United Kingdom) diluted 1/20 in PBS. After extensive washing, slides were incubated for a further 30 min in fluorescein isothiocyanate-conjugated rabbit-anti-mouse IgG antibody. Finally, cells were incubated with 4,6-diamidino-2-phenylindole (DAPI) (1 μg ml−1) to stain nuclear and mitochondrial DNA and mounted in MOWIOL (Harlow Chemical, Kent, United Kingdom) containing 1 mg ml−1 phenylene diamine as an antifading agent. Cell images were captured on an Axioscope 2 (Carl Zeiss, Jena, Germany) by using Scion Image 1.62. Images were processed using Adobe Photoshop 6 (Adobe Systems, Mountain View, CA).
Mitotracker Staining
Bloodstream-form trypanosomes (2 × 106 ml−1) were incubated in HMI-medium containing 100 nM Mitotracker Red CMXROS (Molecular Probes, Eugene, OR) for 30 min at 37°C. Then the cells were washed with HMI-9 and incubated for a further 20 min in the absence of Mitotracker, after which the parasites were fixed for 2 min at 4°C with 0.4% paraformaldehyde (prepared fresh in PBS). The cells were then washed once with PBS and air-dried smears were prepared. The slides were fixed for >10 min in methanol at −20°C, before rehydration for 10 min in PBS, followed by DAPI staining and mounting in MOWIOL, as described above. Mitotracker staining was visualized at 100× by using the tetramethylrhodamine B isothiocyanate channel on an Axioscope 2 fluorescence microscope (Carl Zeiss).
RESULTS
Kinetoplast DNA (kDNA)-encoded COX Complex mRNAs Are Not Governed by Nuclear COX VI mRNA Abundance
The coordination between nuclear- and kinetoplast-encoded components of the respiratory complex was investigated during trypanosome differentiation by using the cytochrome oxidase complex as a model. Initially, the stage-regulated induction of nuclear encoded COX VI and kinetoplast encoded COX I, II, and III was assayed during differentiation to the procyclic form. When initiated with populations highly enriched for stumpy forms, this differentiation is synchronous, allowing the temporal kinetics of nuclear and mitochondrial gene activation to be determined. Figure 1 shows the abundance of each mRNA (COX I, II, III, and VI) at intervals during the first 10 h of differentiation. The transcript abundance of COX I, II, and VI increased significantly during the first 2–4 h of differentiation, approximately matching the induction kinetics of the mRNA for the EP procyclin surface antigen (Roditi et al., 1989). For COX III, the RNA abundance did not increase during differentiation and was somewhat variable between experiments. These results are compatible with previous analyses of the stage regulation of COX complex mRNAs, whereby COX I, II, and VI demonstrate a developmental increase in abundance between bloodstream and procyclic forms (Michelotti and Hajduk, 1987; Tasker et al., 2001), whereas the abundance of COX III transcripts is less stringently regulated (Feagin et al., 1988).
Figure 1.
Expression of nuclear COX VI and kinetoplast-encoded COX I, II, and edited COX III during differentiation to the procyclic form. Populations of stumpy form cells were induced to differentiate in vitro. RNA was prepared at intervals during the following 10 h. The relative loading is indicated by the rRNA (EtBr). Induction of the mRNA for the stage-regulated surface antigen EP procyclin is included as a differentiation control.
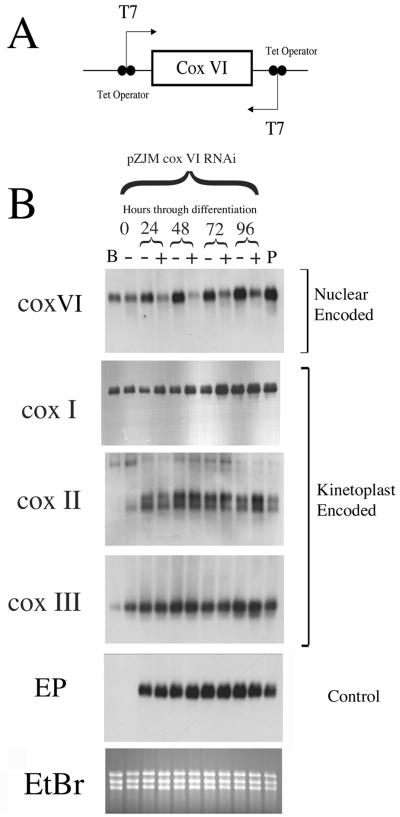
To investigate whether the regulation of kinetoplast-encoded components of COX was linked to the expression of the nuclear-encoded components of that complex, we disrupted the developmental induction of COX VI. Initially, RNA interference technology was used to ablate COX VI mRNA during development to the procyclic form. This technology enables gene-specific silencing in bloodstream and procyclic-form trypanosomes by expression of sense and antisense RNA for a target gene in transgenic parasites (Ngo et al., 1998). Thus, a cultured monomorphic bloodstream cell line that expressed the tetracycline repressor protein and T7 RNA polymerase (Line SMB, a kind gift of Professor George Cross, Rockefeller University; Wirtz et al., 1999) was transfected with the plasmid pZJM COX VI (Figure 2A). This vector comprises the full-length COX VI gene flanked by two opposing T7 promoters, each modified with a tetracycline operator sequence (Wang et al., 2000). When grown in the presence of tetracycline, transcription of each T7 promoter in pZJM COX VI is activated, generating a double-stranded RNA. This is expected to result in gene-specific ablation of COX VI mRNA.
Figure 2.
Transcript ablation for nuclear encoded COX VI has no consequence for differentiation or the induction of kinetoplast-encoded COX I, II, or III. (A) Schematic representation of the RNAi construct pZJMCOXVI. The gene-coding region is integrated between opposing T7 promoters, generating sense and antisense transcripts. (B) Northern blot of COX VI, COX I, II, and III during the asynchronous differentiation of the monomorphic SMB line from bloodstream to procyclic forms. RNA samples were prepared from cells where COX VI RNAi was either not induced (− samples) or induced (+ samples). At the extreme left is shown RNA from control SMB cells untransfected with the pZJM COX VI construct (B) and the COX VI RNAi line grown in the absence of tetracycline (−; 0 h). At the extreme right is shown an established wild-type procyclic line grown in the presence of tetracycline (P).
SMB bloodstream-form cells transfected with pZJMCOXVI were induced to differentiate to procyclic forms by addition of 6 mM cis-aconitate and temperature reduction to 27°C (Czichos et al., 1986). The SMB cell line is monomorphic, i.e., the cell line has a reduced capacity to generate stumpy-form populations after long-term passage in the laboratory and therefore differentiates asynchronously. In consequence, RNA was isolated at 24-h intervals after the initiation of differentiation for a period of 4 d (Figure 2B). Over this time, the differentiation efficiency was assessed both by immunofluorescence for the gain of the procyclic stage-specific surface antigen, EP procyclin (our unpublished data) and by Northern blotting for the appearance of EP procyclin mRNA (Figure 2B, EP). This revealed no difference in the gain of this differentiation marker regardless of the induction of pZJMCOXVI or the presence or absence of tetracycline in the culture medium. In contrast, the abundance of COX VI mRNA was significantly reduced only in the tetracycline-induced cell samples (Figure 2, + lanes), despite the increasing abundance of COX VI mRNA in the uninduced samples (Figure 2, − lanes) during the differentiation time course. This demonstrated that the stage-regulated induction of COX VI mRNA was being efficiently and inducibly prevented in this population by RNAi. The same RNA samples were then hybridized to detect the abundance of mitochondrially encoded cytochrome oxidase mRNAs in each population. In this case, COX I, II, and III each increased in abundance throughout differentiation, and there was no difference between those samples incubated in tetracycline (COX VI mRNA ablated) or without this (COX VI mRNA intact). Thus, ablation of COX VI mRNA in differentiating parasites had no consequences for the induction of mRNAs for mitochondrially encoded components of the same enzyme complex during development to the procyclic form.
Although COX VI RNA was suppressed in the induced RNAi line, mRNA ablation for this gene was incomplete. Therefore, we exploited the absence of a functional respiratory chain in bloodstream forms to generate a phenotypic COX VI null mutant in cultured monomorphic bloodstream forms of the same genetic origin as the pleomorphic cells used in Figure 1. COX VI is a single-copy gene in the T. brucei genome (our unpublished observations). To generate a phenotypic null mutant, we used a nested insertion strategy in which the first allele of this gene was replaced by homologous recombination with a neomycin resistance cassette, whereas the second allele was disrupted by insertion into the COX VI gene of a hygromycin-resistance cassette (ΔCOXVI::NEO/∧COXVI::HYG). This strategy is shown schematically in Figure 3A. Appropriate insertion of the drug-resistance cassettes was confirmed after drug selection by a diagnostic polymerase chain reaction strategy with primers binding to each drug-resistance gene and the COX VI gene flanking sequences (our unpublished observations).
The selected COX VI mutants were then induced to differentiate to procyclic forms in culture. RNA was then prepared over a period of 24 h and assayed for the transcript abundance of components of the cytochrome oxidase complex (Figure 3B). As expected, the COX VI transcript was absent in the null mutants, although a very weak additional band was detected, representing a readthrough transcript generated from integration of the hygromycin cassette into the COX VI coding region (Figure 3A). Although COX VI was ablated, we found no effect on the expression of mitochondrially encoded components of the cytochrome oxidase complex (Figure 3B). Thus, COX I and II significantly increased in abundance early during differentiation, whereas COX III levels were unperturbed or elevated over wild-type procyclic levels. Moreover, the induction of the stage-specific mRNA for EP procyclin (Figure 3B) was also unaffected in the absence of COX VI, as was appearance of its encoded protein on the parasite surface as determined by immunofluorescence (Figure 3C). This conclusively established that stage-regulated mRNA expression of mitochondrially encoded components of the cytochrome oxidase complex is not dependent upon induction of a nuclear-encoded components of this complex, COX VI. Furthermore, unrelated early differentiation events (gain of EP procyclin mRNA and protein) were not perturbed in cells in which COX VI expression was prevented.
Bloodstream Forms Lacking a Kinetoplast Do Not Proliferate In Vitro or In Vivo
To investigate whether differentiation events were perturbed in the absence of a mitochondrial genome, we exploited the ability to isolate DK bloodstream trypanosome lines that lack mitochondrial DNA. Initially, we made use of a preexisting DK line derived by Dr. Kenneth Stuart (University of Washington). This was derived by a selection regime that involved dosing mice harboring trypanosome infections with acriflavine (Stuart, 1971). The resulting line, D2, has been shown to lack kDNA by DAPI staining, by Southern blotting for kDNA (Stuart, 1983), and by the absence of polymerase chain reaction amplification for COX I, II, and III genes (our unpublished observations). Having confirmed the absence of kDNA in the D2 line the ability of these cells to differentiate was assessed in comparison to its kDNA+ parent. However, we found that both lines differentiated very poorly as assessed by the expression of EP procyclin protein. In the case of the parental line, a maximal differentiation of 15% was observed after 48 h (control monomorphic lines routinely express EP procyclin at a level of ∼90% at this time), whereas in the DK line not one differentiating cell was seen in a number of replicate experiments. Although this raised the possibility that the presence of kDNA was required for the initiation of differentiation, the DK line had been subjected to >15 rounds of acriflavine selection in vivo during its isolation (Stuart, 1971). Thus, secondary mutations may have contributed to their differentiation defect.
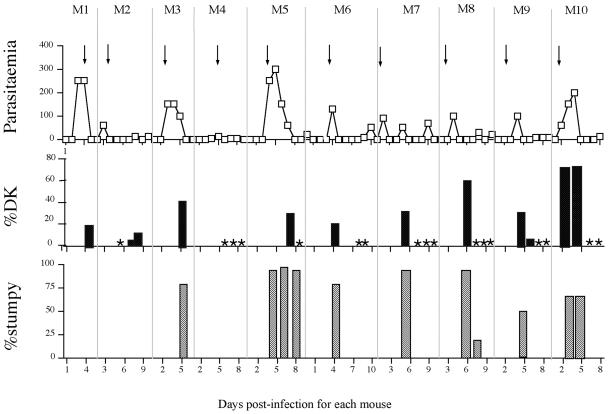
We set out to derive independent DK lines de novo. First, mice infected with a pleomorphic line of T. b. rhodesiense were dosed with 2.5 mg kg−1 acriflavine as the parasitaemia reached 1 × 108 parasites ml−1. This coincided with the point where most parasites transformed from being slender in morphology to being stumpy and was found to generate the highest levels of DK cells (our unpublished observations). Figure 4 shows an acriflavine dosage regime through 10 consecutive mouse passages. In each parasitaemia the point of treatment with acriflavine (arrows) is shown as is the resulting proportion of DK cells in the population (black bars). We found that between 21 and 76% of parasites became DK ∼24 h after acriflavine treatment. However there was no consistent trend in the proportion of DK cells generated, and in each relapse parasitaemia all parasites were consistently found to possess a kinetoplast (Figure 4, asterisks). Indeed, even after >10 rounds of selection in vivo we were unable to derive a stable and heritable population that lacked a kinetoplast.
Figure 4.
Generation of dyskinetoplastid bloodstream forms by acriflavine dosage of infected rodents. Top, parasitaemia achieved for a consecutive series of 10 mouse infections, with each mouse being dosed with 2.5 mg kg−1 acriflavine as the parasitaemia approached 1 × 108 parasites ml−1 (indicated by arrows). Scale, cell no. × 106 parasites/ml. Middle, percentage of DK cells in the population assessed at particular points during each parasitaemia. An asterisk indicates that a search for DK cells was carried out at that time point, but that <0.1% dyskinetoplastid cells was detected. Bottom, percentage of stumpy cells in each population assessed by morphological examination. The time at which each sample was taken, expressed as the number of days postinfection for each mouse (M1–M10) is shown at the bottom.
Induction of dyskinetoplasty by acriflavine treatment of parasites grown in vitro was also attempted. By titrating the acriflavine dosage, 0.001 and 0.0005 μg/ml acriflavine was found to generate a significant level (30–40%) of DK cells in a culture-adapted monomorphic line of T. b. rhodesiense EATRO 2340 after 48-h growth. However, these cells could not be cloned. Of 244 clonal cell lines generated from a cell population containing 18% DK cells, not one line outgrew that lacked a mitochondrial genome, or that showed reduced kDNA content as assessed by DAPI staining. This demonstrated that viable and proliferative dyskinetoplastid lines could not be easily derived either in vivo or in vitro by using acriflavine, despite the generation of significant levels of DK cells.
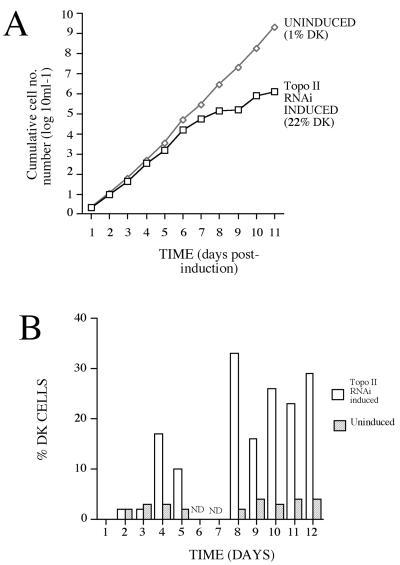
Because acriflavine treatment of bloodstream-form parasites may result in collateral damage to nuclear DNA, an alternative approach to the isolation of bloodstream form DK cells was selected: inducible RNA interference of mitochondrial topoisomerase II. This approach has been shown recently to generate dyskinetoplasty in procyclic forms of T. brucei over a period of 10 d (Wang and Englund, 2001). Although such cells ultimately die, presumably due to the metabolic requirement for mitochondrial function in procyclic forms, such cells are expected to be viable as bloodstream forms. Thus, T. brucei s427 SMB bloodstream-form cells were transfected with the Topo II stem loop RNAi construct (a kind gift of Zefeng Wang and Prof. Paul Englund, Johns Hopkins Medical School; Wang and Englund, 2001). Once established, the transfected cells were grown either in the presence (RNAi+) or absence (RNAi−) of tetracycline and scored for cell growth and the proportion of cells containing a detectable kinetoplast (as assessed by examination of DAPI-stained cells by fluorescence microscopy). Figure 5A demonstrates that over a period of 11 d the RNAi+ line showed reduced population growth with respect to the uninduced cells. Concomitant with this, there was the appearance of DK cells to a level of 32% between 6 and 8 d after induction (Figure 5B). However, the percentage of cells without detectable kinetoplast DNA never exceeded 35% (although there were many cells with apparently diminished kDNA content as assessed by DAPI staining). Moreover, as with acriflavine treatment, the DK cells generated in the RNAi population could not be cloned; from 200 wells isolated by limiting dilution from a population containing 22% DK cells, not one well outgrew that was DK or had detectably reduced kDNA content as assessed by DAPI staining.
Figure 5.
Cell growth and the percentage of DK cells after transcript ablation for mitochondrial topoisomerase II. (A) Growth of cells transfected with the Topo II stem-loop construct, with RNAi either induced (+tetracycline) or not. The percentage of DK cells in each population is shown in each population 11 d after induction. (B) Kinetics of the appearance of DK cells where RNAi for Topo II is induced or not. DK cells are detected 4 d after induction and increased to a maximum of 35% of the population. ND, not done. Note that A and B are derived from independent experiments.
Thus, three different approaches were used to generate de novo clonal lines of DK bloodstream cell: acriflavine treatment both in vitro and in vivo and inducible RNAi of topoisomerase II. In each case, significant levels of DK cells were generated but these were not able to generate a proliferative, clonal population. We conclude the DK cells are either not viable or severely disadvantaged as bloodstream forms both in vitro and in vivo.
Differentiation in Absence of a Mitochondrial Genome
Although we were unable to derive de novo clonal populations of dyskinetoplastid cells, acriflavine dosage of rodents infected with pleomorphic trypanosomes generated high proportions of DK cells (up to 75%). Being generated over a short time period and nonclonal, these cells allow the analysis of differentiation events in a dyskinetoplastid population where possible secondary mutations have not been selected.
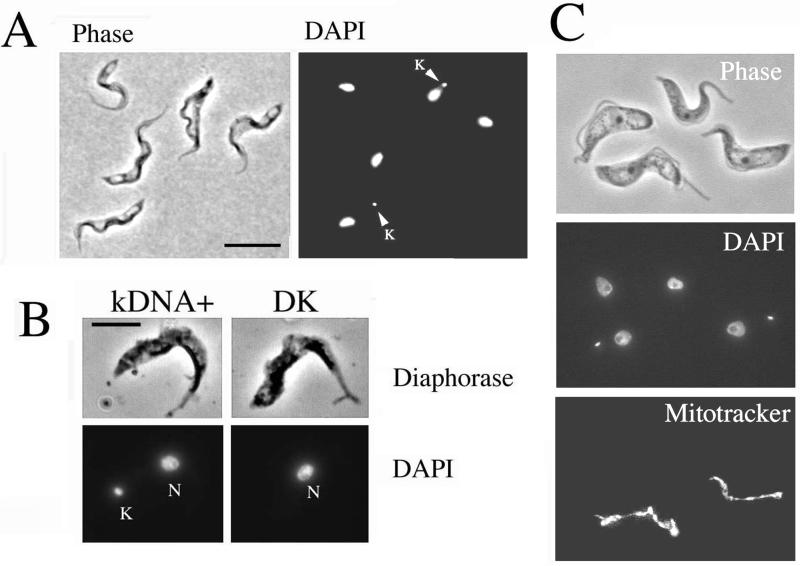
Our first observation regarded the transition from slender to stumpy forms. To investigate whether a mitochondrial genome was required for the generation of stumpy forms the DK cells from rodents infected with pleomorphic trypanosomes were examined for established characteristics of that form. Figure 6A shows a population of cells dosed once with acriflavine 24 h before harvest. The cell population contained many cells (∼50%) that retained a kinetoplast, but in addition there was a significant proportion of cells that were both stumpy in morphology and DK (∼40% of the total cell population). Significantly, these cells were not only morphologically stumpy but also demonstrated positive reactivity for diaphorase, a cytochemical assay for the mitochondrial protein dihydrolipoamide dehydrogenase that is diagnostic for the stumpy form (Figure 6B).
Figure 6.
Dyskinetoplastid cells can develop to stumpy forms. (A) Phase-contrast/DAPI image of parasites 16 h after acriflavine dosage of the host. Cells with (arrowed) and without a kinetoplast are shown. Both normal (kDNA+) and DK cells show stumpy morphology. Bar, 15 μm. (B) Diaphorase staining of a kDNA+ (left) and a dyskinetoplastid cell (right). Both cells are morphologically stumpy and demonstrate clear diaphorase staining along the mitochondrion, a cytochemical characteristic of stumpy forms. Bar, 10 μm. (C) kDNA+ and dyskinetoplastid cells are shown, all with stumpy morphology. Only the kDNA+ cells exhibit detectable mitochondrial staining with Mitotracker Red CMX ROS.
Because stumpy cells are nonproliferative, their kDNA must have been lost before, or at, the final division of the cell that generated them. Such cells might therefore retain sufficient mitochondrial function to enable stumpy production. To establish that these cells did not retain kDNA-dependent mitochondrial activity, the population was stained using Mitotracker. This detects mitochondrial membrane potential, which in bloodstream forms requires protein subunits encoded in both the nucleus and kinetoplast (Nolan and Voorheis, 1992). Figure 6C shows that those cells with an obvious kinetoplast demonstrated a prominent mitochondrial staining with Mitotracker, indicating an extant mitochondrial membrane potential. In contrast, DK cells showed an absence of detectable staining with Mitotracker despite retaining a discrete mitochondrion (as revealed by diaphorase staining; Figure 6B). This established that stumpy cells generated during acriflavine treatment are functionally DK on the basis of the absence of kDNA and a detectable mitochondrial membrane potential. We conclude that a mitochondrial genome is not required for the morphological events of stumpy formation.
Differentiation to Procyclic Forms Reveals a Novel Differentiation Control Point
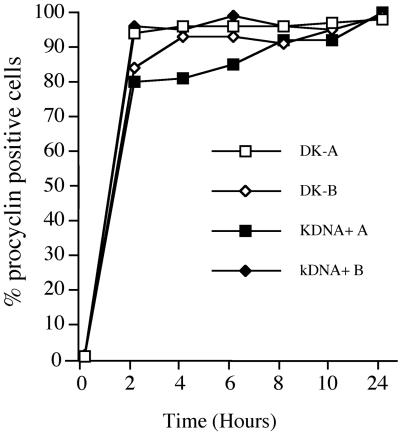
Having demonstrated that morphologically stumpy forms could be generated without a mitochondrial genome, we investigated whether such cells could initiate and progress through differentiation to the procyclic form. Thus, pleomorphic populations enriched (>50%) for DK stumpy-form cells were incubated in SDM-79 medium at 27°C containing 6 mM cis-aconitate. These cells had been derived by a single dose of acriflavine into the rodent host 16 h before harvest. The population was then assessed by immunofluorescence for expression of EP procyclin to determine the respective ability of the DK and kDNA+ cells in the population to express this early marker for differentiation. Figure 7 shows that the kDNA+ and DK cells each initiated differentiation with equivalent kinetics; in each subpopulation 95% of the cells expressed procyclin 4 h after the initiation of differentiation. This established that the kinetoplast was not required for the initiation of differentiation to the procyclic form.
Figure 7.
Differentiation of dykinetoplastid cells. Acriflavine-treated pleomorphic populations with either 74% (DK-A) or 68% (DK-B) dyskinetoplastid cells were induced to differentiate to procyclic forms. The expression of the stage specific antigen EP procyclin was monitored by immunofluorescence for 24 h, for both the DK and kDNA+ cells in each population. For both experiment A and B, each population comprised >80% stumpy forms.
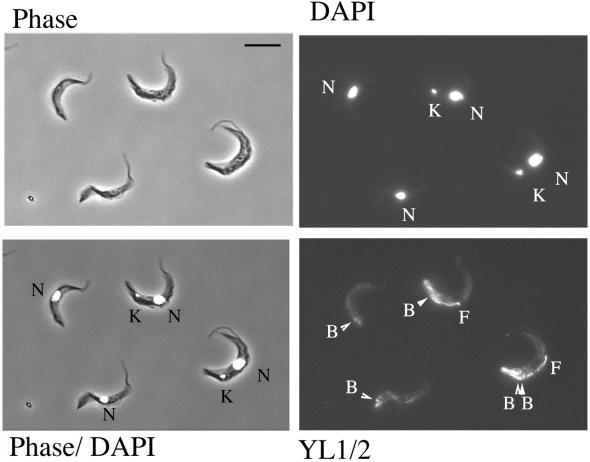
Development from the stumpy to the procyclic form involves a temporal progression of events in which surface antigen exchange is followed by morphological restructuring and reentry into a proliferative cell cycle. Because the most useful marker for these events, the relative position and segregation of the kinetoplast, could not be assessed in the absence of a mitochondrial genome, we examined the cells with the antibody YL1/2. This stains tyrosinated α-tubulin in the trypanosome. Specifically, this antibody labels the posterior tip of cells as microtubule extension gets underway during differentiation and also the position of the basal body (Matthews et al., 1995). This is particularly informative because basal body position closely shadows kinetoplast migration during differentiation, with the basal bodies moving slightly (∼1 μm) anterior to the kinetoplast during differentiation of wild-type cells (Matthews et al., 1995). Also YL1/2 provides an obvious early indication of cell cycle reentry because the newly growing flagellum of the daughter cell contains tyrosinated tubulin and is brightly labeled, whereas the old flagellum, being detyrosinated, is unlabeled (Sherwin et al., 1987). Thus, YL1/2 staining provides an assay of later differentiation events by providing a simultaneous indication of morphological restructuring and cell cycle reentry.
Figure 8 shows cells harvested 14 h after the initiation of differentiation and stained with the antibody YL1/2. Strikingly, we found that the kDNA+ cells in the population underwent extensive morphological restructuring and cell cycle initiation, whereas the DK cells did not. Thus, from an analysis of 1000 cells, 52% of cells with a kinetoplast showed growth of a new daughter flagellum, indicating cell cycle reentry, whereas only 4% of cells without a kinetoplast exhibited new flagellum growth (Figures 8 and 9B). Moreover, the relative position of the basal bodies within the kDNA+ and DK cells types was clearly different: those cells without a kinetoplast exhibited only weak posterior staining of their microtubule cytoskeleton (Figure 8) and a basal body-posterior extension ∼50% of that seen in control cells (Figure 9C). In contrast, the kDNA+ cells in the population, and control cells that had never been exposed to acriflavine, demonstrated a bright labeling at their cell posterior and extensive basal body-posterior extension (Figures 8, YL1/2, and 9C). Apparently, the DK cells do not progress into the first cell cycle after the initiation of differentiation.
Figure 8.
Dyskinetoplastid cells do not reenter into the cell cycle. Cells with a kinetoplast (K) and nucleus (N) show bright labeling with YL1/2 at the posterior end of the cell, a newly growing daughter flagellum (F) and, in one case, segregated basal bodies (lBB). DK cells, in contrast, show limited posterior staining with YL1/2 and no new flagellum growth.
Figure 9.
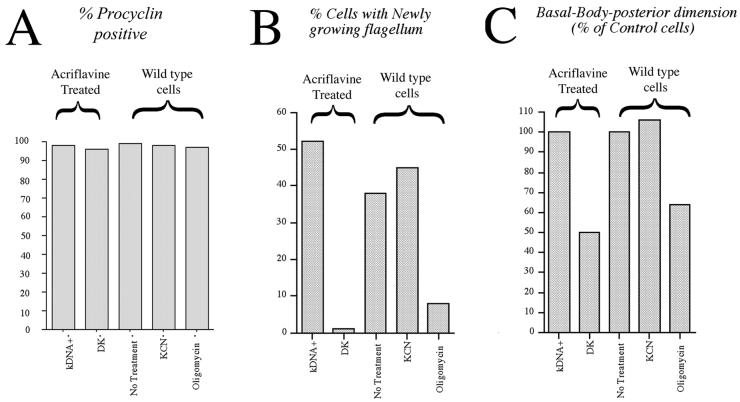
Differentiation and cell cycle reentry for dyskinetoplast cells and cells exposed to metabolic inhibitors. (A) Procyclin expression, assessed by immunofluorescence, 24 h after exposure to differentiation triggers. kDNA+ and DK cells represent, respectively, the cells with or without a detectable kinetoplast in a population of stumpy forms isolated 16 h after exposure of the rodent host to a single dose of acriflavine. KCN represents wild-type stumpy form cells treated with 2 mM KCN throughout differentiation. Oligomycin represents wild-type stumpy form cells treated with 1.25 μg ml−1 oligomycin throughout differentiation. (B) Proportion of cells displaying new flagellum growth as assessed by YL1/2 staining, for the populations in A. (C) Basal Body-posterior dimension for the same cell populations as in A measured after 14 h (acriflavine-derived) or 18 h (wild-type cells). Each is expressed as a percentage of the Basal Body-posterior dimension in control cells at the same time point and represents an analysis 100 cells under each condition. Each set of data represents an average of two replicate experiments.
We considered that the dyskinetoplastid subpopulation may fail to reenter a cell cycle either due to absence of kDNA replication or due to their inability to synthesize a kinetoplast encoded protein. To distinguish these possibilities, wild-type (kDNA+) stumpy cells were differentiated to procyclic forms in the presence of either 2 mM KCN or 1.25 μg/ml oligomycin. These drug regimes inhibit the cytochrome oxidase complex (our unpublished observations) and F1F0 ATPase (Nolan and Voorheis, 1992), respectively, and thereby mimic two metabolic consequences of the absence of kDNA. On exposure of wild-type (kDNA+) cells to these treatments we observed that 2 mM KCN had no effect upon the events of differentiation: the cells gained procyclin (Figure 9A), reentered into a cell cycle with equivalent kinetics to untreated cells (Figure 9B) and repositioned their kinetoplast (Figure 9C). In contrast, exposure of the cells to oligomycin completely mimicked the differentiation response of dyskinetoplastid cells (Figure 9, A–C). Thus, the oligomycin-treated wild-type population gained procyclin but thereafter demonstrated a reduced kinetoplast-posterior repositioning and inability to enter a proliferative cell cycle, the cells arresting with one kinetoplast and one nucleus and without detectable new flagellum growth as assessed by YL1/2 staining. We conclude that in the absence of kDNA, bloodstream form trypanosomes arrest in differentiation after the gain of procyclin but before cell cycle reentry. This suggests the existence of a restriction point in the differentiation program dependent upon the function of the kinetoplast.
DISCUSSION
Differentiation between bloodstream and procyclic forms of African trypanosomes involves a temporally regulated progression of events comprising surface antigen exchange, morphological restructuring, cell cycle reentry, and metabolic adaptation. Herein, we have investigated the interaction between the nucleus and kinetoplast during differentiation between bloodstream and procyclic forms. We found that kinetoplast gene expression was not disrupted by perturbation of the nuclear input to the mitochondrial cytochrome oxidase complex. We also found that transition to stumpy forms was not dependent upon a discrete mitochondrial genome. However, our results provide strong evidence for a requirement of the kinetoplast in the bloodstream and at a specific point during differentiation to the procyclic form.
Initially, we examined the regulation of nuclear and mitochondrial components of the cytochrome oxidase complex during differentiation between bloodstream and procyclic forms. Although previous analyses have examined the respective levels of these transcripts in slender, stumpy, and procyclic forms (Michelotti and Hajduk, 1987), this highly synchronous differentiation allowed the temporal regulation of the respective components of the complex to be compared with other mapped events of differentiation. As reported previously (Feagin et al., 1988), the expression pattern of COX III was not consistently regulated, this being somewhat variable between experiments (our unpublished observations). In contrast, COX I, II, and VI mRNA showed approximately coincident up-regulation soon after the initiation of differentiation, in parallel with the induction of the EP procyclin mRNA. This led us to investigate the potential for regulatory cross talk between mRNAs encoding components of the cytochrome oxidase complex encoded in either the nucleus or kinetoplast.
Cell lines were generated that were ablated for COX VI mRNA by RNAi and by creation of a COX VI phenotypic null mutant. On differentiation of each cell line, COX VI mRNA depletion or ablation had no detectable effect on gain of procyclin or morphological restructuring, demonstrating that there is no requirement for an intact cytochrome oxidase complex during these early events of differentiation. This supports earlier observations where respiratory inhibitors did not effect a number of biochemical events of differentiation (Markos et al., 1989). Moreover, analysis of the relative transcript levels of the kinetoplast-encoded components of the COX complex demonstrated that these were independent of the level of nuclear encoded COX VI. This demonstrates that there is no interorganellar communication that strictly couples mitochondrial COX I, II, and III mRNA abundance to nuclear encoded COX VI RNA levels as these transcripts are induced during this differentiation. This does not, however, exclude possible interorganellar regulation at the protein level or as the cells move to becoming established procyclic forms. This may be significant because the transition to forms with fully active cytochrome-dependent respiration can take some time in vitro (∼72 h; Overath et al., 1986)
In both mammalian and yeast cells with mitochondrial dysfunction a number of nuclear transcripts show altered expression (Poyton and McEwen, 1996). This is best studied in yeast where cells with either mitochondrial DNA damage (rho−) or the absence of a mitochondrial genome (rho0) modulate a retrograde signaling pathway by which nuclear transcript levels are regulated (Parikh et al., 1987; Sekito et al., 2000). To investigate the consequence of mitochondrial disruption on regulated events of the differentiation program, a number of approaches were used to generate dyskinetoplastid trypanosomes. Initially, the ability of DNA intercalating dyes to induce dyskinetoplasty was investigated by growth of bloodstream forms in rodents dosed with acriflavine. Although high levels of DK cells could be generated by this approach, these never outgrew in relapse parasitaemias even after long-term passage through several rounds of selection. Similarly, acriflavine treatment of parasites grown in culture generated dyskinetoplastid cells, but these could not be cloned. As an alternative approach therefore, mitochondrial topoisomerase II was ablated by RNAi in bloodstream forms. In procyclic forms this results in the loss of kinetoplast DNA in up to 80% of cells, although the cells generated are not viable long term (Wang and Englund, 2001). Using the same construct in bloodstream forms also generated dykinetoplastid forms in the induced cell population, although these never accumulated to a level of >35%. Moreover, population growth slowed during the appearance of DK cells and these dyskinetoplastid cells could not be cloned. In consequence, we could not isolate a stable, clonal dykinetoplastid line either in vivo or in vitro.
The requirement for kDNA in bloodstream forms is controversial. Although the kinetoplast encodes components of NADH dehydrogenase (Complex I) and the F1F0 ATPase, bloodstream-form dykinetoplastid cells are expected to be viable because stable populations of DK cells have been generated on several occasions in both T. brucei and T. equiperdum (Stuart, 1971; Hajduk, 1979). However, in these cell populations long-term exposure to DNA-binding agents (acriflavine and ethidium bromide) may have selected for secondary mutations or adaptation that allows their survival in the blood. Furthermore, ablation of a component of the RNA-editing machinery (an RNA ligase) was found recently to be lethal in bloodstream forms (Schnaufer et al., 2001). Although an additional function for this molecule outside of the mitochondrion could not be excluded, these experiments indicated a requirement for mitochondrially encoded proteins at this life cycle stage. Our experiments used both acriflavine treatment and targeted RNAi of a specific component of the kinetoplast replication machinery and in each case generated significant levels of DK cells that did not grow. Although collateral nuclear damage could be associated with acriflavine treatment, when combined with gene-specific mitochondrial topoisomerase II RNAi these experiments provide strong evidence that either kinetoplast DNA is essential in bloodstream forms or that DK cells are severely disadvantaged both in vitro and in vivo.
Although we could not generate new DK lines, we were able to investigate the requirement for the kinetoplast during differentiation steps in the trypanosome life cycle by using pleomorphic trypanosomes dosed with acriflavine. Initially, we examined the DK population to assess whether stumpy forms could be detected. Stumpy generation involves cell cycle arrest initially, followed by morphological and metabolic adaptation (Tyler et al., 1997). Because acriflavine treatment would result in the loss of kDNA during cell division, any stumpy cells must have undergone morphological and metabolic adaptation in the absence of a kinetoplast. Moreover, to confirm that these cells were not merely relying on remnant mitochondrially encoded proteins synthesized before kinetoplast loss, the population was examined with the mitochondrial vital dye Mitotracker. We found cells that were morphologically stumpy and dykinetoplastid in the population at high frequency and further demonstrated that these cells specifically lacked a significant mitochondrial membrane potential as assessed by Mitotracker staining. This suggests a disruption of the function of the F1F0 ATPase, believed to be responsible for mitochondrial import in T. brucei (Nolan and Voorheis, 1992). Thus, the morphological and biochemical events of slender to stumpy transition can occur in the absence of the kinetoplast despite the fact that stumpy cells normally up-regulate transcripts for mitochondrial proteins and exhibit some metabolic adaptation to the procyclic form.
DK stumpy cells were also assayed for their ability to differentiate to the procyclic form. Significantly, we found that the DK cells could express procyclin but specifically arrested before cell cycle reentry. Although these DK cells lack mitochondrially encoded components of the COX complex, treatment of wild-type cells with KCN established that this arrest in differentiation did not reflect the absence of a functional respiratory chain. In contrast, treatment of the cells with oligomycin, an inhibitor of the F1F0 ATPase, mimicked loss of kDNA and resulted in cells that had gained procyclin but did not reenter a proliferative cell cycle. Although there is evidence the F1F0ATPase may be able to function in the absence of its mitochondrially encoded subunit (reviewed in Schnaufer et al., 2002) and dyskinetoplastid lines have been found to exhibit reduced but detectable staining with Mitotracker (Klein et al., 1995; our unpublished observations), a reduced Δψm may restrict differentiation. For example, this may limit mitochondrial import of proteins required during differentiation to procyclic forms. Alternatively, other kinetoplast-encoded proteins may be essential for viability during these stages. For example, it has been suggested that mutants in the RNA-editing machinery are not viable as bloodstream forms due to their inability to generate NADH dehydrogenase (Schnaufer et al., 2001), a complex that contains components encoded in the kinetoplast and that are functionally edited at this life cycle stage (Schneider, 2001).
An additional explanation for the observed effect of kinetoplast loss for differentiation is the existence of a kDNA-dependent cell cycle checkpoint. In the trypanosome cell cycle, the replication and segregation of kDNA is strictly regulated and coordinated with nuclear DNA replication (Sherwin and Gull, 1989). Although previous experiments have demonstrated that inhibiting nuclear DNA replication with aphidicolin does not effect the differentiation process (Matthews and Gull, 1994), differentiation in the absence of kDNA replication has not been examined previously. Thus, kDNA replication may represent a control point without which later events in cell cycle progression are restricted. Indeed, such a checkpoint has been suggested to explain the inability of bloodstream DK bloodstream forms to proliferate when generated either by RNAi for topoisomerase II or by acriflavine (Schnaufer et al., 2002). Although the isolation of proliferative bloodstream DK cells in previous studies demonstrates that this potential checkpoint can be by-passed, these cell lines have been subjected to extreme selection pressure over a long period in acriflavine-treated rodent hosts. In this circumstance, it would not be surprising if cell cycle or metabolic mutants were isolated.
Regardless of whether operating at the metabolic or cell cycle level or both, our results reveal the existence of a novel differentiation checkpoint dependent upon the presence of the kinetoplast. The phenotype of the differentiation defect is such that the cells arrest in the differentiation program after the gain of procyclin but before repositioning of the kinetoplast is complete. This indicates that the checkpoint does not operate on the events that regulate the initiation of differentiation (e.g., signal reception) but instead during progression through the differentiation pathway itself. Dissection of the relationship between the presence of kDNA, metabolic development during the transition to procyclic forms, and reentry into a proliferative cell cycle will be informative in understanding the important control points in trypanosome stage differentiation.
ACKNOWLEDGMENTS
We thank Professor Ken Stuart for the gift of the D2 dyskinetoplastid line and Dr. Zefeng Wang and Professor Paul Englund for providing the plasmid constructs pZJM and the Topo II stem-loop construct. We are also grateful to Dr. Andrè Schneider for helpful and constructive reading of the manuscript at short notice. This work was supported by the Biotechnology and Biological Sciences Research Council and the Wellcome Trust. K.M. holds a Wellcome Trust University Award.
Abbreviations used:
- COX
cytochrome oxidase
- DK
dyskinetoplastid
- RNAi
RNA interference
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0266. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0266.
REFERENCES
- Bienen EJ, Maturi RK, Pollakis G, Clarkson Ab., Jr Non-cytochrome mediated mitochondrial ATP production in bloodstream form Trypanosoma brucei brucei. Eur J Biochem. 1993;216:75–80. doi: 10.1111/j.1432-1033.1993.tb18118.x. [DOI] [PubMed] [Google Scholar]
- Bienen EJ, Saric M, Pollakis G, Grady RW, Clarkson AB., Jr Mitochondrial development in Trypanosoma brucei brucei transitional bloodstream forms. Mol Biochem Parasitol. 1991;45:185–192. doi: 10.1016/0166-6851(91)90085-k. [DOI] [PubMed] [Google Scholar]
- Brown RC, Evans DA, Vickerman K. Changes in oxidative metabolism and ultrastructure accompanying differentiation of the mitochondrion in Trypanosoma brucei. Int J Parasitol. 1973;3:691–704. doi: 10.1016/0020-7519(73)90095-7. [DOI] [PubMed] [Google Scholar]
- Czichos J, Nonnengaesser C, Overath P. Trypanosoma brucei: cis-aconitate and temperature reduction as triggers of synchronous transformation of bloodstream to procyclic trypomastigotes in vitro. Exp Parasitol. 1986;62:283–291. doi: 10.1016/0014-4894(86)90033-0. [DOI] [PubMed] [Google Scholar]
- Feagin JE, Abraham JM, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Feagin JE, Stuart K. Developmental aspects of uridine addition within mitochondrial transcripts of Trypanosoma brucei. Mol Cell Biol. 1988;8:1259–1265. doi: 10.1128/mcb.8.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk SL. Demonstration of kinetoplast DNA in dyskinetoplastic strains of Trypanosoma equiperdum. Science. 1976;191:858–859. doi: 10.1126/science.1251198. [DOI] [PubMed] [Google Scholar]
- Hajduk SL. Dyskinetoplasty in two species of trypanosomatids. J Cell Sci. 1979;35:185–202. doi: 10.1242/jcs.35.1.185. [DOI] [PubMed] [Google Scholar]
- Klein KG, Olson CL, Engman DM. Mitochondrial heat shock protein 70 is distributed throughout the mitochondrion in a dyskinetoplastic mutant of Trypanosoma brucei. Mol Biochem Parasitol. 1995;70:207–209. doi: 10.1016/0166-6851(95)00013-q. [DOI] [PubMed] [Google Scholar]
- Klingbeil MM, Drew ME, Liu Y, Morris JC, Motyka SA, Saxowsky TT, Wang Z, Englund PT. Unlocking the secrets of trypanosome kinetoplast DNA network replication. Protist. 2001;152:255–262. doi: 10.1078/1434-4610-00066. [DOI] [PubMed] [Google Scholar]
- Markos A, Blahuskova A, Kalous M. Metabolic differentiation of bloodstream forms of Trypanosoma brucei brucei into procyclic forms in hemin-depleted medium and in the presence of respiratory inhibitors. Folia Parasitol Praha. 1989;36:301–306. [PubMed] [Google Scholar]
- Matthews K, Gull K. Identification of stage specific and differentiation enriched transcripts during transformation of the African trypanosome from its bloodstream to procyclic form. Mol Biochem Parasitol. 1998;95:81–95. doi: 10.1016/s0166-6851(98)00100-5. [DOI] [PubMed] [Google Scholar]
- Matthews KR. Developments in Differentiation of. Trypanosoma.brucei. Parasitol Today. 1999;15:76–80. doi: 10.1016/s0169-4758(98)01381-7. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Gull K. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J Cell Biol. 1994;125:1147–1156. doi: 10.1083/jcb.125.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR, Sherwin T, Gull K. Mitochondrial genome repositioning during the differentiation of the African trypanosome between life cycle forms is microtubule mediated. J Cell Sci. 1995;108:2231–2239. doi: 10.1242/jcs.108.6.2231. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Hajduk SL. Developmental regulation of trypanosome mitochondrial gene expression. J Biol Chem. 1987;262:927–932. [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan DP, Voorheis HP. The mitochondrion in bloodstream forms of Trypanosoma brucei is energized by the electrogenic pumping of protons catalyzed by the F1F0-ATPase. Eur J Biochem. 1992;209:207–216. doi: 10.1111/j.1432-1033.1992.tb17278.x. [DOI] [PubMed] [Google Scholar]
- Overath P, Czichos J, Haas C. The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur J Biochem. 1986;160:175–182. doi: 10.1111/j.1432-1033.1986.tb09955.x. [DOI] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr. 1994;26:179–191. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol. 1988;31:203–216. doi: 10.1016/0166-6851(88)90150-8. [DOI] [PubMed] [Google Scholar]
- Roditi I, Clayton C. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol Biochem Parasitol. 1999;103:99–100. doi: 10.1016/s0166-6851(99)00124-3. [DOI] [PubMed] [Google Scholar]
- Roditi I, et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Domingo GJ, Stuart K. Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol. 2002;32:1071–1084. doi: 10.1016/s0020-7519(02)00020-6. [DOI] [PubMed] [Google Scholar]
- Schneider A. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int J Parasitol. 2001;31:1403–1415. doi: 10.1016/s0020-7519(01)00296-x. [DOI] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Phil Trans R Soc Lond B Biol Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, et al. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated α-tubulin in the microtubules of Trypanosoma brucei brucei. J Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K. Mitochondrial DNA of an African trypanosome. J Cell Biochem. 1983;23:13–26. doi: 10.1002/jcb.240230103. [DOI] [PubMed] [Google Scholar]
- Stuart K, Allen TE, Heidmann S, Seiwert SD. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD. Evidence for the retention of kinetoplast DNA in an acriflavine-induced dyskinetoplastic strain of Trypanosoma brucei which replicates the altered central element of the kinetoplast. J Cell Biol. 1971;49:189–195. doi: 10.1083/jcb.49.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker M, Wilson J, Sarkar M, Hendriks E, Matthews K. A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol Biol Cell. 2000;11:1905–1917. doi: 10.1091/mbc.11.5.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker M, Timms M, Hendriks E, Matthews K. Cytochrome oxidase subunit VI of Trypanosoma brucei is imported without a cleaved presequence and is developmentally regulated at both RNA and protein levels. Mol Microbiol. 2001;39:272–285. doi: 10.1046/j.1365-2958.2001.02252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens AG, Van Hellemond JJ. Differences in energy metabolism between Trypanosomatidae. Parasitol Today. 1998;14:265–272. doi: 10.1016/s0169-4758(98)01263-0. [DOI] [PubMed] [Google Scholar]
- Tyler KM, Matthews KR, Gull K. The bloodstream differentiation-division of Trypanosoma brucei studied using mitochondrial markers. Proc R Soc Lond B Biol Sci. 1997;264:1481–1490. doi: 10.1098/rspb.1997.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Englund PT. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–483. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]