Abstract
Background
The high degree of sequence heterogeneity found in Hepatitis C virus (HCV) isolates, makes robust nucleic acid-based assays difficult to generate. Polymerase chain reaction based techniques, require efficient and specific sequence recognition. Generation of robust primers capable of recognizing a wide range of isolates is a difficult task.
Results
A position weight matrix (PWM) and a consensus sequence were built for each region of HCV and subsequently assembled into a whole genome consensus sequence and PWM. For each of the 10 regions, the number of occurrences of each base at a given position was compiled. These counts were converted to frequencies that were used to calculate log odds scores. Using over 100 complete and 14,000 partial HCV genomes from GenBank, a consensus HCV genome sequence was generated along with a PWM reflecting heterogeneity at each position. The PWM was used to identify the most conserved regions for primer design.
Conclusions
This approach allows rapid identification of conserved regions for robust primer design and is broadly applicable to sets of genomes with all levels of genetic heterogeneity.
Background
Genetic heterogeneity is a hallmark of RNA viruses in general, and the hepatitis C virus (HCV) in particular, due to the lack of fidelity of viral RNA-dependent RNA polymerases [1,2]. In HCV, this genetic diversity has been organized into six major genotypes and numerous subtypes (over 80). Isolates of the same genotype have an average DNA sequence identity of 95%, but different genotypes have DNA sequence identity close to 65% on average [2-5].
Nucleic acid-based assays, such as the polymerase chain reaction (PCR), the ligase chain reaction (LCR), nucleic acid sequence-based amplification (NASBA), branched chain DNA (bDNA) and sequence analysis itself, rely on the efficient hybridization of oligonucleotides to the targeted sequence. Mismatches between the oligonucleotides and the targeted nucleic acid can affect duplex stability and may compromise the ability of a system to amplify and detect the targeted sequences. Numerous factors determine the effect of mismatches, including: the length of the oligonucleotide, the nature and position of the mismatches, the temperature of hybridization, the presence of co-solvents and the concentrations of oligonucleotides, as well as monovalent and divalent cations [6].
The sequence heterogeneity of HCV challenges efficient detection with nucleic acid-based assays. PCR is widely used for the detection of HCV specific nucleic acids due to its sensitivity. Generally speaking however, effective primers require the genotype of the sample to be known in advance and even then will often be less than 100% effective due to minor variations in the isolates.
Design of robust primers to maximize success with unsequenced isolates (i.e. clinical samples), is a common challenge facing the molecular virologists. A number of software products exist to facilitate primer selection with defined genomes. Many factors are considered in these programs, for example, melting temperature of primers, avoiding primer dimers, avoiding self-complementary primers etc (e.g., Primer Premier [7], Primer3 [8], PRIDE [9]). These algorithms deal mostly with a single template or a small number of sequences. Little effort has been made to handle large number of heterogeneous variants of a given genome.
A large number of HCV related sequences have been deposited in GenBank, making genome wide comparison of all different HCV genotypes and subtypes possible. In this report, more than 100 complete and 14,000 partial sequences deposited in GenBank (Release 129, April 15, 2002) have been used to generate a genome wide consensus sequence and Position Weight Matrix (PWM) for the HCV genome. A PWM based approach for identifying highly conserved regions is proposed which should aid in robust primer design for nucleic acid-based assays. This approach is general enough to be used to optimize any set of genomes with a high degree of heterogeneity.
Results and Discussion
Aligning genomes and generating a position weight matrix(PWM)
One HCV genome (D90208) was used as a template and was separated into pieces based on known gene boundaries. All complete and partial sequences that contained a given region were collected by TBLASTN [10] against HCV sequences from the GenBank non-redundant nucleotide sequence database (nt). An alignment was then made for each part of the genome using ClustalW [11]. A weight score for each position in each fragment was calculated and a PWM was created for that fragment. A whole genome PWM was created by joining the individual PWMs. Finally a 25-bp window, (representative of a typical primer), was walked through the genome/PWM to identify the most conserved locations for primer design.
Due to the extreme genetic heterogeneity of the HCV genome and the nature and large number of complete and partial sequences in the public database, a direct genome wide sequence alignment was not feasible. The approach taken, to break the HCV genome into 10 pieces according to the gene boundaries, proved to be successful. HCV sequence D90208 was chosen arbitrarily as the template sequence and the number of sequences included for alignment of each region is indicated in Table 1.
Table 1.
Sequence numbers for each region that was used to construct the whole genome PWM
| Region | Start | Stop | # of Sequences |
| 5' UTR | 1 | 329 | 1333 |
| CORE | 330 | 899 | 1818 |
| E1 | 900 | 1475 | 3830 |
| E2(p7) | 1476 | 2564 | 3792 |
| NS2 | 2565 | 3407 | 1496 |
| NS3 | 3408 | 5300 | 520 |
| NS4A | 5301 | 5462 | 277 |
| NS4B | 5463 | 6245 | 345 |
| NS5A | 6246 | 7586 | 1571 |
| NS5B | 7587 | 9413 | 1914 |
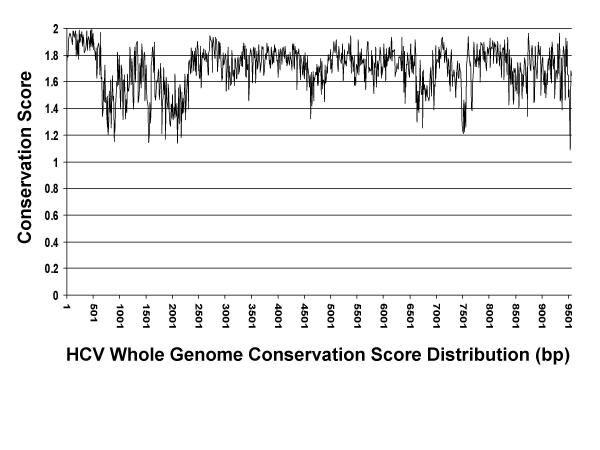
Some regions of the HCV genome share only 50 percent identity across strains. Figure 1 shows a plot of conservation score using a 10-bp window for the whole HCV genome. Region 1–350, which corresponds to the 5' UTR is very conserved across all strains while region 1860–2230 corresponding to the E2 protein, is very heterogeneous. In addition, third position wobble causes mismatching at virtually every third base (in the coding region), leading as expected, to less identity at the DNA level [12]. In the process of collecting sequences for each HCV region, using a nucleotide level comparison algorithm like BLASTN, a lot of sequence entries will be missed. To solve this problem, a protein level comparison algorithm TBLASTN was used via a six-frame translation. Different stringency scores were used to ensure that as many sequences as possible were retrieved. A sequence was chosen for alignment (for a given region) if it shared at least 50% identity over a 30 amino acid stretch or 65 % identity over a 20 amino acid stretch, or over 90% identity over a 10 amino acid stretch with the template sequence. These cutoffs were chosen following inspection of the blast hits for the different regions. Only 4.9% of the available sequences were discarded due to failure to meet the aforementioned criteria.
Figure 1.
HCV genome conservation score distribution.
For each regional alignment, flanking sequences were trimmed prior to generating the PWM. The genome wide PWM was created by combining all individual PWMs (see additional file 1). Insertions (represented by '-' in additional file 1) were added to the template sequence only if greater than 1% of the sequences contained this insertion. This was done to reduce the inclusion of spurious insertions that are caused by sequencing errors or that exist in only a single isolate. A consensus sequence was derived by picking the most frequently occurring base at each position.
Choosing a conserved region for optimized primer design
Using the PWM, the most conserved stretches were rapidly identified making possible the design of robust primers based on the criteria described in Methods. The 25-bp segments in Table 2 and Table 3 are listed by positions in the genome. The higher the odds score, the more conserved the region. Figure 2 shows samples from the final PWM; one 10-bp region in NS5B has a very low conservation score (A), a second 10-bp region shows a very high conservation score (B). This approach allows rapid identification of the most conserved regions of the genome with no regard for self-complementarity of primers, optimizing melting temperature, avoiding primer dimers, etc. Once potential regions of interest are identified, other primer design algorithms can then be used to ensure that self-complementarity etc. will not be a problem. This two step strategy for designing robust primers can be applied to any set of genomes with a high degree of heterogeneity such as viruses, bacterial genes etc. Once a specific sequence has been identified, partially degenerate or multiple oligonucleotides can easily be generated as deemed appropriate for the particular application. Empirical validation of all primers is still prudent.
Table 2.
Suggested forward primer regions based on HCV whole genome PWM
| Primer Start | Primer End | Conservation Score | Primer Start | Primer End | Conservation Score | Primer Start | Primer End | Conservation Score |
| 7 | 31 | 1.804 | 465 | 489 | 1.936 | 5672 | 5696 | 1.821 |
| 8 | 32 | 1.805 | 470 | 494 | 1.938 | 6263 | 6287 | 1.74 |
| 32 | 56 | 1.94 | 471 | 495 | 1.939 | 6302 | 6326 | 1.775 |
| 33 | 57 | 1.94 | 559 | 583 | 1.716 | 6317 | 6341 | 1.732 |
| 40 | 64 | 1.954 | 560 | 584 | 1.736 | 6318 | 6342 | 1.734 |
| 54 | 78 | 1.957 | 580 | 604 | 1.873 | 6323 | 6347 | 1.78 |
| 93 | 117 | 1.911 | 581 | 605 | 1.873 | 6356 | 6380 | 1.786 |
| 94 | 118 | 1.912 | 582 | 606 | 1.873 | 6376 | 6400 | 1.728 |
| 106 | 130 | 1.962 | 600 | 624 | 1.89 | 6424 | 6448 | 1.744 |
| 107 | 131 | 1.962 | 610 | 634 | 1.757 | 6431 | 6455 | 1.77 |
| 141 | 165 | 1.956 | 611 | 635 | 1.779 | 6542 | 6566 | 1.736 |
| 142 | 166 | 1.956 | 612 | 636 | 1.779 | 6556 | 6580 | 1.707 |
| 143 | 167 | 1.957 | 619 | 643 | 1.798 | 6976 | 7000 | 1.708 |
| 165 | 189 | 1.865 | 707 | 731 | 1.614 | 7018 | 7042 | 1.748 |
| 168 | 192 | 1.866 | 1297 | 1321 | 1.782 | 7081 | 7105 | 1.862 |
| 178 | 202 | 1.897 | 1298 | 1322 | 1.782 | 7097 | 7121 | 1.922 |
| 204 | 228 | 1.841 | 1302 | 1326 | 1.766 | 7098 | 7122 | 1.925 |
| 212 | 236 | 1.871 | 1303 | 1327 | 1.804 | 7249 | 7273 | 1.66 |
| 213 | 237 | 1.881 | 1402 | 1426 | 1.719 | 7250 | 7274 | 1.686 |
| 214 | 238 | 1.881 | 1403 | 1427 | 1.733 | 7251 | 7275 | 1.686 |
| 215 | 239 | 1.882 | 1404 | 1428 | 1.733 | 7288 | 7312 | 1.68 |
| 218 | 242 | 1.893 | 1447 | 1471 | 1.765 | 7568 | 7592 | 1.782 |
| 219 | 243 | 1.893 | 1448 | 1472 | 1.795 | 7569 | 7593 | 1.782 |
| 220 | 244 | 1.899 | 1449 | 1473 | 1.794 | 7581 | 7605 | 1.829 |
| 221 | 245 | 1.9 | 1754 | 1778 | 1.587 | 7587 | 7611 | 1.902 |
| 222 | 246 | 1.901 | 1755 | 1779 | 1.626 | 7689 | 7713 | 1.734 |
| 225 | 249 | 1.922 | 2511 | 2535 | 1.676 | 7879 | 7903 | 1.792 |
| 237 | 261 | 1.922 | 2572 | 2596 | 1.796 | 7895 | 7919 | 1.812 |
| 238 | 262 | 1.922 | 2692 | 2716 | 1.856 | 7985 | 8009 | 1.849 |
| 263 | 287 | 1.952 | 2715 | 2739 | 1.928 | 8015 | 8039 | 1.804 |
| 264 | 288 | 1.954 | 2746 | 2770 | 1.789 | 8016 | 8040 | 1.803 |
| 337 | 361 | 1.833 | 2747 | 2771 | 1.791 | 8034 | 8058 | 1.835 |
| 345 | 369 | 1.849 | 2748 | 2772 | 1.792 | 8035 | 8059 | 1.852 |
| 351 | 375 | 1.883 | 2820 | 2844 | 1.928 | 8040 | 8064 | 1.878 |
| 364 | 388 | 1.934 | 3247 | 3271 | 1.728 | 8089 | 8113 | 1.88 |
| 381 | 405 | 1.918 | 3308 | 3332 | 1.806 | 8090 | 8114 | 1.88 |
| 387 | 411 | 1.899 | 3328 | 3352 | 1.851 | 8159 | 8183 | 1.872 |
| 395 | 419 | 1.929 | 3329 | 3353 | 1.871 | 8192 | 8216 | 1.794 |
| 405 | 429 | 1.914 | 3330 | 3354 | 1.87 | 8233 | 8257 | 1.791 |
| 406 | 430 | 1.914 | 3439 | 3463 | 1.591 | 8234 | 8258 | 1.811 |
| 407 | 431 | 1.937 | 3607 | 3631 | 1.729 | 8295 | 8319 | 1.807 |
| 408 | 432 | 1.936 | 3703 | 3727 | 1.852 | 8312 | 8336 | 1.728 |
| 422 | 446 | 1.923 | 3886 | 3910 | 1.779 | 8726 | 8750 | 1.739 |
| 443 | 467 | 1.908 | 3892 | 3916 | 1.762 | 8893 | 8917 | 1.686 |
| 444 | 468 | 1.908 | 4955 | 4979 | 1.788 | 8926 | 8950 | 1.816 |
| 447 | 471 | 1.907 | 5049 | 5073 | 1.898 | 8962 | 8986 | 1.74 |
| 448 | 472 | 1.907 | 5186 | 5210 | 1.751 | 8963 | 8987 | 1.749 |
| 451 | 475 | 1.909 | 5187 | 5211 | 1.774 | 9048 | 9072 | 1.642 |
| 452 | 476 | 1.922 | 5198 | 5222 | 1.801 | 9114 | 9138 | 1.717 |
| 453 | 477 | 1.922 | 5354 | 5378 | 1.84 | 9180 | 9204 | 1.794 |
| 454 | 478 | 1.922 | 5355 | 5379 | 1.843 | 9202 | 9226 | 1.745 |
| 455 | 479 | 1.922 | 5499 | 5523 | 1.811 | 9315 | 9339 | 1.826 |
| 456 | 480 | 1.921 | 5636 | 5660 | 1.724 | 9475 | 9499 | 1.704 |
| 464 | 488 | 1.929 | 5657 | 5681 | 1.678 |
To ensure optimal polymerization, the 3' end and the penultimate position were required to be G or C with frequencies of ≥0.98 and the upstream position, (3' -2), a G or C with a frequency of ≥0.90 or alternatively an A or T with a frequency of ≥0.95.
Table 3.
Suggested reverse primer regions based on HCV whole genome PWM.
| Primer Start | Primer End | Conservation Score | Primer Start | Primer End | Conservation Score | Primer Start | Primer End | Conservation Score |
| 30 | 54 | 1.94 | 467 | 491 | 1.938 | 3352 | 3376 | 1.822 |
| 31 | 55 | 1.94 | 470 | 494 | 1.938 | 3630 | 3654 | 1.808 |
| 55 | 79 | 1.956 | 471 | 495 | 1.939 | 3726 | 3750 | 1.684 |
| 63 | 87 | 1.942 | 474 | 498 | 1.938 | 5043 | 5067 | 1.876 |
| 77 | 101 | 1.929 | 475 | 499 | 1.931 | 5072 | 5096 | 1.835 |
| 116 | 140 | 1.972 | 476 | 500 | 1.926 | 5209 | 5233 | 1.818 |
| 117 | 141 | 1.972 | 477 | 501 | 1.923 | 5210 | 5234 | 1.792 |
| 129 | 153 | 1.954 | 478 | 502 | 1.891 | 5221 | 5245 | 1.763 |
| 164 | 188 | 1.865 | 487 | 511 | 1.918 | 5377 | 5401 | 1.754 |
| 165 | 189 | 1.865 | 488 | 512 | 1.918 | 5378 | 5402 | 1.732 |
| 166 | 190 | 1.865 | 493 | 517 | 1.89 | 5522 | 5546 | 1.745 |
| 187 | 211 | 1.9 | 494 | 518 | 1.89 | 5659 | 5683 | 1.695 |
| 188 | 212 | 1.891 | 572 | 596 | 1.805 | 5680 | 5704 | 1.882 |
| 191 | 215 | 1.89 | 582 | 606 | 1.873 | 5695 | 5719 | 1.866 |
| 197 | 221 | 1.878 | 603 | 627 | 1.811 | 6286 | 6310 | 1.782 |
| 201 | 225 | 1.846 | 604 | 628 | 1.77 | 6325 | 6349 | 1.815 |
| 209 | 233 | 1.868 | 605 | 629 | 1.77 | 6340 | 6364 | 1.79 |
| 235 | 259 | 1.923 | 633 | 657 | 1.897 | 6341 | 6365 | 1.746 |
| 236 | 260 | 1.922 | 634 | 658 | 1.864 | 6379 | 6403 | 1.648 |
| 237 | 261 | 1.922 | 642 | 666 | 1.743 | 6454 | 6478 | 1.783 |
| 238 | 262 | 1.922 | 691 | 715 | 1.542 | 6579 | 6603 | 1.838 |
| 241 | 265 | 1.915 | 730 | 754 | 1.484 | 7120 | 7144 | 1.868 |
| 242 | 266 | 1.914 | 856 | 880 | 1.58 | 7121 | 7145 | 1.868 |
| 243 | 267 | 1.913 | 1297 | 1321 | 1.782 | 7272 | 7296 | 1.794 |
| 244 | 268 | 1.912 | 1320 | 1344 | 1.893 | 7273 | 7297 | 1.754 |
| 245 | 269 | 1.912 | 1325 | 1349 | 1.862 | 7311 | 7335 | 1.674 |
| 260 | 284 | 1.953 | 1326 | 1350 | 1.834 | 7591 | 7615 | 1.856 |
| 261 | 285 | 1.953 | 1425 | 1449 | 1.762 | 7712 | 7736 | 1.821 |
| 286 | 310 | 1.924 | 1426 | 1450 | 1.729 | 7902 | 7926 | 1.813 |
| 287 | 311 | 1.922 | 1439 | 1463 | 1.716 | 8038 | 8062 | 1.878 |
| 360 | 384 | 1.936 | 1470 | 1494 | 1.67 | 8057 | 8081 | 1.912 |
| 387 | 411 | 1.899 | 1471 | 1495 | 1.62 | 8058 | 8082 | 1.912 |
| 403 | 427 | 1.914 | 1777 | 1801 | 1.532 | 8092 | 8116 | 1.864 |
| 407 | 431 | 1.937 | 2534 | 2558 | 1.737 | 8112 | 8136 | 1.833 |
| 418 | 442 | 1.894 | 2715 | 2739 | 1.928 | 8256 | 8280 | 1.833 |
| 428 | 452 | 1.914 | 2769 | 2793 | 1.918 | 8293 | 8317 | 1.806 |
| 429 | 453 | 1.914 | 2770 | 2794 | 1.907 | 8916 | 8940 | 1.839 |
| 430 | 454 | 1.914 | 2771 | 2795 | 1.894 | 8985 | 9009 | 1.684 |
| 445 | 469 | 1.908 | 3270 | 3294 | 1.759 | 9203 | 9227 | 1.742 |
| 452 | 476 | 1.922 | 3331 | 3355 | 1.854 | 9498 | 9522 | 1.574 |
| 466 | 490 | 1.936 | 3351 | 3375 | 1.84 |
To ensure optimal polymerization, the 3' end and the penultimate position were required to be G or C with frequencies of ≥0.98 and the upstream position, (3' -2), a G or C with a frequency of ≥ 0.90 or alternatively an A or T with a frequency of ≥ 0.95.
Figure 2.
Comparison of two 10-bp regions in NS5B: the first with a very low conservation score (A), the second with a very high conservation score (B). Conservation scores were calculated by taking the average of the highest log odds score for each position (see Methods). The sequence shown on top of the matrices is the consensus sequence.
Methods
Databases and Resources
Genbank Release 129 was downloaded from ftp://ncbi.nlm.nih.gov. Pairwise alignment TBLASTN [13] was used to determine whether or not two sequences share similarity. ClustalW [11] was used for multiple sequence alignment. All non-commercial softwares used in this study were written in PERL 5.0.
Construction of alignment
All HCV related sequences were extracted from GenBank (Release 129) by using keyword HCV or Hepatitis C. D90208 was chosen as the organizing template for its fully annotated genome in the GenBank. (Other organizing HCV genomes yielded virtually identical consensus sequences and PWM profiles.) The genomes were separated into 10 regions according to D90208's annotation: 5' UTR, core, E1, E2(P7), NS2, NS3, NS4A, NS4B, NS5A, NS5B. The DNA sequences for each of these regions were retrieved and used for TBLASTN analysis against all HCV sequences. If a sequence shared 50% identity over 90-bp (30 amino acids), 65% identity over 60-bp (20 amino acids) or 90% over 30-bp (10 amino acids) with the query template region, it was considered to contain part of the corresponding gene from a HCV genome in that region, and therefore was used for multiple sequences alignments of this region. For each region, a multiple alignment was done using ClustalW. Alignment was manually curated to eliminate obvious false alignments due to bad sequence quality or inappropriate BLAST hits.
Construction of PWM
A PWM and a consensus sequence were built for each region of HCV and subsequently assembled into a whole genome consensus sequence and PWM. For each of the 10 regions, the number of occurrences of each base at a given position was compiled. These counts were converted to frequencies that were used to calculate log odds scores. The odds score is the frequency observed divided by the theoretical frequency expected (i.e., the background frequency of the base, usually averaged over the genome ~0.25/base). For example, if the base frequency is 0.79 and the estimated background frequency is 0.25, the odds score would be 0.79/0.25 = 3.16. Finally, odds scores were converted to log odds scores by taking the logarithm base 2.
Wi,j = log2(Fi,j/Pi,)
Where
Wi,j is the scoring matrix value of base i in position j
Fi,j is the frequency of base i in position j, Pi is the background frequency of base i
As the logarithm of zero is infinity, a zero occurrence of a particular base in the matrix creates a problem. In this case, a large negative log odds score may be used at such a position in a scoring matrix. A formula proposed by Hertz and Stormo [14] was used instead in our calculations.
Wi,j = log2 [(Ci,j + Pi)/{(N + 1)Pi}] ≈ log2(Fi,j/Pi,)
Where Ci,j is the count of base i in position j, N is the total number of sequences.
Choosing a conserved region for primer design
By sliding 25 bp window (representing average primer length) incrementally along the genome in 1-bp intervals, an average of the highest log odds scores for each position (either A, C, G or T) was calculated to generate the overall degree of conservation (conservation score) for this 25-bp region.
![]()
where L is the length of the region (25-bp in this case).
For PCR applications (or those involving polymerization, where homology at the 3' end of the primer is most critical), it is recommended that the 3' end and the penultimate position be G or C with frequencies of ≥0.98. It is also beneficial if the previous position (3' -2) is a G or C with a frequency of ≥0.90 or alternatively, an A or T with a frequency of ≥0.95. Regions that contain insertions, should in general, be avoided.
Authors' contributions
PQ carried out the data analysis. PQ, XC, LW, JG and BM participated in the design of the study. PQ and BM drafted the manuscript.
All authors read and approved the final manuscript.
Supplementary Material
HCV whole genome PWM. The first line is the consensus sequence and the second line is the template sequence, D90208.
Contributor Information
Ping Qiu, Email: ping.qiu@spcorp.com.
Xiao-Yan Cai, Email: xiao-yan.cai@spcorp.com.
Luquan Wang, Email: luquan.wang@spcorp.com.
Jonathan R Greene, Email: jonathan.greene@spcorp.com.
Bruce Malcolm, Email: bruce.malcolm@spcorp.com.
References
- Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GL. Hepatitis C virus genotypes and quasispecies. Am J Med. 1999;107:21S–26S. doi: 10.1016/S0002-9343(99)00376-9. [DOI] [PubMed] [Google Scholar]
- Kato N. Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb Comp Genomics. 2000;5:129–151. doi: 10.1089/omi.1.2000.5.129. [DOI] [PubMed] [Google Scholar]
- Kato N. Molecular virology of hepatitis C virus. Acta Med Okayama. 2001;55:133–159. doi: 10.18926/AMO/32025. [DOI] [PubMed] [Google Scholar]
- Forns X, Bukh J. The molecular biology of hepatitis C virus. Genotypes and quasispecies. Clin Liver Dis. 1999;3:693–716. doi: 10.1016/s1089-3261(05)70234-8. [DOI] [PubMed] [Google Scholar]
- Wetmur JG, Sninsky JJ, In Innis MA, Gelfand DH, Sninsky JJ. PCR strategies. New York: Academic Press; 1995. [Google Scholar]
- Singh VK, Mangalam AK, Dwivedi S, Naik S. Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques. 1998;24:318–319. doi: 10.2144/98242pf02. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Haas S, Vingron M, Poustka A, Wiemann S. Primer design for large scale sequencing. Nucleic Acids Res. 1998;26:3006–3012. doi: 10.1093/nar/26.12.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HJ, Lau JY, Lauder IJ, Shi N, Lai CL, Hollinger FB. The hepatitis C virus genome: a guide to its conserved sequences and candidate epitopes. Virus Res. 1993;30:27–41. doi: 10.1016/0168-1702(93)90013-D. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HCV whole genome PWM. The first line is the consensus sequence and the second line is the template sequence, D90208.