Abstract
Background
Baculovirus genomes encode a gene called very late expression factor 1 (VLF-1) that is a member of the integrase (Int) family of proteins. In this report we describe the binding properties of purified Autographa californica multiple capsid nucleopolyhedrovirus (AcMNPV) VLF-1 to a number of different DNA structures including homologous regions. In addition, its enzymatic activity was examined.
Results
VLF-1 was expressed in a recombinant baculovirus as a fusion with both HA and HIS6 tags and its binding activity to different DNA structures was tested. No binding was evident to single and double strand structures, very low binding was observed to Y-forks, more binding was observed to three-way junctions, whereas cruciform structures showed high levels of binding. VLF-1 binding was affected by divalent cations; optimal binding to three-way junctions and cruciforms was 2 and 0 mM MgCl2, respectively. Homologous region (hr) sequences was also examined including oligomers designed to expose the hr palindrome as a hairpin, linear double strand, or H-shaped structure. Efficient binding was observed to the hairpin and H-shaped structure. No topoisomerase or endonuclease activity was detected. Sedimentation analysis indicated that *VLF-1 is present as a monomer.
Conclusions
An HA- and HIS-tagged version of AcMNPV VLF-1 showed structure-dependent binding to DNA substrates with the highest binding affinity to cruciform DNA. These results are consistent with the involvement of VLF-1 in the processing of branched DNA molecules at the late stages of viral genome replication. We were unable to detect enzymatic activity associated with these complexes.
Background
Baculoviruses contain double-stranded, circular, covalently closed DNA genomes of 100–180 kb [1,2]. A distinctive feature of their genomes is the presence of homologous regions (hrs) located at a number of positions. In the best characterized baculovirus, Autographa californica multiple capsid nucleopolyhedrovirus (AcMNPV), the hr repeat units contain about 70-bp with an imperfect 30-bp palindrome located near the center, and are repeated two to eight times at each of eight locations around the genome. Hrs have been implicated both as transcriptional enhancers and origins of DNA replication for a number of baculoviruses [3-9]. Hr-containing plasmid DNA transfected into AcMNPV-infected cells appears to replicate as high molecular weight concatemers containing a number of copies of the plasmid [7]. Evidence also suggests that the AcMNPV genome may replicate via a rolling circle mechanism that results in large concatemeric DNA intermediates that are resolved into genome length sizes [10].
All baculovirus genomes sequenced to date encode a gene called very late expression factor 1 (VLF-1) that was originally identified using an AcMNPV temperature sensitive mutant that failed to produce occlusion bodies at the non permissive temperature [11]. VLF-1 is a member of the integrase (Int) family of proteins which are a large group of enzymes that includes many site-specific DNA recombinases [12]. They are found in a variety of organisms including viruses where they are involved in the integration and excision of viral genomes and decatenation of newly replicated viral chromosomes. The baculovirus VLF-1 gene has been demonstrated to be required for very late gene expression and evidence suggests that it binds to the polyhedrin promoter region [13]. Therefore, it is likely to be involved in stimulating the baculovirus RNA polymerase to express high levels of polyhedrin which is required for occlusion body formation. The AcMNPV vlf-1 gene may also encode an essential function associated with a putative integrase/resolvase activity [14] and this activity may be involved in the processing of concatemeric replicative intermediates into monomeric DNAs. Members of the int family encode four conserved amino acids (R, H, R, Y) that are involved in the active site and catalyze DNA rearrangements [15]. Baculovirus VLF-1 sequences align with three of these amino acids (R, R and Y but not H). Mutagenesis of the tyrosine residue, that forms a covalent bond with DNA during recombination reactions in int homologs, resulted in the inability of mutant AcMNPV to produce infectious virions, but did not affect activation of very late promoters [14]. In addition, VLF-1 has been found to be associated with viral nucleocapsids [14], which indicates that it could be involved in genome processing concomitantly with DNA packaging. Therefore, it has been suggested that AcMNPV VLF-1 may be involved in both transcriptional activation of very late genes and in the processing of viral DNA.
In this report we describe the binding properties of purified AcMNPV VLF-1 to a number of different DNA structures which are predicted to mimic the late replicative and recombination intermediates including those containing hr sequences. In addition, the ability of VLF-1 to cleave such structures was also examined.
Results
Expression and purification of HAHIS-VLF-1 (*VLF-1)
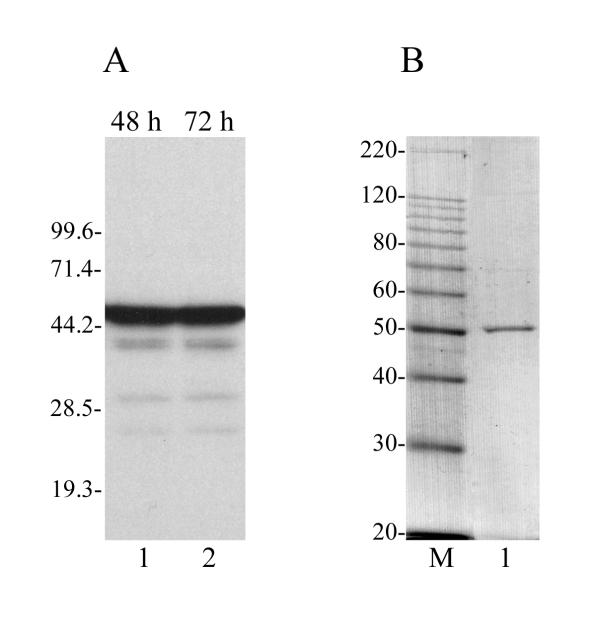
In order to facilitate the purification and characterization of VLF-1, the VLF-1 open reading frame (ORF) was fused at its N-terminus with the HA epitope and a HIS6 tag and overexpressed in a recombinant baculovirus under the control of the polyhedrin promoter. The recombinant virus is called vfbHAHISVLF-1 and the recombinant protein HAHIS-VLF-1 is designated *VLF-1 in this report. The HA epitope allowed monitoring of VLF-1 by Western blot analysis, whereas the HIS6 tag allowed purification of VLF-1 using Ni-NTA resin. *VLF-1 was purified from infected Sf9 cells by sequential liquid chromatography on Ni-NTA-agarose, DEAE-Toyopearl, and heparin-Sepharose columns. Fig. 1A shows a Western blot of extracts of infected cells at 48 and 72 h p.i. and Fig. 1B shows a Coomassie stained gel of purified *VLF-1 demonstrating that the *VLF-1 preparation is highly purified. The predicted size of VLF-1 is about 44 kDa. With the addition of the HA and HIS tags, *VLF-1 is predicted to have a mass of about 47 kDa. A slightly higher apparent molecular mass of *VLF-1 under SDS-PAGE (~50 kDa) (Fig. 1) might be caused by the basic nature of AcMNPV VLF-1 which has estimated pI value equal to 9.16.
Figure 1.
Expression and purification of *VLF-1. (A) Expression of *VLF-1 in infected Sf9 cells analyzed by SDS-12% PAGE followed by Western blotting with the monoclonal antibody HA.11. Lane 1, 48 h p.i.; lane 2, 72 h p.i.. (B) Purified *VLF-1 analyzed by SDS-10% PAGE followed by staining with Coomassie brilliant blue. Lane 1, *VLF-1, 100 ng; lane M, 10-kDa protein ladder. The molecular masses of protein markers (in kilodaltons) are shown on the left.
Binding of *VLF-1 to DNA structures
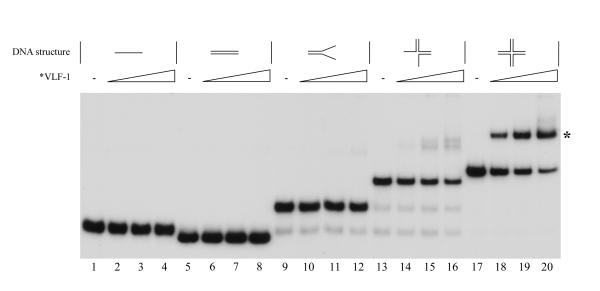
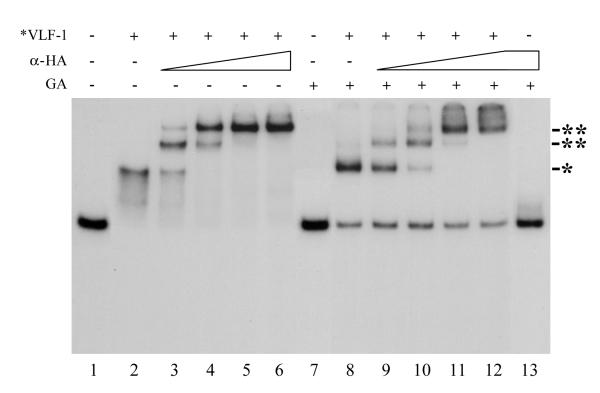
VLF-1 is homologous to the tyrosine recombinases that are typified by phage λ integrase. This family of recombinases is involved in site specific recombination e.g. the integration and excision of the phage λ genome into and out of the host genome. Since baculovirus genomes may replicate via complex branched concatemeric intermediates, we investigated whether VLF-1 might be involved in the resolution of structures that could result from DNA replication and recombination. Using electrophoretic mobility shift analyses, we tested the ability of *VLF-1 to bind different DNA structures (Fig. 2). Although there was no evidence that *VLF-1 could bind to linear or double-stranded DNA (lanes 1–8), trace amounts of a shifted band was detected at the highest *VLF-1 concentration for the Y-shaped DNA structure (lanes 9–12). Higher binding was observed for three-way structure (lanes 13–16). In contrast, significant levels of binding were found to cruciform structures with the majority of the labeled DNA shifted at the highest *VLF-1 concentration (lanes 17–20). Therefore VLF-1 appears to preferentially bind the cruciform-like structures. To confirm that *VLF-1 binding was responsible for the radioactive band shift, we incubated the pre-formed protein-cruciform DNA complexes with the monoclonal antibody HA.11 prior the electrophoresis. We found that anti-HA antibodies caused the protein-cruciform DNA band to supershift (Fig. 3, lanes 3–6 and 9–12). This result indicated that *VLF-1 is a part of the protein-cruciform DNA complexes observed under EMSA, because the anti-HA antibodies interact specifically with *VLF-1 in extracts from the infected cells (Fig. 1A) and do not interact with the DNA probe alone (Fig. 3, lane 13). We routinely fixed the protein-DNA complexes with 0.5% glutaraldehyde prior the electrophoresis. This resulted in more distinct radioactive bands in the gel (compare lanes 2 and 8 in Fig. 3). The general conclusions that could be drawn from the experiments with the samples treated with glutaraldehyde and untreated samples were the same (compare lanes 1–6 and lanes 7–12 in Fig. 3).
Figure 2.
Binding of *VLF-1 to different DNA conformations as determined by electrophoretic mobility shift assays. Reaction mixtures (10 μl) containing 0.004 pmol 32P-labeled oligonucleotide probe and increasing amounts of *VLF-1 were incubated for 20 min at 23°C, fixed with 0.5% glutaraldehyde, and analyzed by electrophoresis in a native 6% polyacrylamide gels as described in Materials and Methods. Reactions shown in lanes contained the following DNA probes: lanes 1 to 4, linear single-stranded DNA (substrate I); lanes 5 to 8, linear double-stranded DNA (substrate II); lanes 9 to 12, forked DNA (pseudo-Y form, substrate III); lanes 13 to 16, three-way junction DNA (substrate IV); and lanes 17 to 20, cruciform DNA (X-shaped form, substrate V). The following amounts of *VLF-1 were added to the reaction mixtures: lanes 2, 6, 10, 14, and 18, 12.5 ng; lanes 3, 7, 11, 15, and 19, 25 ng; lanes 4, 8, 12, 16, and 20, 50 ng. No protein was added to the reactions shown in lanes 1, 5, 9, 13, and 17. The position of a complex of *VLF-1 with cruciform DNA is indicated by asterisk.
Figure 3.
The effect of monoclonal antibody HA.11 and glutaraldehyde fixation on electrophoretic mobility of *VLF-1 complexes with cruciform DNA. Reaction mixtures (10 μl) containing 0.004 pmol 32P-labeled cruciform DNA (substrate V) and 40 ng of *VLF-1 were preincubated for 30 min at 23°C. No *VLF-1 was added to the reactions shown in lanes 1, 7, and 13. After preincubation, the monoclonal antibody HA.11 (α-HA) was added at the following final dilutions: lanes 3 and 9, 1:6,000; lanes 4 and 10, 1:2,000; lanes 5 and 11, 1:600; and lanes 6, 12, and 13, 1:200, and the incubation was continued for 10 min. The reactions shown in lanes 7–13 were fixed with 0.5% glutaraldehyde prior electrophoresis, whereas those shown in lanes 1 to 6 were analyzed by electrophoresis in a native 6% polyacrylamide gel without fixation with glutaraldehyde. The position of the *VLF-1 complexes with cruciform DNA is indicated by an asterisk. The supershifted bands formed in the presence of monoclonal antibody HA.11 are indicated by double asterisks.
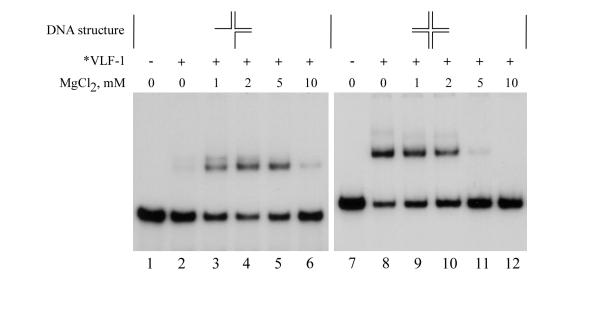
The magnesium optima for binding to the three-way and cruciform structures was then examined (Fig. 4). It was found that, whereas *VLF-1 binding to the three-way structure was markedly stimulated by 1–5 mM MgCl2 and had an optimum at 2 mM, binding to the cruciform was inhibited by 1–10 mM MgCl2. Although the magnesium ions are not required for enzymatic activity of resolvases typified by phage λ integrase (27), our data suggest that divalent cations may efficiently modulate binding of *VLF-1 to some DNA substrates. The opposite effect of MgCl2 on *VLF-1 binding to the three-way and cruciform structures (Fig. 4) suggests also a Mg2+-dependent adjustment of protein and DNA structures upon interaction.
Figure 4.
Effect of magnesium ions on binding of *VLF-1 to three-way junction DNA and cruciform DNA. Reaction mixtures (10 μl) containing 0.004 pmol 32P-labeled oligonucleotide probe and 35 ng of *VLF-1 were incubated for 20 min at 23°C, fixed with 0.5% glutaraldehyde, and analyzed by electrophoresis in a native 6% polyacrylamide gel as described in Materials and Methods. Reactions shown in lanes 1–6 contained three-way junction DNA (substrate IV), those shown in lanes 7–12 contained cruciform DNA (substrate V). No protein was added to the reactions shown in lanes 1 and 7. The final concentrations of MgCl2 (mM) were as indicated.
Binding of *VLF-1 to homologous regions
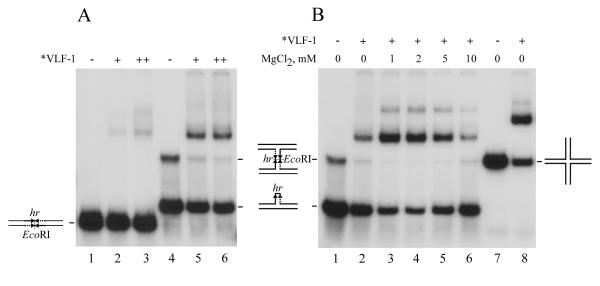
A distinctive feature of baculovirus genomes is the presence of homologous regions (hrs) distributed at a number of locations around the genome. In many baculoviruses, hrs are composed of imperfect palindrome sequences that resemble the binding sites of site-specific recombinases [16]. In order to determine if *VLF-1 could bind to AcMNPV hr sequences, we synthesized oligonucleotides with a consensus hr sequence that included the central imperfect palindrome and about 20 nt of flanking sequences on both ends [17]. The annealing of the hr-containing oligonucleotide to a full size complementary oligonucleotide would form a linear double-stranded structure, whereas its annealing to a complemetary oligonucleotide lacking the 30-nt central palindrome would produce a hairpin-like structure or a double Y (H-shaped) structure (Fig. 5). The presence of the H-shaped structure in the latter substrate was confirmed by a control experiment in which we observed that digestion with EcoRI endonuclease resolved the H-shaped molecules into products with a mobility corresponding to the hairpin structures (data not shown). We found that there was only minimal *VLF-1 binding to the double-stranded hr consensus sequence (Fig. 5A lanes 2, 3). In contrast, *VLF-1 interaction with the hairpin and H-shaped structures was more efficient, with *VLF-1 binding preferentially to the H-shaped molecules (Fig. 5A, lanes 5, 6). The binding to the hairpin and H-shaped structures was stimulated by MgCl2 and had an optimum of 2 mM (Fig. 5B, lanes 1–6). However, *VLF-1 binding to the hr-containing structures at the optimal conditions did not exceed its binding to the non-hr cruciform DNA (compare lanes 4 and 8 in Fig. 5B). This result suggests that under the conditions that we employed, the hr sequence does not serve as a specific determinant for the VLF-1 binding to DNA.
Figure 5.
Binding of *VLF-1 to hr-containing DNA analyzed by electrophoretic mobility shift assay. Reaction mixtures (10 μl) containing 0.004 pmol 32P-labeled oligonucleotide probe and *VLF-1 were incubated for 20 min at 23°C, fixed with 0.5% glutaraldehyde, and analyzed by electrophoresis in a native 6% polyacrylamide gel as described in Materials and Methods. (A) Reactions shown in lanes 1 to 3 contained linear double-stranded hr DNA (substrate VI), those in lanes 4–6 contained a mixture of hairpin hr DNA (substrate VII) and H-shaped (double Y-form) hr DNA (substrate VIII). *VLF-1 was added to the reaction mixtures in the following amounts: lanes 2 and 5, 25 ng; lanes 3 and 6, 50 ng. No protein was added to the reactions shown in lanes 1 and 4. No MgCl2 (B) Reactions shown in lanes contained the following DNA probes: lanes 1 to 6, a mixture of hairpin hr DNA (substrate VII) and H-shaped (double Y-form) hr DNA (substrate VIII); lanes 7 and 8, cruciform DNA lacking hr sequence (substrate V). *VLF-1 (35 ng) was added to the reactions shown in lanes 2 to 6, and 8. No protein was added to the reactions shown in lanes 1 and 7. The final concentrations of MgCl2 in the reaction mixtures were 1 mM (lane 3), 2 mM (lane 4), 5 mM (lane 5), and 10 mM (lane 6). No MgCl2 was added to the reactions shown in lanes 1, 2, 7, and 8.
Testing for DNA cleavage and topoisomerase activity
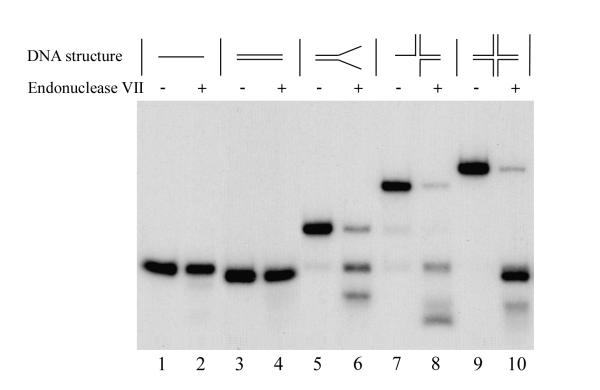
The phage λ int protein has topoisomerase activity in addition to its recombinase function. However, in contrast to the sequence specificity of the recombinase action, the topoisomerase activity is non specific [18]. In order to investigate the activities of *VLF-1, we tested its ability to cleave cruciform DNA and we also examined its topoisomerase activity. Before analysis of the cleavage activity, we tested our DNA substrates to determine if they were accessible to cleavage with bacteriophage T4 endonuclease VII which efficiently cleaves a variety of branched DNA structures [19]. As shown in Fig. 6, T4 endo VII cleaved Y, three-way, and Holliday-like structures. This indicated that our substrates were suitable for the assay of this structure-specific endonuclease. We focused our experiments on the cruciform structure since *VLF-1 demonstrated maximal binding to this substrate (Fig. 2). We examined four cruciform preparations that differed in the oligonucleotide that was 5' end-labeled. Despite varying the enzyme concentration in the range that showed saturation of the DNA substrate under EMSA, we could detect no cleavage of any strand in the cruciforms (data not shown). In addition, we were unable to detect endonuclease activity in the *VLF-1 preparation using hr-containing substrates at different assay conditions described in Materials and Methods. Furthermore, although infected cell extracts showed high levels of topoisomerase activity in assays employing superhelical plasmid DNA, we could not correlate this activity to *VLF-1 (data not shown).
Figure 6.
Hydrolysis of different DNA conformations with phage T4 endonuclease VII. Reaction mixtures (10 μl) containing 0.004 pmol 32P-labeled oligonucleotide probe, 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 15% glycerol, 100 μg of BSA per ml, and 2 mM DTT were incubated for 20 min at 37°C in the presence of phage T4 endonuclease VII (20 units) (lanes 2, 4, 6, 8, and 10) or in its absence (lanes 1, 3, 5, 7, and 9). The reactions were terminated and the samples were processed for electrophoresis in a native 6% polyacrylamide gel as described in Materials and Methods. Reactions shown in lanes contained the following DNA probes: lanes 1 and 2, linear single-stranded DNA (substrate I); lanes 3 and 4, linear double-stranded DNA (substrate II); lanes 5 and 6, forked DNA (pseudo-Y form, substrate III); lanes 7 and 8, three-way junction DNA (substrate IV); and lanes 9 and 10, cruciform DNA (X-shaped form, substrate V).
It has been found that modification of the N-terminus of phage λ integrase affected protein-protein interactions [20]. Because *VLF-1 was tagged with both HIS6 and HA sequences, we considered that these changes at the N-terminus may have inhibited the enzymatic or binding activity of VLF-1. In order to investigate this possibility, we purified VLF-1 from a virus called vpolhHisVlf1 in which a HIS-tagged version of VLF-1 under the control of the polyhedrin promoter was substituted for the wt VLF-1 gene. This virus is replication competent as evidenced by its production of budded virus at wt levels [14] indicating that if VLF-1 is involved in the production of mature viral genomes, the genome is correctly processed. We purified this version of VLF-1 and tested it in the endonuclease assay using the cruciform structures, but again, no activity was detected. In addition, when tested in a topoisomerase assay with superhelical plasmid DNA, no activity could be correlated with this protein.
Sedimentation analysis of *VLF-1 in glycerol gradients
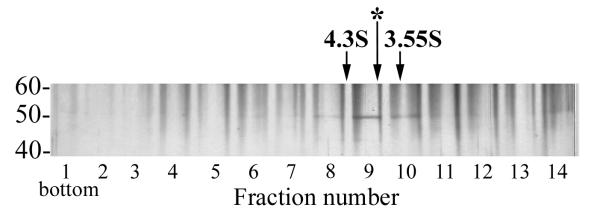
To obtain more information about the molecular structure of *VLF-1, we analyzed its sedimentation in a 15 to 30% glycerol gradient. To prevent nonspecific aggregation of protein during centrifugation, potassium chloride at a final concentration of 0.4 M was added to the solutions used for the gradients. In the glycerol gradients, *VLF-1 sedimented slower than BSA (4.3 S, 66 kDa), but faster than ovalbumin (3.55 S, 45 kDa) (the position marked by the asterisk in Fig. 7). The sedimentation coefficient of *VLF-1 of about 3.9 S corresponds to a globular protein of approximately 50 kDa. Since *VLF-1 has a predicted molecular mass of 47 kDa (including the HA and HIS tags), this indicates that the purified *VLF-1 is present as a monomer.
Figure 7.
Sedimentation analysis of *VLF-1. Purified *VLF-1 (10 μg in 60 μl) was layered over a 15 to 30% glycerol gradient and centrifuged in an SW 50.1 rotor at 48,000 rpm and 4°C for 22 h. The gradient was fractionated from the bottom into 14 fractions, and 5-μl portions from each fraction were analyzed by SDS-10% PAGE followed by silver staining. The position of the standards centrifuged in separate tubes is shown by arrows: BSA, 66 kDa, 4.3 S (fraction 8.45) and ovalbumin, 45 kDa, 3.55 S (fraction 9.86). The position of *VLF-1 (fraction 9.25) is indicated by the asterisk.
Discussion
VLF-1 has homology to members of the phage λ integrase family of tyrosine recombinases and evidence suggests that the VLF-1 plays two independent roles in baculovirus infection cycle. These include a role as a transcription factor for the high level expression of very late genes polyhedrin and p10 [13,21] which is independent of the conserved tyrosine residue. Mutagenesis of the conserved tyrosine, although not affecting very late gene expression, results in an inability of the mutant to produce viable virus. This suggests that VLF-1 may also be involved in genome replication [14]. In particular, it may be involved in the resolution of complex branched concatenated structures that may be produced during genome replication [7,10]. In agreement with this theory, the purified HA- and HIS-tagged VLF-1 showed a structure-dependent binding to DNA substrates. Our data indicates that VLF-1 has the highest binding affinity for cruciform structures. These structures would likely mimic recombination intermediates (Fig. 2) which is consistent with VLF-1 being involved in resolution of these structures. *VLF-1 was also shown to bind two different hr-containing substrates that would form the hairpin-like and H-shaped structures (Fig. 5). Hrs (homologous regions) appear to be a common feature of many baculoviruses and vary from imperfect palindromes to imperfect direct repeats in different viruses [1]. Although they appear to act as enhancers of early gene transcription and replication origins (reviewed in [1]), they could also act as signals for genome processing. However, in our experiments when the hr sequences were inserted into DNA substrates, they did not appear to specifically affect *VLF-1 binding.
Although we clearly showed that VLF-1 is capable of binding to branched DNA structures consistent with a role in genome processing, we were unable to detect enzymatic activity associated with these complexes. If VLF-1 is involved in genome processing as we suspect, there are a number of reasons that might explain this result. It is possible that it might require another viral or host protein for activity. Phage λ integrase requires two host (E. coli) encoded proteins in order to carry out recombination in vitro [22]. It may be site-specific and it is possible that we have not employed a compatible DNA sequence in our assays. It could require superhelicity of its DNA substrate in a manner similar to eukaryotic topoisomerase I which has a preference for supercoiled DNA ([23] and reviewed in [24]). It is also possible that we have not identified the correct conditions for its enzymatic activity or it may become inactivated during our purification protocol.
Conclusions
An HA- and HIS-tagged version of AcMNPV VLF-1 showed structure-dependent binding to DNA substrates with the highest binding affinity for cruciform DNA which mimics a structure common to recombination intermediates. These results are consistent with the involvement of VLF-1 in the processing of branched DNA molecules at the late stages of viral genome replication. Although we showed the VLF-1 is capable of binding to branched DNA structures, we were unable to find enzymatic activity associated with these complexes.
Methods
Chemicals and enzymes
Radiolabeled nucleotide [γ-32P]ATP was from Perkin-Elmer, and T4 polynucleotide kinase was from New England Biolabs, bacteriophage T4 endonuclease VII was the kind gift of Dr. B. Kemper [25].
Cell culture
Spodoptera frugiperda 9 (Sf9) cells were cultured in Sf900II serum-free media (Invitrogen, Inc.), penicillin G (50 units/ml), streptomycin (50 μg/ml, Whittaker Bioproducts), and fungizone (amphotericin B, 375 ng/ml, Flow Laboratories) as previously described [26].
Transfer plasmids and virus construction
Recombinant baculovirus vfbHAHISVlf-1 for overexpression of an AcMNPV His6-tagged VLF-1 (orf77) under the control of the polyhedrin promoter was produced as previously described [27]. The transfer plasmids were constructed as follows: The plasmid pHSEpiHisVLF-1 [28] was a gift from Lois Miller. It was digested with BglII, treated with calf intestinal phosphatase and modified using two oligonucleotides that were phosphorylated by treatment with polynucleotide kinase and ATP, gel purified, annealed [29], and inserted into the BglII site. The oligo names and sequences are:
5bamhahisc – GATCGGGATC CGACCATGAG CTCCCGATAC CCATACGACG TCCCAGATTA CGCCCGGCAT CATCATCATC ATCATCACC
3bamhahisc – GATCGGTGAT GATGATGATG ATGCCGGGCG TAATCTGGGA CGTCGTATGG GTATCGGGAG CTCATGGTCG GATCCC.
This resulted in the creation of an ATG translation start site followed by a HA epitope and a HIS tag upstream of the ORF, but downstream of a new BamHI site (see [27]). Clones were screened using BamHI digestion (the correct orientation is about 70 bp smaller than the reverse orientation), and presumptive correct plasmids were sequenced to confirm the correct orientation of the insert. The primer used for confirming the sequence of the altered region was called AcVLF250 and has the coordinates 64681–64700 (TCCAACGAGTACGACATGTC) [30]. The ORF was then removed by digestion with BamHI and NotI, gel purified, and inserted into a pFastbac1 vector (Gibco-BRL) digested with the same enzymes and gel purified. This plasmid was then used to produce the recombinant baculovirus vfbHAHISVlf-1 using the Bac-to-Bac Baculovirus Expression system (Gibco-BRL) following the manufacturer's instructions.
The virus vpolhisvlf-1 [14] was a gift from Dr. L. Passarelli. It contains the VLF-1 gene expressed from the polyhedrin promoter and is His tagged and produces wt levels of budded virions. However, in contrast to vfbHAHISVlf-1, the VLF-1 that it expresses lacks an HA tag.
DNA substrates
All oligomers were synthesized and PAGE purified by QIAGEN Operon, Inc. The oligonucleotides to produce different structures are identical to those used by [31]. They include the following (61–63)-mers:
4X12-1: 5'GACGCTGCCGAATTCTACCAGTGCCTTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC3'
4X12-2: 5'TGGGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAATGTAATCGTCAAGCTTTATGCCGTT3'
4X12-3: 5'CAACGGCATAAAGCTTGACGATTACATTGCTAGGACATGCTGTCTAGAGGATCCGACTATCGA3'
4X12-4a: 5'ATCGATAGTCGGATCCTCTAGACAGCATGTCCTAGCAAGGCACTGGTAGAATTCGGCAGCGT3'
4X12-5: 5'AACGGCATAAAGCTTGACGATTACATTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCCA3'
The oligonucleotide 4X12-2 was 5' end-labeled using T4 polynucleotide kinase and [γ-32P]ATP. For the various DNA conformations the labeled oligonucleotide was annealed with a 1.3-fold excess of unlabeled oligonucleotides in the following combinations: single-stranded DNA (substrate I), none; double-stranded DNA, 4X12-5 (substrate II); for a pseudo-Y fork 4X12-1(substrate III); for a three-way junction, 4X12-1 plus 4X12-3 (substrate IV); for a cruciform, 4X12-1 plus 4X12-3 plus 4X12-4 (substrate V). The cruciforms (substrates V) with any one of four oligonucleotides, 4X12-1, 4X12-2, 4X12-3 or 4X12-4, labeled with 32P at the 5' end were prepared in the same manner.
The oligonucleotides that were used to examine binding to homologous repeat (hr) sequences are:
80-mer v-hr1: 5'GGGGTTTGTTTTTCAAAACTAAACTCGCTTTACGAGTAGAATTCTACTTGTAACGCACAATCAAGGGATGATGTCAGGGG3'
80-mer v-hr2: 5'CCCCTGACATCATCCCTTGATTGTGCGTTACAAGTAGAATTCTACTCGTAAAGCGAGTTTAGTTTTGAAAAACAAACCCC3'
50-mer v-hr3: 5'CCCCTGACATCATCCCTTGATTGTGAGTTTAGTTTTGAAAAACAAACCCC
The oligonucleotide v-hr1 was labeled with 32P at 5' end and annealed with v-hr2 to form the linear double-stranded hr containing substrate (substrate VI), or with v-hr3 to form the hairpin hr DNA (substrate VII) and double forked hr DNA (substrate VIII).
Purification of HAHIS-VLF-1 (*VLF-1)
Sf9 cells at a density of 1.5 × 106/ ml in shaker flasks were infected with the recombinant baculovirus vfbHAHISVlf-1 at a multiplicity of infection (MOI) of about 4, and incubated with shaking for 72 h at 28°C. *VLF-1 (HAHIS-VLF-1) was purified routinely from 100-ml cultures of infected cells by liquid chromatography sequentially on Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen), DEAE-Toyopearl 650 (TosoHaas), and heparin-Sepharose CL-6B (Amersham Pharmacia Biotech) columns. The infected cells were pelleted by centrifugation for 5 min at 500 × g and resuspended in 8 ml of lysis buffer containing 50 mM Tris-HCl (pH 8.5), 200 mM KCl, 1% Nonidet P-40, 5 mM 2-mercaptoethanol and a set of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 2 μg of E64 per ml). After extraction for 15 min at 4°C on a rotating shaker, the preparation was clarified by centrifuged at 30,000 × g for 30 min. The supernatant was loaded onto a Ni-NTA-agarose column (0.8 ml), equilibrated with buffer A (20 mM Tris-HCl [pH 8.5], 0.5 M KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol, 20 mM imidazole). The column was washed successively with 10 ml of buffer A, 2.5 ml of buffer B (20 mM Tris-HCl [pH 8.5], 1 M KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol, 20 mM imidazole), 1.5 ml of buffer A, and finally with 2 ml of buffer C (20 mM Tris-HCl [pH 8.5], 75 mM KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol) containing 20 mM imidazole.
Protein was eluted from the column with 4 ml of buffer C containing 150 mM imidazole. The sample was loaded at a rate of 4 ml per h onto a DEAE-Toyopearl column (0.5 by 2.5 cm) equilibrated with buffer D (10 mM Tris-HCl [pH 7.5], 20% [vol/vol] glycerol, 1 mM dithiothreitol [DTT], 1 mM EDTA) containing 75 mM KCl. The column was washed successively with 1 ml of buffer D containing 75 mM KCl, 3 ml of buffer D containing 100 mM KCl, and protein was then eluted with 3 ml of the same buffer containing 240 mM KCl. The sample was loaded on a 0.5-ml column of heparin-Sepharose equilibrated with buffer D containing 240 mM KCl. The column was washed with 2 ml of buffer D containing 240 mM KCl, 3 ml of the same buffer containing 280 mM KCl, and *VLF-1 was then eluted with sequential 1-ml portions of buffer D containing KCl in final concentrations of 0.31, 0.35, 0.40, and 0.5 M. Proteins from each fraction were analyzed by SDS-10% PAGE followed by staining with Coomassie brilliant blue. The fractions collected at 0.35 and 0.4 M KCl were combined or dialyzed separately against buffer E (10 mM Tris-HCl [pH 7.5], 50% [vol/vol] glycerol, 1 mM DTT, 0.2 mM EDTA), and stored at -20°C for periods of 1 to 2 months or at -80°C for long-term storage. Protein concentrations were determined by SDS-PAGE followed by optical densitometry of the gel stained with Coomassie brilliant blue. Bovine serum albumin (BSA) loaded in different amounts on separate lanes of the same gel was used for generation of the calibration curve.
PAGE and Western blotting
SDS-(10 or 12%) PAGE was performed as described by Laemmli [32]. Gels were either fixed and stained, or electrophoretically transferred to PVDF-Plus transfer membranes (Micron Separations Inc) using a Trans-blot SD semidry transfer cell (Bio-Rad) according to the manufacturer's guidelines. Western blots were probed with 1:1,000 dilution of monoclonal antibody HA.11 (BAbCO), washed, incubated with a 1:3,000 dilution of goat anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad Laboratories), and developed using ECL detection reagents (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Assays for endonuclease activity of *VLF-1
Reaction mixtures (10 μl) contained 0.004 pmol of substrate V (cruciform DNA) with one of four 62-mers labeled with 32P at 5' end, 20 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2, 100 μg of BSA per ml, and 2 mM DTT. In some experiments, the substrate V was replaced with a mixture of hairpin hr DNA (substrate VII) and double forked (double Y-form) hr DNA (substrate VIII) (total 0.008 pmol), 25 mM Tris-HCl buffer was used at pH 7.5 or pH 8.8, and the mixtures contained no monovalent salt or 100 mM KCl. Purified *VLF-1 (40 or 80 ng in 3 μl of buffer D) was added to a final volume of 10 μl. After incubation for 1 h at 37°C, reactions were terminated by chilling on ice and adding 7 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% each bromophenol blue and xylene cyanol). After heating for 10 min at 85°C, a 6-μl portion of each reaction was loaded onto a 15% polyacrylamide-8 M urea slab gel (17 by 14.7 by 0.08 cm). Electrophoresis was performed in TBE (Tris-borate-EDTA) buffer at 450 V for 1.5 to 2 h until the bromophenol blue migrated 3 cm above the bottom of the gel. The gel was transferred onto a polymer support and exposed to X-ray film at -80°C with an intensifying screen.
Topoisomerase activity of *VLF-1 was assayed as previously described for the baculovirus replication factor LEF-1 [27].
Assay conditions for bacteriophage T4 endonuclease VII
Reaction mixtures containing 0.004 pmol of 32P-labeled oligonucleotide probe (substrates I to V), 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 100 μg of BSA per ml, and 2 mM DTT were assembled on ice. Bacteriophage T4 endonuclease VII (20 units in 3 μl of buffer D) (the gift of Prof. Börries Kemper) was added to a final volume of 10 μl. After incubation for 20 min at 37°C, reactions were terminated by the addition of EDTA to 15 mM and SDS to 0.5%, and the samples were treated with proteinase K (100 μg/ml) for 30 min at 37°C. Half of each reaction mixture was loaded onto a 6% polyacrylamide (acrylamide-bisacrylamide, 60:1) slab gel (6 by 10 by 0.075 cm) prepared in a buffer containing 20 mM HEPES (pH 8.0) and 0.1 mM EDTA. Electrophoresis was performed at 80 V in the same buffer until the bromophenol blue migrated 12 mm above the bottom of the gel. The gel was transferred onto Whatman DE81 paper, dried under vacuum, and then exposed to X-ray film at -80°C with an intensifying screen
Electrophoretical mobility shift assay
Reaction mixtures (10 μl) contained 0.004 pmol of 32P-labeled oligonucleotide probe (substrates I to VIII), 20 mM HEPES (pH 8.0), 50 mM KCl, 100 μg of BSA per ml, and 2 mM DTT. Various amounts of the purified *VLF-1 were added in 3 μl of buffer E. The assembled reaction mixtures were incubated for 20 min on 23°C. For fixation, 0.5 μl of 10% glutaraldehyde was then added to each mixture, and the incubation was continued for 30 min. Half of each reaction mixture was loaded onto a 6% polyacrylamide (acrylamide-bisacrylamide, 60:1) slab gel (6 by 10 by 0.075 cm) prepared in a buffer containing 20 mM HEPES (pH 8.0) and 0.1 mM EDTA. Electrophoresis was performed at 80 V in the same buffer until the bromophenol blue migrated 5 to 10 mm above the bottom of the gel. The gel was transferred onto Whatman DE81 paper, dried under vacuum, and then exposed to X-ray film at -80°C with an intensifying screen.
Sedimentation in glycerol gradients
Linear 15 to 30% glycerol gradients were prepared in buffer F (0.4 M KCl, 10 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM EDTA). Purified *VLF-1 (10 μg in 60 μl), after dialysis against buffer F containing 5% glycerol, was layered over 4.8 ml of a the gradient prepared in nitrocellulose tubes. Ovalbumin (45 kDa, 3.55 S) and BSA (66 kDa, 4.3 S) (0.2 mg of each) were centrifuged in individual tubes as sedimentation standards. After centrifugation in the SW 50.1(Beckman) rotor at 48,000 rpm and 4°C for 22 h, the gradients were fractionated from the bottom with a peristaltic pump. The presence of *VLF1 was monitored by SDS-10% PAGE followed by silver staining.
Authors' contributions
GFR constructed the recombinant virus and maintained, cultured, and infected the cells for protein expression. VSM purified the protein and designed and carried out the experiments. Both authors contributed equally to writing the manuscript and have read and approved it.
Acknowledgments
Acknowledgments
We thank Dr. Kazuhiro Okano for participation in some experiments. The gift of reagents from Drs. Lois Miller, Lorena Passarelli, and Boris Kemper are gratefully acknowledged. This research was supported by grants from the NIH (GM9982536) to G.F.R. and from the Russian Foundation for Basic Research (00-04-49237) to V.S.M.. This is Technical Report No. 11933 from the Oregon State University Agricultural Experiment Station.
Contributor Information
Victor S Mikhailov, Email: Victor.Mikhailov@proxima.idb.ac.ru.
George F Rohrmann, Email: rohrmanng@orst.edu.
References
- Hayakawa T, Rohrmann GF, Hashimoto Y. Patterns of genome organization and content in lepidopteran baculoviruses. Virology. 2000;278:1–12. doi: 10.1006/viro.2000.0668. [DOI] [PubMed] [Google Scholar]
- Herniou EA, Luque T, Chen X, Vlak JM, Winstanley D, Cory JS, O'Reilly DR. Use of whole genome sequence data to infer baculovirus phylogeny. J Virol. 2001;75:8117–8126. doi: 10.1128/JVI.75.17.8117-8126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Summers MD. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J Virol. 1986;60:215–223. doi: 10.1128/jvi.60.1.215-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Gonzalez MA, Summers MD. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J Virol. 1986;60:224–229. doi: 10.1128/jvi.60.1.224-229.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MN, Bjornson RM, Pearson GD, Rohrmann GF. The Autographa californica baculovirus genome: Evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- Pearson MN, Bjornson RM, Ahrens C, Rohrmann GF. Identification and characterization of a putative origin of DNA replication in the genome of a baculovirus pathogenic for Orgyia pseudotsugata. Virology. 1993;197:715–725. doi: 10.1006/viro.1993.1647. [DOI] [PubMed] [Google Scholar]
- Leisy DJ, Rohrmann GF. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology. 1993;196:722–730. doi: 10.1006/viro.1993.1529. [DOI] [PubMed] [Google Scholar]
- Pearson MN, Rohrmann GF. Lymantria dispar nuclear polyhedrosis virus homologous regions: characterization of their ability to function as replication origins. J Virol. 1995;69:213–221. doi: 10.1128/jvi.69.1.213-221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MN, Rohrmann GF. Characterization of a baculovirus-encoded ATP-dependent DNA ligase. J Virol. 1998;72:9142–9149. doi: 10.1128/jvi.72.11.9142-9149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DI, Volkman LE. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch Virol. 1997;142:2107–2113. doi: 10.1007/s007050050229. [DOI] [PubMed] [Google Scholar]
- McLachlin JR, Miller LK. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol. 1994;68:7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaro M, Landy A. λ integrase and the λ int family. In: Craig N, Craigie R, Gellert M, Lambowitz A, editor. Mobile DNA II. Washington DC: ASM press; 2002. [Google Scholar]
- Yang S, Miller LK. Activation of baculovirus very late promoters by interaction with very late factor 1. J Virol. 1999;73:3404–3409. doi: 10.1128/jvi.73.4.3404-3409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Miller LK. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology. 1998;245:99–109. doi: 10.1006/viro.1998.9152. [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul D, van Duyne G. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- Van Duyne G. A structural view of tyrosine recombinase site-specific recombination. In: Craig N, Craigie R, Gellert M, Lambowitz A, editor. Mobile DNA II. Washington DC: ASM press; 2002. pp. 93–117. [Google Scholar]
- Kool M, Ahrens CH, Vlak JM, Rohrmann GF. Replication of baculovirus DNA. J Gen Virol. 1995;76:2103–2118. doi: 10.1099/0022-1317-76-9-2103. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Nash H. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc Natl Acad Sci U S A. 1979;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K, Kemper B, Hays J, Weisberg RA. T4 endonuclease VII cleaves holliday structures. Cell. 1982;29:357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Jessop L, Bankhead T, Wong D, Segall A. The amino terminus of bacteriophage lambda integrase is involved in protein-protein interactions during recombination. J Bacteriol. 2000;182:1024–1034. doi: 10.1128/JB.182.4.1024-1034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlin JR, Yang S, Miller LK. A baculovirus mutant defective in PKIP, a protein which interacts with a virus-encoded protein kinase. Virology. 1998;246:379–391. doi: 10.1006/viro.1998.9210. [DOI] [PubMed] [Google Scholar]
- Nash H, Robertson C. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981;256:9246–9253. [PubMed] [Google Scholar]
- Camilloni G, Di Martino E, Di Mauro E, Caserta M. Regulation of the function of eukaryotic DNA topoisomerase I: topological conditions for inactivity. Proc Natl Acad Sci U S A. 1989;86:3080–3084. doi: 10.1073/pnas.86.9.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Pottmeyer S, Kemper B. T4 endonuclease VII resolves cruciform DNA with nick and counter-nick and its activity is directed by local nucleotide sequence. J Mol Biol. 1992;223:607–615. doi: 10.1016/0022-2836(92)90977-R. [DOI] [PubMed] [Google Scholar]
- Harwood SH, Li L, Ho PS, Preston AK, Rohrmann GF. AcMNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology. 1998;250:118–134. doi: 10.1006/viro.1998.9334. [DOI] [PubMed] [Google Scholar]
- Mikhailov VS, Rohrmann GF. The Baculovirus Replication Factor LEF-1 is a DNA Primase. J Virol. 2002;76:2287–2297. doi: 10.1128/jvi.76.5.2287-2297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JC, Wilson JA, Miller LK. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol. 1998;72:10197–10206. doi: 10.1128/jvi.72.12.10197-10206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Second. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- Goldstein JN, Weller SK. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J Virol. 1998;72:8772–8781. doi: 10.1128/jvi.72.11.8772-8781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]