Abstract
Objectives: Clinical prediction rules have been advocated as a possible mechanism to enhance clinical judgment in diagnostic, therapeutic, and prognostic assessment. Despite renewed interest in the their use, inconsistent terminology makes them difficult to index and retrieve by computerized search systems. No validated approaches to locating clinical prediction rules appear in the literature. The objective of this study was to derive and validate an optimal search filter for retrieving clinical prediction rules, using the National Library of Medicine's medline database.
Design: A comparative, retrospective analysis was conducted. The “gold standard” was established by a manual search of all articles from select print journals for the years 1991 through 1998, which identified articles covering various aspects of clinical prediction rules such as derivation, validation, and evaluation. Search filters were derived, from the articles in the July through December issues of the journals (derivation set), by analyzing the textwords (words in the title and abstract) and the medical subject heading (from the MeSH Thesaurus) used to index each article. The accuracy of these filters in retrieving clinical prediction rules was then assessed using articles in the January through June issues (validation set).
Measurements: The sensitivity, specificity, positive predictive value, and positive likelihood ratio of several different search filters were measured.
Results: The filter “predict$ OR clinical$ OR outcome$ OR risk$” retrieved 98 percent of clinical prediction rules. Four filters, such as “predict$ OR validat$ OR rule$ OR predictive value of tests,” had both sensitivity and specificity above 90 percent. The top-performing filter for positive predictive value and positive likelihood ratio in the validation set was “predict$.ti. AND rule$.”
Conclusions: Several filters with high retrieval value were found. Depending on the goals and time constraints of the searcher, one of these filters could be used.
Clinical prediction rules, otherwise known as clinical decision rules, are tools designed to assist health care professionals in making decisions when caring for their patients. They comprise variables, obtained from the history, physical examination, and simple diagnostic tests, that show strong predictive value.1 The use of these rules to assist in decision making relevant to diagnosis, treatment, and prognosis has been the subject of increasing discussion over the past 15 years.
Wasson et al.2 brought clinical prediction rules to the forefront in a seminal article on their evaluation, validation, and application to medical practice. Twelve years later, Laupacis et al.,1 building on the work of Wasson et al., raised awareness of this topic by reviewing the quality of published reviews and suggesting further modifications of methodological standards.
With the advent of managed care and evidence-based medicine, interest in easily administered and valid rules that are applicable in various clinical settings has increased.
A recent addition to the JAMA Users' Guides to the Medical Literature3 again highlights the use of clinical decision rules. As the article illustrates, the National Library of Medicine's medline database is often used to locate articles that discuss the derivation, validation, and use of such rules. Because of inconsistent use of terminology in describing clinical prediction rules, the rules are difficult to index and retrieve by computerized search systems.
In 1994, Haynes et al.4 developed optimal search strategies for locating clinically sound studies in medline by determining sensitivities, specificities, precision, and accuracy for multiple combinations of terms and medical subject headings (from the MeSH thesaurus) and comparing them with results of a manual review of the literature, which provided the “gold standard.” They showed that medline retrieval of these studies could be enhanced by utilizing combinations of indexing terms and textwords.
In 1997, van der Weijden et al.5 furthered the research called for in the article by Haynes et al. by determining the performance of a diagnostic search filter combined with use of disease terms (e.g., urinary tract infections) and content terms (e.g., erythrocyte sedimentation rate, dipstick). This study, comparing filter with gold standard, again confirmed that the combination of MeSH terms with textwords resulted in higher sensitivity than the use of subject headings alone.
A second study, published in 1994 by Dickersin et al.,6 found that sensitivities of search strategies for a specific study type, randomized clinical trials, also benefited from the use of textwords and truncation. The present work has incorporated the findings of these studies and has again extended the analytic survey of search strategies for clinical studies, by Haynes et al., to include clinical prediction rules.
Methods
This study was designed with two phases, derivation and validation (Figure 1▶). Initially, a gold standard was determined to identify clinical prediction rules, through a manual review of print journals from 1991 to 1998. In the derivation phase, the performance of the search filters in terms of accuracy measures was determined. In the validation phase, a different set of journal articles for the same years was used to assess the external validity of the derived filters. Medline, accessed through OVID Technologies (New York, NY) Web interface, was used in both phases. The comparison (filter vs. gold standard) yielded values for sensitivity, specificity, positive predictive value, and positive likelihood ratio, as shown in Figure 2▶.
Figure 1.
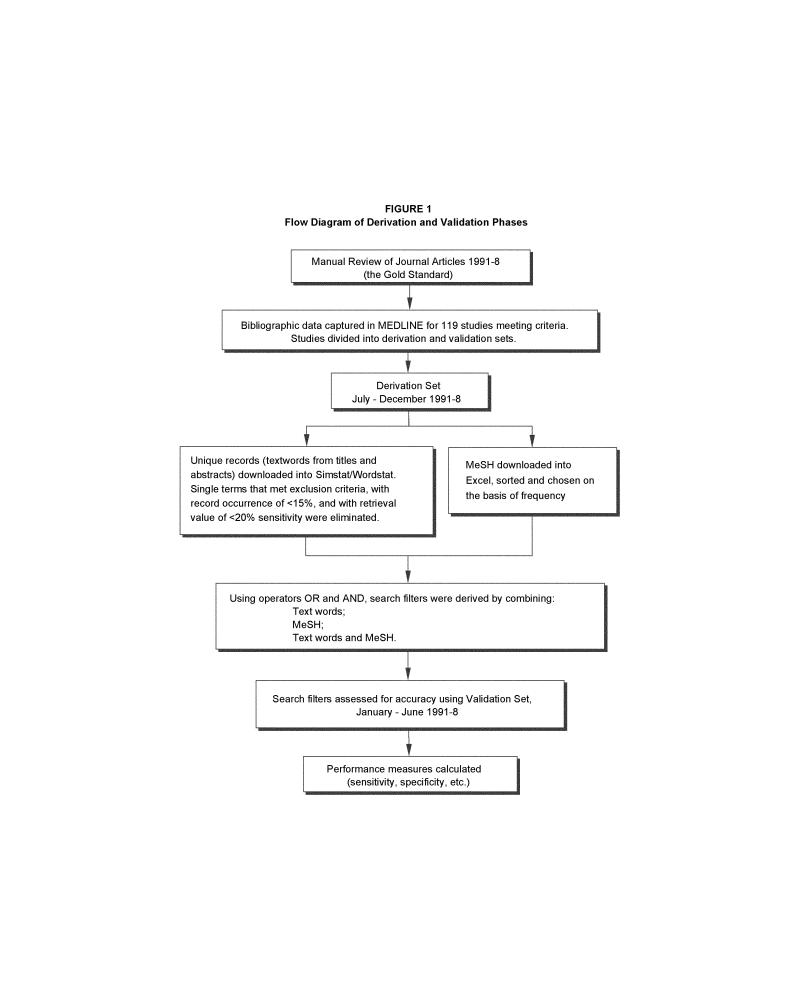
Flow diagram of derivation and validation phases.
Figure 2.
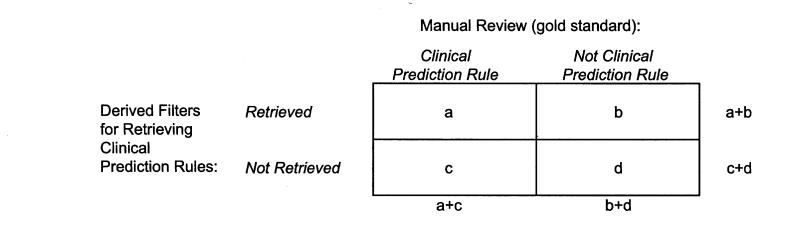
Calculation of performance measures.Sensitivityequals a/(a+c), the proportion of articles with clinical prediction rules that were retrieved by filter. Specificityequals d/(b+d), the proportion of articles without clinical prediction rules that were not retrieved by filter. Positive predictive valueequals a/(a+b), the proportion of retrieved articles that contained clinical prediction rules. Positive likelihood ratio equals (a/[a+c])/(b/[b+d]), the ratio of sensitivity to 1–specificity, which was the proportion of desired articles retrieved compared with the proportion of undesired articles retrieved.
Gold Standard
The foundation of the data set consisted of 34 reports on clinical prediction rules found by Laupacis et al.1 in their manual review of the literature from Jan 1, 1991 through Dec 31, 1994. Building on this work, all articles of every issue of the same four journals— Annals of Internal Medicine, BMJ, JAMA, and New England Journal of Medicine—from Jan 1, 1995 through Dec 31, 1998 were manually reviewed to identify clinical prediction rules. To ensure a sufficient data pool, two additional journals—Annals of Emergency Medicine and the Journal of General Internal Medicine—were reviewed from Jan 1, 1991 through Dec 31, 1998.
The reviewers used the Laupacis definition of clinical prediction rules as the standard and criteria for inclusion in the present study; that is, studies that contained a “prediction-making tool that included three or more variables obtained from the history, physical examination, or simple diagnostic tests and that either provided the probability of an outcome or suggested a diagnostic or therapeutic course of action”1 were selected. This included articles that derived, evaluated, or validated clinical prediction rules.
The initial review identified 211 potential rules. These were independently read by a librarian and a physician, both with expertise in evidence-based medicine. Disagreements were resolved by completion of a third review, based on the Laupacis definition, followed by discussion and consensus.
Eighty-five articles met the criteria for inclusion. These, plus the 34 reports from the Laupacis study, resulted in a data set of 119 articles on clinical prediction rules—63 in the derivation set (July through December 1991 to 1998) and 56 in the validation set (January through June 1991 to 1998). This longitudinal division allowed the search filter to incorporate changes in indexing and terminology over time.
Of the 63 studies in the derivation set, 27 (43 percent) described the development of a particular clinical prediction rule, 8 (13 percent) involved the validation of a previously derived rule, and 20 (32 percent) described both development and validation of a clinical prediction rule. Of the 56 studies in the validation set of this report, 19 (34 percent) included the development of a clinical prediction rule, 8 (14 percent) involved validation of a previously derived rule, and 21 (38 percent) included both development and validation of a clinical prediction rule.
Both phases used articles from the six reviewed journals. In the derivation phase, all 10,877 articles from July through December 1991 to 1998 were used; the remaining 10,878 articles (January through June 1991 to 1998) were used for the validation phase.
Comments, letters, editorials, and animal studies were excluded from the total number of articles, as in the study by Boynton et al.7
Derivation
Titles and abstracts of the reports in the derivation set were downloaded from medline and imported as 63 unique records into SimStat statistical software (Provalis Research, Montreal, Canada). WordStat, the content-analysis module for SimStat, was used to calculate the word frequencies and record occurrences for every textword (word used in the title or abstract of articles) of the 63 records. Word frequency is equal to the total number of times each word was used in the 63 records. Record occurrence is equal to the total number of unique records that contained each word.
It is possible for different words to have the same word frequency but different record occurrences, as shown in Table 1▶. Words that retrieved more records were considered higher impact terms with the potential for greater retrieval. The record occurrence value was used as the first filter in the derivation process. Textwords occurring in less than 15 percent of the records were excluded. This reduced the total number of words from 1,458 to 169.
Table 1.
▪ Content Analysis of Retrieval Value
| Term | Word Frequency | Record Occurrence |
|---|---|---|
| Survival | 40 | 8 (13%) |
| Variables | 40 | 23 (37%) |
An exclusion dictionary, created in WordStat, filtered out terms that were common to many studies, leaving those terms that were unique to clinical prediction rules. Terms in the following categories were excluded: adverbs, articles, conjunctions, prepositions, pronouns, and words for anatomic parts, demographics, locations (e.g., emergency room, hospital), measurements (e.g., positive, high), named populations (e.g., physicians, nurses), numbers, analytic methods (e.g., multivariate, statistical), and study types (e.g., cohort, trials) as well as single- or two-letter abbreviations. Since the goal was to retrieve all clinical prediction rules, specific diagnoses, treatment procedures, and outcomes (e.g., mortality, hospitalization) were also excluded. Filtration through the exclusion dictionary reduced the 169 terms to 66. To accommodate variations in grammatical form and maximize retrieval potential, terms were truncated using the symbol $. For example, predict$ retrieves predicted, prediction, and predicts.
The retrieval value of each textword was determined using accuracy measures. Of the 66 textwords , a sensitivity of at least 20 percent and a positive predictive value of at least 1.5 percent were necessary for a word to remain in further consideration. This narrowed the set to 22 terms. Sensitivity was defined as the proportion of clinical prediction rules in the gold standard that were retrieved using a given filter (Figure 2▶). Positive predictive value was the proportion of retrieved articles that contained clinical prediction rules.
The MeSH terms, used to index the 63 reports, were considered separately. Subject headings were downloaded from medline into Microsoft Excel. They were sorted alphabetically and then filtered using the exclusion dictionary. Based on highest frequency, four headings were included: Decision Support Techniques, Predictive Value of Tests, Logistic Models, and Risk Factors.
The list of search terms totaled 26 (22 single textwords and 4 MeSH terms). Using the search operators AND and OR, two-term combinations were created. All 650 possible combinations were searched in the derivation set, and accuracy measures of these combinations were calculated (Figure 2▶). Based on the performance measures of the two-term combinations (e.g., high sensitivity), 18 search strategies were developed using three or more combinations of textwords and MeSH terms. Therefore, using the derivation set, a total of 694 search filters were assessed for usefulness in retrieving clinical prediction rules.
Validation
The filters from the derivation phase were searched in medline using the journal articles from the validation set. Accuracy measures (Figure 2▶) and 95 percent confidence intervals were calculated.
Results
The filter “predict$ OR clinical$ OR outcome$ OR risk$” yielded the highest sensitivity, 98.4 percent. Top filters ranked by sensitivity are listed in Table 2▶. The single term with the greatest sensitivity (78.6 percent) in the validation set was “predict$.” The filter with the highest specificity (99.97 percent) in the validation set was “predict$.ti. AND rule$,” although the sensitivity was 16.1 percent. Of single textwords or MeSH terms, “Decision Support Techniques” yielded the highest specificity (99.5 percent), followed by the single term “rule$” (99.3 percent).
Table 2.
▪ Performance Measures for Search Filters with Highest Sensitivities
| Search Filters* | Sensitivity (95% CI) | Specificity (95% CI) | Positive Predictive Value (95% CI) | Positive Likelihood Ratio (95% CI) |
|---|---|---|---|---|
| Predict$ OR Clinical$ OR Outcome$ OR Risk$ | ||||
| Derivation set | 98.4% (92.4–99.9) | 54.9% (54.0–55.9) | 1.3% (1.0–1.6) | 2.2 (2.1–2.3) |
| Validation set | 98.2% (91.5–99.9) | 55.2% (54.3–56.2) | 1.1% (0.9–1.5) | 2.2 (2.1–2.3) |
| 1—Validat$ OR Predict$.ti. OR Rule$ | ||||
| 2—Predict$ AND (Outcome$ OR Risk$ OR Model$) | ||||
| 3—(History OR Variable$ OR Criteria OR Scor$ OR Characteristic$ OR Finding$ OR Factor$) AND (Predict$ OR Model$ OR Decision$ OR Identif$ OR Prognos$) | ||||
| 4—Decision$ AND (Model$ OR Clinical$ OR Logistic Models/) | ||||
| 5—Prognostic AND (History OR Variable$ OR Criteria OR Scor$ OR Characteristic$ OR Finding$ OR Factor$ OR Model$) | ||||
| 6—OR 1–5* | ||||
| Derivation set | 96.8% (89.9–99.5) | 85.2% (84.6–85.9) | 3.7% (2.8–4.7) | 6.6 (6.2–7.0) |
| Validation set | 98.2% (91.5–99.9) | 86.1% (85.4–86.7) | 3.5% (2.7–4.6) | 7.0 (6.6–7.5) |
| Clinical$ OR Predict$ OR Outcome$ OR | ||||
| Validat$ OR Rule$ OR Predict$.ti. OR | ||||
| Decision Support Techniques/ | ||||
| Derivation set | 95.2% (87.6–98.8) | 61.9% (60.9–62.8) | 1.4% (1.1–1.9) | 2.5 (2.4–2.6) |
| Validation set | 96.4% (88.7–99.4) | 62.4% (61.4–63.3) | 1.3% (1.0–1.7) | 2.6 (2.4–2.7) |
| Predict$ OR Risk$ | ||||
| Derivation set | 93.7% (85.4–98.0) | 77.7% (76.9–78.5) | 2.4% (1.8–3.1) | 4.2 (3.9–4.5) |
| Validation set | 87.5% (76.8–94.4) | 78.1% (77.3–78.8) | 2.0% (1.5–2.7) | 4.0 (3.6–4.4) |
| Predict$ OR Clinical$ OR Outcome$ | ||||
| Derivation set | 93.7% (85.4–98.0) | 62.5% (61.6–63.4) | 1.4% (1.1–1.9) | 2.5 (2.3–2.7) |
| Validation set | 94.6% (86.1–98.6) | 62.9% (62.0–63.8) | 1.3% (1.0–1.7) | 2.6 (2.4–2.7) |
| Predict$ OR Clinical$ OR Outcome$ OR Rule$ | ||||
| Derivation set | 93.7% (85.4–97.9) | 62.1% (61.2–63.0) | 1.4% (1.1–1.8) | 2.5 (2.3–2.6) |
| Validation set | 94.6% (86.1–98.6) | 62.7% (61.7–63.6) | 1.3% (1.0–1.7) | 2.5 (2.4–2.7) |
*Numbers represent separately entered search statements. A slash (/) indicates a term searched as a subject heading.
Four filters were found to have both sensitivity and specificity greater than 90 percent; these are listed in Table 3▶. Using the top two filters in Table 3▶, searches were conducted using PubMed (the National Library of Medicine's medline retrieval service) to assess retrieval for a specific example. Similarly to the example described in the JAMA guide for clinical decision rules, the filters were combined with the MeSH term—Ankle Injuries—and limited to human studies in English published from 1995 through 2000. These searches yielded 55 and 67 articles, respectively.
Table 3.
▪ Performance Measures for Search Filters with High-accuracy Measures
| Search Filters* | Sensitivity (95% CI) | Specificity (95% CI) | Positive Predictive Value (95% CI) | Positive Likelihood Ratio (95% CI) |
|---|---|---|---|---|
| Filters with both sensitivity and specificity above 90%: | ||||
| 1—Validat$ OR Predict$.ti. OR Decision Support Techniques/ OR Rule$ OR Predictive Value of Tests/ | ||||
| 2—Predict$ AND (Clinical$ OR Identif$) | ||||
| 3—OR 1–2* | ||||
| Derivation set | 92.1% (83.3–97.0) | 94.2% (93.7–94.6) | 8.5% (6.5–10.9) | 15.8 (14.3–17.6) |
| Validation set | 91.1% (81.3–96.6) | 94.2% (93.8–94.6) | 7.5% (5.7–9.8) | 15.7 (14.1–17.6) |
| Predict$ OR Validat$ OR Rule$ OR | ||||
| Predictive Value of Tests/ | ||||
| Derivation set | 92.1% (83.3–97.0) | 92.8% (92.3–93.3) | 6.9% (5.3–8.9) | 12.8 (11.6–14.1) |
| Validation set | 91.1% (81.3–96.6) | 93.0% (92.5–93.5) | 6.3% (4.8–8.3) | 13.0 (11.7–14.5) |
| 1—Validat$ OR Predict$.ti. OR Decision Support Techniques/ OR Rule$ OR Predictive Value of Tests/ | ||||
| 2—Predict$ AND (Variable$ OR Factor$ OR Model$ OR Develop$ OR Sensitivit$ OR Clinical$ OR Independent$ OR Prospective OR Identif$) | ||||
| 3—Clinical$ AND (Independent$ OR Model$ OR Sensitivit$) | ||||
| Derivation set | 92.1% (83.3–97.0) | 90.8% (90.2–91.3) | 5.5% (4.2–7.1) | 10.0 (9.1–11.0) |
| Validation set | 92.9% (83.7–97.7) | 91.0% (90.5–91.6) | 5.1% (3.9–6.7) | 10.3 (9.4–11.4) |
| 4—OR 1–3* | ||||
| Predict$ OR Validat$ | ||||
| Derivation set | 90.5% (81.2–96.0) | 93.4% (93.0–93.9) | 7.4% (5.7–9.6)` | 13.8 (12.4–15.4) |
| Validation set | 85.7% (74.7–93.1) | 93.6% (93.1–94.0) | 6.4% (4.8–8.5) | 13.3 (11.7–15.1) |
| Top filters for positive predictive value and likelihood ratios: | ||||
| Decision Support Techniques/ AND Predictive Value of Tests/ | ||||
| Derivation set | 4.8% (1.2–12.4) | 100% (99.97–100.00) | 100% (31.0–100.0) | Undefined |
| Validation set | 1.8% (0.1–8.5) | 99.96% (99.90–99.99) | 20.0% (1.1–70.1) | 48 (5–425) |
| Decision Support Techniques/ AND Predict$.ti. | ||||
| Derivation set | 3.2% (0.5–10.1) | 100% (99.97–100.00) | 100% (19.8–100.00) | Undefined |
| Validation set | 1.8% (0.1–8.5) | 99.95% (99.89–99.98) | 16.7% (0.9–63.5) | 39 (5–326) |
| Predict$.ti. AND Rule$ | ||||
| Derivation set | 9.5% (4.0–18.8) | 99.99% (99.94–100.00) | 85.7% (42.0–99.2) | 1,030 (126–8,431) |
| Validation set | 16.1% (8.1–27.4) | 99.97% (99.91–99.99) | 75.0% (42.8–93.3) | 580 (161–2,085) |
*Numbers represent separately entered search statements. A slash (/) indicates a term searched as a subject heading.
Of the 55 articles retrieved, 26 (47 percent) discussed clinical prediction rules; of the 67 articles, 28 (42 percent) discussed clinical prediction rules. In contrast, the filter with the highest sensitivity but a lower positive predictive value, “predict$ OR clinical$ OR outcome$ OR risk$,” retrieved 345 articles, of which 29 (8 percent) discussed prediction rules.
Filters with the highest positive predictive value and positive likelihood ratios (Table 3▶) had low sensitivities. Of the three shown in Table 3▶, “predict$.ti. and rule$” performed most consistently in both derivation and validation sets. Of single terms, “validat$” and “rule$” were top performers. “Validat$” had a positive predictive value of 23.5 percent and a positive likelihood ratio of 59.3 in the validation set. “Rule$” had a positive predictive value of 19.1 percent and a positive likelihood ratio of 45.6.
Discussion
The goal of this study was to develop and validate an optimal search filter for the retrieval of clinical prediction rules. It was realized, during the course of this study, that one filter could not address the needs of the researcher, clinician, and educator simultaneously. For researchers concerned with maximum retrieval of studies containing clinical prediction rules, a search filter with high sensitivity would be advisable (Table 2▶). As noted previously, the filter with the highest sensitivity captured 98 percent of such studies.
A researcher may want the highest sensitivity to avoid missing a valuable article, whereas a busy clinician may be willing to sacrifice some sensitivity for a higher positive predictive value, resulting in fewer nuisance articles (those that do not contain clinical prediction rules). Several filters performed well for both sensitivity and specificity and had higher positive predictive values (Table 3▶). Depending on the level of searching expertise and the medline search system available, the user may find one of these four filters a good starting point.
For the medical educator who wants to quickly retrieve a clinical prediction rule for illustrative purposes, the use of “predict$.ti. AND rule$” would be a wise choice, yielding three relevant articles for every four articles retrieved (positive predictive value 75.0 percent in the validation set). Although such a search will not yield a comprehensive list of articles containing clinical prediction rules (i.e., the sensitivity is low), it is useful for educators who want to quickly find an article with a clinical prediction rule for educational demonstration. Computer database searching is integral to the practice of evidence-based medicine. Optimal retrieval of the best evidence is based on the formulation of a well-defined question, which includes population, intervention, comparison and outcome, and its translation into a searchable strategy. This was evidenced by the search for ankle injuries, which resulted in an increase in the positive predictive value, from less than 10 percent to over 40 percent. The number of nuisance articles may be reduced when a specific disease, therapy, intervention, or outcome—or a combination of these—is incorporated into the search, as needed.
Conclusion
While the lack of standard nomenclature in articles describing clinical prediction rules and the current vocabulary used for indexing do not offer a simple mechanism for their retrieval, several validated filters perform quite well. The choice of the filter is dependent on the goal of the searcher.
Acknowledgments
The authors thank Carol Lefebvre, Information Specialist at the UK Cochrane Centre, for suggesting that the software SimStat/WordStat, being investigated by colleagues Victoria White and Julie Glanville (NHS Centre for Reviews and Dissemination, University of York), might be suitable for this project.
This work was supported in part by a grant received through the New York State/United University Professions, Joint LaborManagement Committees, Individual Development Awards Program.
References
- 1.Laupacis A, Sekar N, Stiell IG. Clinical prediction rules: a review and suggested modifications of methodological standards. JAMA. 1997;277:488–94. [PubMed] [Google Scholar]
- 2.Wasson JH, Sox HC, Neff RK, Goldman L. Clinical prediction rules: applications and methodological standards. N Engl J Med. 1985;313:793–9. [DOI] [PubMed] [Google Scholar]
- 3.McGinn TG, Guyatt GH, Wyer PC, Naylor CD, Stiell IG, Richardson WS. Users' guides to the medical literature, XXII: How to use articles about clinical decision rules. Evidence-based Medicine Working Group. JAMA. 2000;284:79–84. [DOI] [PubMed] [Google Scholar]
- 4.Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in medline. J Am Med Inform Assoc. 1994;1:447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Weijden T, Ijzermans CJ, Dinant GJ, van Duijn NP, de Vet R. Buntinx F. Identifying relevant diagnostic studies in medline. The diagnostic value of the erythrocyte sedimentation rate (ESR) and dipstick as an example. Fam Pract. 1997;14:204–8. [DOI] [PubMed] [Google Scholar]
- 6.Dickersin K, Scherer R, Lefebvre C. Identifying relevant studies for systematic reviews. BMJ. 1994;309:1286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boynton J, Glanville J, McDaid D, Lefebvre C. Identifying systematic reviews in medline: developing an objective approach to search strategy design. J Info Sci. 1998;24:137–57. [Google Scholar]


