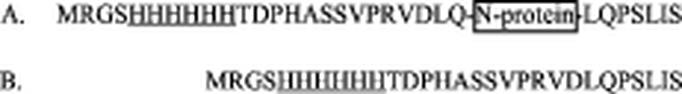Abstract
Two types of porcine reproductive and respiratory syndrome virus (PRRSV) have been reported, the European type (EU PRRSV) and the North American type (US PRRSV). We developed a dual enzyme-linked immunosorbent assay (ELISA) for the simultaneous detection and differentiation of serum antibodies directed against either of the two PRRSV types. This tandem PRRS ELISA is based on affinity-purified recombinant nucleocapsid protein expressed in Escherichia coli. Sensitivity and specificity were assessed by using the IDEXX HerdChek PRRS ELISA and the indirect immunofluorescence assay as reference tests. A total of 1,571 sera originating from the United States, Europe, and two PRRS-free countries, i.e., Switzerland and New Zealand, were used for validation of the tandem PRRS ELISA. The new test performed at least as well as the reference tests in regard to sensitivity (0.94 for the US PRRS ELISA and 0.93 for the EU PRRS ELISA) and specificity (0.96 for the US PRRS ELISA and 0.99 for the EU PRRS ELISA). Positive sera were correctly differentiated in 582 of 591 cases, indicating a high differentiation capability of this dual ELISA. The robustness and repeatability of the test were assessed and found to be appropriate for diagnostic applications. Taken together, the data indicate that the tandem PRRS ELISA described here is the first differentiation ELISA for PRRSV serology based on recombinant antigen. It is convenient with respect to antigen production, and it is reliable, economical, and highly sensitive and specific. Thus, it is considered to be a powerful tool for routine diagnostics, epidemiological surveys, and outbreak investigations.
Porcine reproductive and respiratory syndrome (PRRS) causes major economic losses in the pig industry worldwide. It was first reported in 1987 in the United States, where animals of affected herds showed reproductive failure and respiratory distress. In 1990 and 1991 a similar disease emerged in Europe and spread very quickly through almost the whole continent (1). The viral etiology of PRRS was first shown in The Netherlands in 1991 by Wensvoort and coworkers (29), who isolated Lelystad virus (LV), which is considered the prototype of the European PRRS virus (EU PRRSV). In 1992 the first isolate of PRRSV was identified in the United States and designated ATCC VR-2332 (3). This virus is referred to as the prototype of North American PRRSV (US PRRSV). During the early 1990s it was supposed that each of the two virus types occurred only in the region from where it originated, namely, either Europe or the United States. This changed in 1996, when the use of a live attenuated US PRRSV-based vaccine was registered in several European countries. In 1997 Danish researchers reported the spread of vaccine virus from vaccinated to PRRS-free herds, which was accompanied by occasional clinical disease (4). To date, both PRRSV types can be found in European swine herds as a result of either infection, vaccination, or both. In addition, EU PRRSV was recently reported to circulate in North American swine herds (11), although in the United States only vaccines derived from US PRRSV are allowed. Interestingly, until now there has been no evidence for PRRSV infections in Switzerland (5), and therefore, Switzerland is considered free from PRRS. To maintain this status, reliable tools for monitoring, import controls, outbreak investigations, and follow-up studies are necessary. Furthermore, for epidemiological reasons it is highly desirable to differentiate antibodies against EU and US PRRSVs in any case of seropositivity.
PRRSV is a small, enveloped virus containing a 15-kb positive-strand RNA genome and is classified within the family Arteriviridae, order Nidovirales (6). The viral genome includes the eight open reading frames (ORFs) 1a, 1b, 2, 3, 4, 5, 6, and 7. ORFs 1a and 1b encode nonstructural proteins, whereas ORFs 2 to 7 code for structural proteins (23). Major structural proteins are the nucleocapsid protein (N-protein) (∼15 kDa), the membrane protein M (∼19 kDa), and the envelope glycoprotein GP5 (∼26 kDa), which are encoded by ORF 7, ORF 6, and ORF 5, respectively. The three additional glycoproteins GP2, GP3, and GP4 represent minor structural proteins (8).
Isolation of EU PRRSV is laborious, as consistent propagation of the virus is obtained only in primary porcine monocytes or macrophages, whereas most US PRRSV strains can also be isolated in either MARC-145 or CL2621 cells, which represent subclones of the monkey kidney cell line MA104 (15). Recently, we found that EU PRRSV can also be adapted to these cells (C. Egli, unpublished data).
Several serological tests for routine diagnostics of PRRS have been developed. The immunoperoxidase monolayer assay (IPMA) (28) and the indirect immunofluorescence assay (IFA) (31) are based on the specific binding of antibodies to viral antigens present in infected porcine alveolar macrophages or MA104-derived cells. Additional tests that have been described represent either indirect (2, 7, 27) or blocking (24) enzyme-linked immunosorbent assays (ELISAs) for which antigen derived from infectious virus is used. Further development of the blocking ELISA to permit differentiation of antibodies against the two types of PRRSV has also been reported (25). The common disadvantage of these tests is the need to propagate PRRSV in cell culture, which is time-consuming and expensive. Furthermore, handling of infectious virus in PRRS-free countries such as Switzerland is restricted to containment facilities. These drawbacks can be overcome by using recombinant antigen. Thus, in 1997 we reported an indirect ELISA for the detection of antibodies against EU PRRSV which is based on viral N-protein expressed in insect cells (10). The performance of this test, which has a sensitivity of 1 and a specificity of 0.96, was superior to that of any other test available at that time. However, it has three limitations: (i) it is not appropriate for the detection of antibodies against US PRRSV, as it is based on antigen derived from EU PRRSV only; (ii) the recombinant N-protein is unstable upon long-term storage; and (iii) high background reactivity frequently is observed, particularly with field sera derived from sows. ELISAs based on bacterially expressed recombinant N-protein of US PRRSV have been developed recently (9, 30). Also, an indirect ELISA (Checkit-PRRS) including recombinant N-protein of both EU and US PRRSVs is commercially available (Dr. Bommeli AG, Bern, Switzerland). It is considered to allow detection of antibodies against both types of PRRSV. This is also the case for the HerdChek PRRS antibody ELISA (IDEXX, Westbrook, Maine), which presumably contains one or more antigens specific for both virus types. However, these two commercial tests do not allow discrimination between US and EU PRRSV-derived antibodies.
Here we report the development and the validation of an indirect ELISA based on recombinant N-protein expressed in Escherichia coli, which allows the simultaneous detection and differentiation of antibodies against EU PRRSV and US PRRSV.
MATERIALS AND METHODS
Cells and virus strains.
The US PRRSV vaccine strain VR-2332 (Boehringer-Ingelheim, St. Joseph, Mo.) was passaged once in the monkey kidney cell line MARC-145. The EU PRRSV strain L3, kindly provided by T. Drew (Veterinary Laboratories Agency, Weybridge, United Kingdom), was propagated in porcine alveolar macrophages which had been collected from 6-week-old specific-pathogen-free piglets by lung lavage. It was passaged twice and grown until a cytopathic effect was observed.
Serological tests.
The IDEXX PRRS HerdChek ELISA (IDEXX ELISA) was performed as recommended by the manufacturer. The IFA was carried out as described previously (31).
Serum samples.
Porcine sera were obtained from the United States, New Zealand, and Europe.
The National Veterinary Services Laboratory (Ames, Iowa) provided 80 serum samples. These samples had been tested in the IFA and defined as either positive or negative. Two of these sera had been experimentally raised against the LV isolate. An additional 156 sera were provided by S. Joo and J. E. Collins (University of Minnesota, St. Paul). A set of 60 sera among them originated from pigs experimentally infected with one of eight different US PRRSV strains bled at different times after infection. The remaining 96 sera, including 40 sera which had tested positive in the IFA, represented American field samples.
D. Fichtner (Federal Research Centre for Virus Diseases of Animals, Insel Riems, Germany) provided 51 experimental and field serum samples which had been tested by IFA or IPMA. An additional 33 field sera originated from Bavaria, Germany, and were a gift of H. Gerbermann, Landesuntersuchungsamt für das Gesundheitswesen Südbayern, Oberschleissheim, Germany. They had tested positive in the IDEXX ELISA.
B. Haas (Federal Research Centre for Virus Diseases of Animals, Tübingen, Germany) kindly provided 451 sera, which had been tested in the course of routine diagnosis of PRRS by IPMA. An additional 50 sera tested by IPMA originated from the United Kingdom and were supplied by T. Drew (Veterinary Laboratories Agency). All European sera dated from 1991 to 1995, before vaccination was registered in the European Union, whereas the American sera originated from between 1995 and 1997.
As negative sera, a total of 748 porcine field sera were chosen. Of these, 248 samples had been collected in Switzerland in 1985 and therefore were considered PRRS negative, and 248 samples had been collected and tested negative in the context of an epidemiological study in Switzerland (5). Finally, 252 samples originated from New Zealand, which is considered to be free of PRRS (21).
In addition, we used a total of 19 sera obtained after experimental infection of two specific-pathogen-free pigs with either of the two PRRSV types (10; M. A. Hofmann, unpublished data). Pig EU8787 was infected intranasally with LV; pig US364 was infected in the same way with VR-2332. Blood samples were taken at different times up to day 42 after infection. Preimmune sera EU8787/d0 and US364/d0 served as negative controls in the tandem PRRS ELISA, whereas the sera EU8787/d42 and US364/d42, collected at slaughter, served as positive controls. The latter sera were also used in Western blot analyses (see below).
Porcine sera seropositive for foot-and-mouth disease virus, classical swine fever virus, porcine teschovirus 1, porcine circovirus 2, swine vesicular disease virus, porcine parvovirus, pseudorabies virus, African swine fever virus, porcine transmissible gastroenteritis coronavirus, and porcine epidemic diarrhea virus were taken from our serum collection.
RNA extraction, amplification, and cloning.
Viral RNA was extracted with Trizol (Life Technologies) from supernatants of cell cultures infected with either VR-2332 or L3. The RNA was reverse transcribed by using the oligonucleotide RT-ORF7 as described by Oleksiewicz et al. (22) and modified by Egli et al. (12), and the resulting cDNA was amplified by PCR with Pfu polymerase (Stratagene) and primers USncp_L_PstI (5′AAAACTGCAGATGCCAAATAAAAACGGCAAG3′) and USncp_R_PstI (5′AAAACTGCAGTGCTGAGGGTGATGCTGTG3′) or EUncp_L_PstI (5′AAAACTGCAGATGGCCGGTAAAAACCAGAG3′) and EUncp_R_PstI (5′AAAACTGCAGACTTGCACCCTGACTGGCG3′), respectively. The forward (L) primers contained a PstI restriction site (underlined) immediately upstream of the ATG (boldface) which codes for the first amino acid residue of the N-protein, and the reverse primers (R) contained a PstI site (underlined) immediately downstream of the termination codon (boldface). They were designed according to the respective published sequences for PRRSV VR-2332 (GenBank accession no. U87392) and PPRSV LV (GenBank accession no. M96262). The PCR products were excised after agarose gel electrophoresis and purified by using the QIAquick gel extraction kit (Qiagen). The DNA fragments were then cleaved with PstI, purified by chromatography on an S-400 MicroSpin column (Amersham Bioscience), and ligated into the PstI site of the expression plasmid pQE31 (Qiagen). The ligation reaction products were transfected into Top 10 E. coli cells (Invitrogen). The resulting plasmids were named pQE31_USncp_PstI and pQE31_EUncp_PstI, respectively. The sequences of the respective inserts were confirmed by DNA sequencing with external primers pQE_L and pQE_R as suggested by the manufacturer.
Protein expression.
For protein expression the plasmids pQE31_USncp_PstI, pQE31_EUncp_PstI, and pQE31 were transfected into M15(pRep4) E. coli cells (Qiagen), and then 250 ml of Luria-Bertani medium containing 25 μg of kanamycin per ml and 50 μg of ampicillin per ml was inoculated with 5 ml of overnight culture and incubated for approximately 3 h at 37°C on a horizontal shaker. When the optical density (OD) at 600 nm reached 0.8, expression of recombinant protein was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (Promega) at a final concentration of 2 mM. After further incubation for 5 h, the bacteria were harvested by centrifugation at 4,000 × g for 15 min at 4°C. To extract denatured N-protein, the cell pellets were resuspended in 10 ml of lysis buffer (150 mM NaCl, 200 mM Na2HPO4, 10 mM Tris-HCl, 6 M guanidine hydrochloride, pH 8.0) and incubated for 30 min at room temperature before they were sonicated five times for 5 s each at 300 W. To obtain native N-protein, the cell pellet was resuspended in buffer lacking guanidine hydrochloride and sonicated immediately. Cell debris was removed by centrifugation at 11,000 × g for 30 min, and the supernatant was collected and filtered through a 0.2-μm-pore-size Minisart RC 15 syringe filter (Sartorius, Göttingen, Germany).
Affinity chromatography.
Nickel chelate affinity chromatography was performed on an AEKTA fast protein liquid chromatography apparatus (Amersham Bioscience) by using prepacked 1-ml HiTrap chelating columns (Amersham Bioscience). Each column was loaded with 6 ml of the lysate and consecutively washed with 15 volumes of lysis buffer, 20 volumes of washing buffer I (same as lysis buffer but at pH 6.8), and 15 volumes of washing buffer II (same as lysis buffer but at pH 6.5). Elution was obtained with a linear gradient of 0 to 500 mM imidazole in washing buffer II. Fractions of 3 ml each were collected and further analyzed for the content of viral N-protein. Pooled fractions containing N-protein were stored at 4°C. To obtain mock antigen, lysates of E. coli cells transfected with pQE31 DNA were processed in parallel using the same protocol as for the N-protein, and the corresponding fractions were collected.
Protein analysis.
For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent Western blot analysis, 10-μl aliquots of the affinity chromatography peak fractions were diluted in 1 ml of phosphate-buffered saline (PBS). To remove the guanidine hydrochloride, the protein was precipitated by addition of 1 ml of 10% trichloroacetic acid. The precipitates were washed with ice-cold ethanol and resuspended in 20 μl of 2× SDS-PAGE sample buffer (16). Separation of the proteins by SDS-15% PAGE and Western blot analysis using nitrocellulose membranes were performed essentially as described before (20). After electrophoresis, proteins were visualized by staining with Coomassie blue. Before blotting, the gel was equilibrated for 2 h in 25 mM Tris-HCl-192 mM glycine-0.1% SDS-6 M urea (pH 8.3) and then blotted by using 25 mM Tris-HCl-192 mM glycine-20% methanol (pH 8.3) as transfer buffer. High-titer anti-PRRSV sera US364/d42 and EU8787/d42 served as primary antibodies. Horseradish peroxidase-conjugated rabbit anti-swine immunoglobulin (Dako, Glostrup, Denmark) was used as the second antibody, and the immune complexes were visualized by chemiluminescence with the ECL Plus detection system (Amersham Bioscience).
ELISA.
The purified antigens termed rUS-N-protein, rEU-N-protein, and mock antigen were applied to 96-well microtiter plates (Maxisorb Immunoplates; Nunc, Roskilde, Denmark) in 0.1 M carbonate buffer, pH 9.6. Optimal antigen and serum concentrations were determined by checkerboard titration of strongly positive sera US364/d42 and EU8787/d42 in twofold dilution steps from 1:5 to 1:80 and antigen dilutions from 1:1,600 to 1:52,600. The microtiter plates were coated as follows. Alternating columns of eight wells were coated with 100 μl of mock antigen, rUS-N-protein, and rEU-N-protein per well and left overnight at 4°C. The plates were then blocked for 1 h at 37°C with 200 μl of 10% (wt/vol) dry skim milk in PBS per well. Plates were washed three times with PBS-0.01% (vol/vol) Tween 20 (pH 7.5) and stored at −20°C after addition of 100 μl of PBS per well.
Sera were clarified by centrifugation for 10 min at 14,000 × g and then diluted 1:5 in dilution buffer (1% [wt/vol] dry skim milk in PBS-0.5% [vol/vol] Tween 20). Portions of 100 μl of each diluted serum were transferred to wells coated with the respective antigens. On each plate positive (EU8787/d42 and US364/d42) and negative (EU8787/d0 and US364/d0) control sera for both virus types were included. Serum EU8787/d42 was diluted 1:2 in serum EU8787/d0 before it was further diluted 1:5 as described above. After addition of the sera, the plates were incubated for 90 min at 37°C and then washed three times with PBS-0.01% (vol/vol) Tween 20 (pH 7.5). Subsequently, 100 μl of protein G-horseradish peroxidase conjugate (Zymed, San Francisco, Calif.) diluted 1:4,000 was added to each well and the plates were incubated for 45 min at 37°C. Washing was performed as described above before the addition of 200 μl of substrate {2 mM ABTS [2,2′-azino-di-(3-ethylbenzthiazolinsulfonate)], 6.9 g of NaH2PO4H2O per liter, 76% [vol/vol] 0.1 M acetic acid, 24% [vol/vol] 0.1 M sodium acetate, 0.01% [vol/vol] H2O2}. After 15 to 20 min, when the OD at 405 nm of the positive control sera had reached approximately 1.5, the plates were analyzed on a Multiskan RC photometer (Labsystems, Helsinki, Finland). In order to standardize the assay, for each serum the ΔODs (OD for well containing positive antigen minus OD for well containing mock antigen) with respect to either of the two positive antigens were calculated separately and expressed as percent reactivity by using the following formulas: % reactivityUS ELISA = 100 × [(ΔODsample on US antigen − ΔODUS364/d0 on US antigen)/(ΔODUS364/d42 on US antigen − ΔODUS364/d0 on US antigen)] and % reactivityEU ELISA = 100 × [(ΔODsample on EU antigen − ΔODEU8787/d0 on EU antigen)/(ΔODEU8787/d42 on EU antigen − ΔODEU8787/d0 on EU antigen)]. For samples scoring positive in both the EU and US ELISAs, a reactivity ratio (r) was calculated according to following formula: r = log(% reactivityUS ELISA/% reactivityEU ELISA).
Data analyses.
Performance and optimal cutoff values for the ELISA were determined by using the TwoGraph-ROC (13) functions of the CMDT software package (http://city.vetmed.fu-berlin.de/∼mgreiner/CMDT/cmdt.htm).
Validation.
In order to assess the intertest repeatability of the tandem PRRS ELISA, one set of sera was tested 10 times by three different persons on different days. The set included one weakly positive serum for each of the two virus types (US Ames 42 and EU Tübingen 1166-2). In addition, on the same ELISA plate the positive control sera US364/d42 and EU8787/d42 were titrated after serial twofold dilution in PRRS-negative serum from 1:2 to 1:128. Intratest repeatability was assessed by testing 1:16 dilutions of both positive control sera 10 times at different positions on a single ELISA plate. Mean values, standard deviations, and coefficients of variation (CVs) were calculated for each serum sample.
In another set of experiments, the influence of various factors on the ELISA procedure was examined in order to estimate test robustness. Thus, incubation time (−50% and +50%), incubation temperature (25 and 42°C), intensity of washing of the plates (two times and four times), and precision of pipetting of the reagents (−10% and +10%) were varied.
Cross-reactivity of antibodies against other important viral diseases of pigs was tested using positive sera against foot-and-mouth disease virus, classical swine fever virus, porcine teschovirus 1, porcine circovirus 2, swine vesicular disease virus, porcine parvovirus, pseudorabies virus, African swine fever virus, porcine transmissible gastroenteritis coronavirus, and porcine epidemic diarrhea virus.
RESULTS
Cloning of the ORF 7 gene.
For cDNA synthesis and amplification of the N-protein, appropriate oligonucleotide primers containing a PstI restriction site for subsequent cloning into the expression vector were designed. RNA extracted from cells infected with either of the two PRRSV types was used for reverse transcription and PCR. The respective cDNA fragments containing the sequence for the N-protein were cloned into the PstI site of plasmid pQE31. This prokaryotic vector is designed for expression of proteins N-terminally fused to a polyhistidine sequence, which allows affinity purification of the respective fusion protein on nickel-charged Sepharose resin. Sequencing of the expression plasmids confirmed the expected sequences encoding the respective viral N-proteins. A total of 31 additional vector-derived amino acid residues, including the hexahistidine tag, extend the N-protein at both ends (Fig. 1A). A polypeptide composed of 29 amino acid residues, including the polyhistidine tag, was expressed from the empty vector pQE31 (Fig. 1B). This polypeptide served as mock antigen (negative control) in the ELISA and was isolated by using the same extraction and purification protocol as for N-protein.
FIG. 1.
Recombinant proteins used in the tandem PRRS ELISA. (A) N-proteins of US PRRSV (124 amino acids) and EU-PRRSV (129 amino acids), C- and N-terminally fused to pQE31 vector-derived sequences, are shown. The hexahistidine tag used for nickel chelate affinity chromatography is underlined. (B) Polypeptide composed of 29 amino acid residues expressed from empty pQE31 vector plasmid and used as mock antigen in the tandem PRRS ELISA.
Expression and extraction of recombinant N-protein.
IPTG-induced expression of recombinant N-protein in E. coli was evaluated by using different expression times and IPTG concentrations. The parameters yielding the largest amount of recombinant protein as determined by SDS-PAGE were chosen (data not shown). Bacterial cells were disrupted by sonication, and both the soluble and insoluble fractions were analyzed for the content of native recombinant N-protein. For both rUS-N-protein and rEU-N-protein, approximately 90% of the protein was found in the insoluble fraction, whereas only about 10% was soluble (data not shown). By lysing the cells with guanidine hydrochloride before sonication, a high concentration of denatured recombinant protein was obtained in a soluble form.
Purification of N-protein.
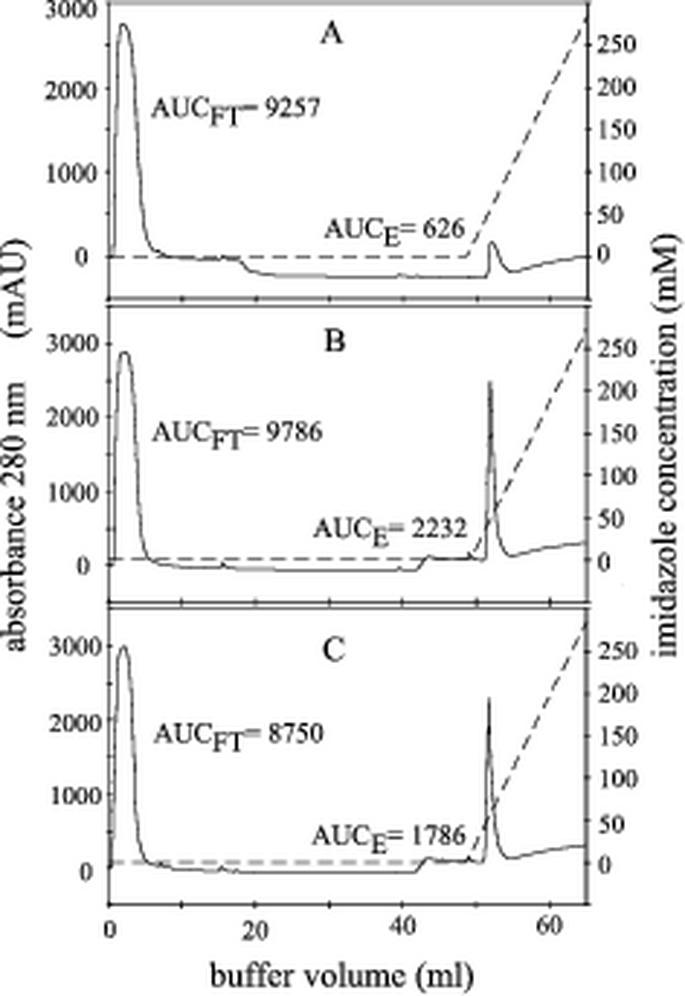
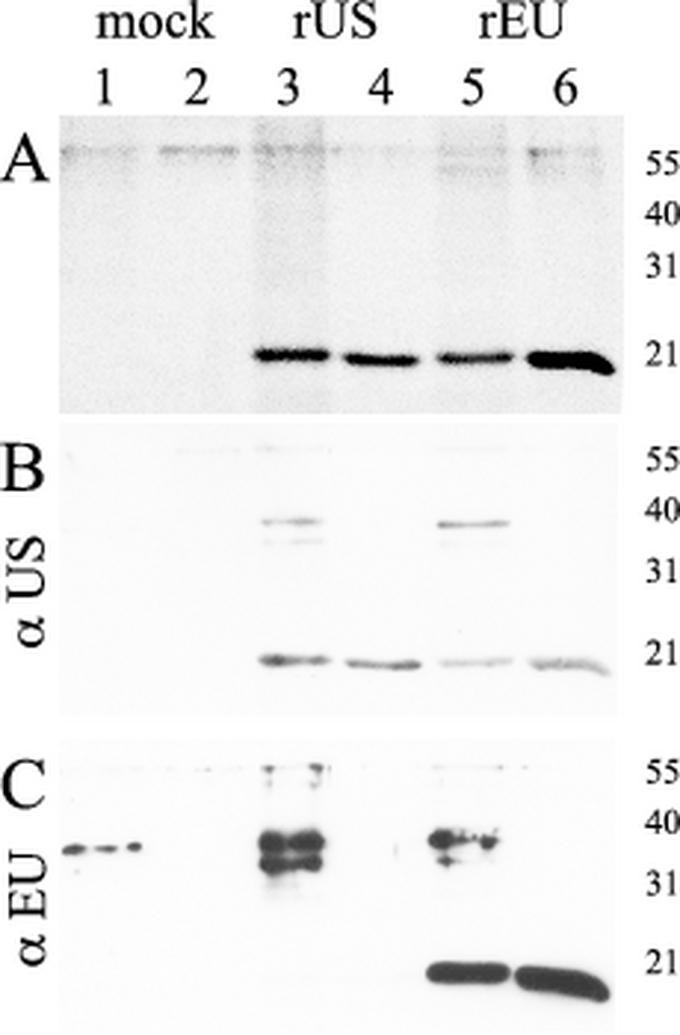
Affinity purification of recombinant protein on nickel-chelate Sepharose was carried out under either native or denaturing conditions in the presence of 6 M guanidine hydrochloride. As shown in Fig. 2, for purification of denatured protein, a well-defined peak was obtained shortly after the onset of the elution gradient for both types of N-protein (Fig. 2B and C). For mock antigen, a peak was observed at the same position, but the amount of protein reflected by the area under the curve was approximately three times smaller (Fig. 2A). Bacterial lysates as well as peak fractions obtained after elution were analyzed by SDS-PAGE and Western blotting. As shown in Fig. 3A for both PRRSV types, SDS-PAGE followed by Coomassie blue staining revealed the presence of a protein slightly below 21 kDa in size. This is in good agreement with the calculated molecular mass of approximately 17 kDa for both N-proteins. Corresponding bands were not visible for mock antigen, which is expected to be a polypeptide of approximately 3.5 kDa and thus not in the resolution range of the 15% polyacrylamide gel used. Western blot analysis with pig serum US364/d42 (Fig. 3B) or EU8787/d42 (Fig. 3C) or monoclonal anti-His antibody (data not shown) proved the identity of the respective protein bands. Additional proteins of approximately 38 and 35 kDa were detected by the pig sera in bacterial lysates but not in the eluate. Interestingly, US364/d42 detected not only the homologous antigen rUS-N-protein (Fig. 3B, lanes 3 and 4) but also the heterologous rEU-N-protein (Fig. 3B, lanes 5 and 6). In contrast, EU8787/d42 detected the rEU-N-protein (Fig. 3C, lanes 5 and 6) but not the rUS-N-protein (Fig. 3C, lanes 3 and 4). Peak fractions of the eluates were pooled and stored at 4°C in elution buffer. Based on photometric measurement of protein concentrations in the eluted fractions, it was calculated that at least 640 μg of purified recombinant N-protein could be obtained per ml of bacterial culture. The recombinant protein was stored in elution buffer and proved to be stable for at least 6 months as concluded from SDS-PAGE and reactivity in the ELISA (data not shown).
FIG. 2.
Nickel chelate affinity purification of denatured recombinant proteins. Lysates containing either mock antigen (A), rUS-N-protein (B), or rEU-N-protein (C) were adsorbed on HiTrap nickel chelate columns. Elution of recombinant protein was obtained with a linear imidazole gradient (dashed line, right y axis). The absorbance at 280 nm measured at the outlet of the column is given in arbitrary milliunits (mAU) (solid line, left y axis). The relative protein contents of flowthrough (FT) and elution (E) calculated by integration of the respective peak areas (AUCFT and AUCE) are given.
FIG. 3.
Analysis of affinity-purified recombinant N-proteins by SDS-PAGE and Western blot analysis. (A) Coomassie blue staining of an SDS-15% polyacrylamide gel. Crude cell lysates (lanes 1, 3, and 5) and affinity-purified proteins (lanes 2, 4, and 6) for mock antigen (lanes 1, 2), rUS-N-protein (lanes 3, 4), and rEU-N-protein (lanes 5, 6) are shown. (B and C) Western blot analysis was performed for the same samples with either US PRRSV-specific antiserum US364/d42 (B) or EU PRRSV-specific antiserum EU8787/d42 (C). The sizes of marker proteins in kilodaltons are indicated.
Evaluation of the tandem PRRS ELISA.
Initially, native recombinant N-protein obtained after sonication of bacterial cells under nondenaturing conditions was evaluated as antigen to establish an indirect ELISA. For both virus types, native N-protein reacted strongly with the homologous experimental PRRSV-positive sera but showed about 40% cross-reactivity with the respective heterologous pig sera (data not shown). In contrast, N-protein isolated under denaturing conditions showed a much lower cross-reactivity. Based on this finding, the ELISA was established and evaluated with denatured antigen.
Checkerboard titration of positive, negative, and respective heterologous sera against increasing antigen dilutions resulted in the highest reactivity and lowest cross-reactivity with sera diluted 1:5 and antigens applied at a concentration of approximately 250 ng of protein per well. Thus, N-protein obtained from one round of purification (3 ml) was sufficient to coat approximately 20,000 plates. Mock antigen was applied at the same dilutions as recombinant N-protein.
Validation of the tandem PRRS ELISA.
For PRRS serology, no method is officially recognized as a “gold standard.” Therefore, we decided to use two established serological tests, the IDEXX ELISA and the IPMA-IFA, to validate the tandem PRRS ELISA. The true status for each of the American, German, and United Kingdom sera was defined by matching results obtained in at least two of the three tests.
A total of 1,571 sera were used for the validation. Initially, 748 sera considered to be negative, as they originated from New Zealand and Switzerland, were tested in the tandem PRRS ELISA. A reactivity of less than 5% was determined for 96% of the sera in the US ELISA and for 97% of the sera in the EU ELISA. Therefore, we selected a preliminary threshold at 5% reactivity. Negative samples with a reactivity of more than 5% in either of the two ELISAs were tested in the IDEXX ELISA, but none of them scored positive.
Sera derived from regions where PRRSV is endemic scored as follows. Sixty-two of the 234 American pig sera were negative in at least two of the three tests (IDEXX ELISA, IPMA-IFA, and tandem PRRS ELISA) and thus were assigned to the negative sera, whereas 172 sera were positive in at least two tests and represented the US PRRS-positive sera. Of 615 European pig sera originating from Germany and the United Kingdom, 132 scored negative and 456 scored positive in at least two tests and were divided into negative and EU PRRS-positive sera, respectively. Overall, 27 of the European sera were excluded from the study: 17 samples reacted unspecifically in either the IPMA-IFA or the tandem PRRS ELISA (strong reaction with mock antigen) and show contradictory results in the two other tests, and 10 sera were excluded due to evident bacterial contamination.
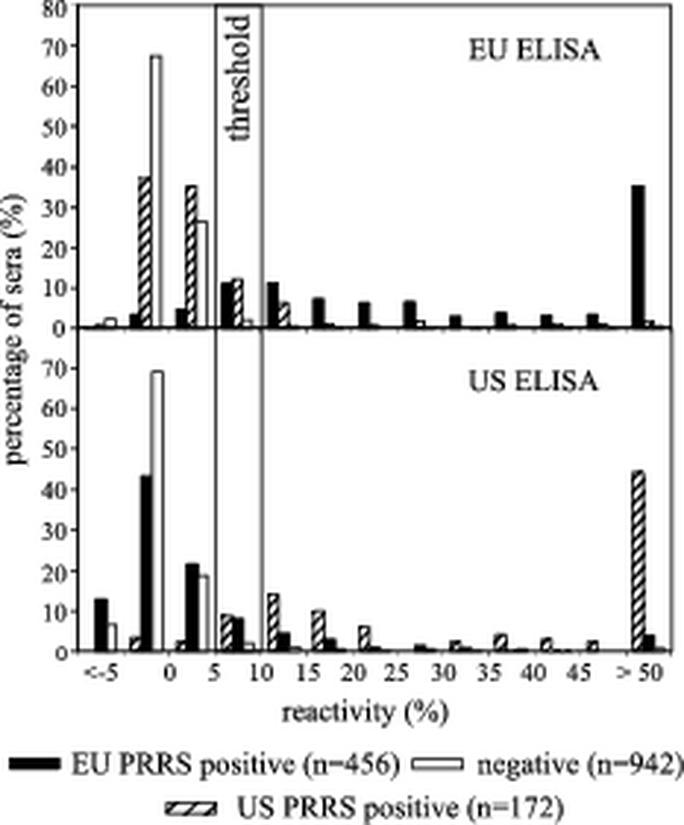
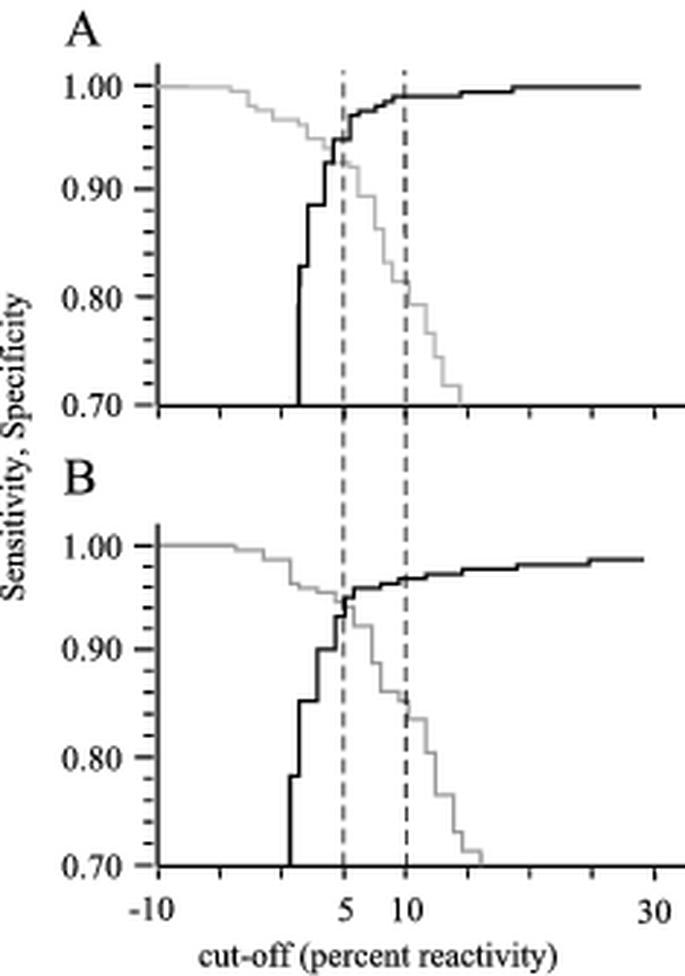
The reactivity of negative, EU PRRS-positive, and US PRRS-positive sera in both the US and EU ELISAs is shown in Fig. 4. With the cutoff set at 5% reactivity, the tandem PRRS ELISA revealed sensitivities of 0.94 (US ELISA) and 0.93 (EU ELISA). The calculated specificities were 0.95 (US ELISA) and 0.97 (EU ELISA). These data were subsequently used to generate TwoGraph-ROC curves (Fig. 5). For the US ELISA, the 5% cutoff correlated with a value of approximately 0.95 for both, sensitivity and specificity. For the EU ELISA, equal sensitivity and specificity was obtained at a calculated cutoff value of 3.8%. However, the analysis shows that a cutoff of 5% is reasonable for both ELISAs. An upper cutoff was set at 10% to define a threshold range.
FIG. 4.
Validation of the tandem PRRS ELISA. The reactivities of a total of 1,571 negative pig sera, EU PRRS-positive pig sera, and US PRRS-positive pig sera were tested in both the EU PRRS ELISA (top panel) and the US PRRS ELISA (bottom panel). The distribution of sera based on their reactivities is shown. Cutoff values at 5 and 10% reactivity are indicated. Reactivity of <5%, negative for PRRSV antibodies; reactivity of ≥5% and <10%, questionable for PRRSV antibodies; reactivity of ≥10%, positive for PRRSV antibodies.
FIG. 5.
TwoGraph-ROC analysis for tandem PRRS ELISA. The correlation between cutoff, sensitivity (gray), and specificity (black) is displayed for the EU ELISA (A) and the US ELISA (B). The graphs were calculated by using the data obtained from testing the 1,571 sera used for validation (Fig. 4) and the CMDT software package. Lower and upper cutoff values of 5 and 10% are indicated with dashed lines.
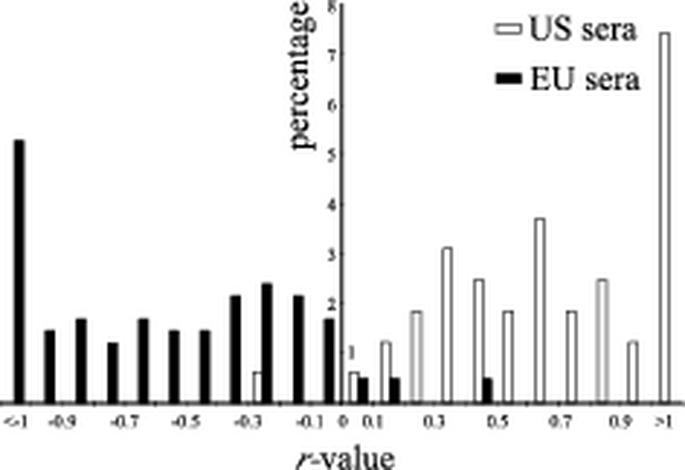
Positive control sera EU8787/d42 and US364/d42 as well as 76% of the EU PRRS sera and 70.5% of the US PRRS sera scoring positive in the tandem PRRS ELISA (Fig. 4) reacted with the homologous antigen (>5% reactivity) but not with the heterologous antigen (<5% reactivity). However, 24% of the EU PRRS-positive sera and 29.5% of the US PRRS-positive sera showed a reactivity of >5% in both the EU and US ELISAs. For each of these double-positive samples an r value, representing the log10 of the ratio obtained by dividing the reactivity observed in the US ELISA by the reactivity observed in the EU ELISA, was calculated (see Materials and Methods). Thus, r values of >0 represent higher reactivity in the US ELISA, and r values of <0 represent higher reactivity in the EU ELISA. The distribution of double-positive sera according to their respective r values is shown in Fig. 6. Only one US PRRS-positive and six EU PRRS-positive sera reacted more strongly with the heterologous than with the homologous antigen. Interestingly, four European sera reacted clearly positive (>10% reactivity) in the US ELISA but were negative in the EU ELISA (data not shown).
FIG. 6.
Potential of differentiation of the tandem PRRS ELISA. The distribution of sera scoring positive in both the EU ELISA and US ELISA is shown according to the calculated r values. The percentage of sera with respect to the total number of positive sera in each test is given.
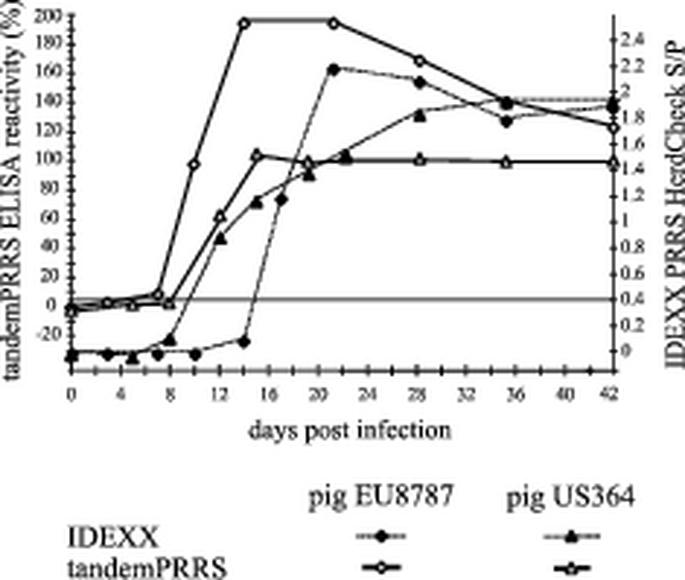
The sensitivity of the tandem PRRS ELISA was additionally assessed by testing sera obtained from pigs experimentally infected with either US PRRSV (pig US364) or EU PRRSV (pig EU8787). These sera were also tested in the IDEXX ELISA (Fig. 7). The tandem PRRS ELISA first detected antibodies in pig EU8787 on day 7 postinfection, whereas the IDEXX ELISA first reacted positively on day 20 postinfection. Pig US364 seroconverted on day 12 according to both ELISAs. IFA was performed with EU8787 serum and yielded the first positive reactions on day 20 postinfection (10).
FIG. 7.
Comparison of the sensitivities of IDEXX ELISA and tandem PRRS ELISA. Sera from two experimental PRRSV-infected pigs were collected at different times postinfection and tested in both ELISAs. Pigs US364 and EU8787 were infected intranasally with VR-2332 and LV, respectively. The horizontal line indicates the cutoff values for both tests.
To exclude reaction of antibodies against other porcine viruses, a set of sera positive for nine different viruses was examined. None of them reached cutoff reactivity (data not shown).
Repeatability and robustness of the tandem PRRS ELISA.
The results obtained in the intertest repeatability experiment show maximal CVs of 16.1 for serum US Ames 42 and 17.9 for serum EU8787/d42, diluted 1:8 (data not shown). Intratest repeatability revealed CVs of 16.4 for the US ELISA and 12.5 for the EU ELISA (data not shown). Robustness studies involving changes introduced in the test procedure never resulted in a higher deviation than the CV observed in the repeatability assays (data not shown).
Performance of the tandem PRRS ELISA.
The sera obtained from the United Kingdom, Germany, and the United States were used to determine the performance of the tandem PRRS ELISA in comparison to the IPMA-IFA and the IDEXX ELISA (Table 1). Using the American sera, the sensitivity and specificity of the US ELISA were determined by alternating selection of one of the other two tests (IPMA-IFA and IDEXX ELISA) as a reference; accordingly, the sera from the United Kingdom and Germany were tested in the EU ELISA (Table 1). For both sets of sera the performance of the IDEXX ELISA in comparison to the IPMA-IFA was also assessed. When the IPMA-IFA was set as a reference, both the IDEXX ELISA and the tandem PRRS ELISA performed almost equally well in detection of antibodies against US PRRSV, with sensitivities of 0.74 and 0.75, respectively, and a specificity of 0.94 for both assays. Comparable results were obtained in detection of EU PRRSV antibodies, with the exception that the tandem PRRS ELISA proved to be more specific (0.80) than the IDEXX ELISA (0.70). When the IDEXX ELISA was set as a reference, the tandem PRRS ELISA was clearly superior to IPMA-IFA the detection of antibodies against US PRRSV. However, when the European sera were tested in the same setup, the IPMA-IFA showed a higher sensitivity (0.88) than the tandem PRRS ELISA (0.83) but a poor specificity of 0.63, versus 0.79 for the tandem PRRS ELISA. Comparison of IPMA-IFA and the IDEXX ELISA with the tandem PRRS ELISA as a reference resulted in sensitivities of at least 0.93 for both tests, but the specificities, particularly that of the IPMA-IFA, were low (0.50 and 0.64).
TABLE 1.
Comparison of sensitivities and specificities of the tandem PRRS ELISA, IDEXX ELISA, and IPMA-IFA for the detection of antibodies against US PRRSV and EU PRRSV
| Origin of sera | Test | Sensitivity and specificity with the following reference testa:
|
|||||
|---|---|---|---|---|---|---|---|
| US or EU ELISAb
|
IDEXX
|
IPMA-IFA
|
|||||
| Se | Sp | Se | Sp | Se | Sp | ||
| United States | US ELISA | 0.94 | 0.86 | 0.74 | 0.94 | ||
| IDEXX | 0.94 | 0.87 | 0.75 | 0.94 | |||
| IPMA-IFA | 0.99 | 0.50 | 0.86 | 0.64 | |||
| Germany and United Kingdom | EU ELISA | 0.83 | 0.79 | 0.85 | 0.80 | ||
| IDEXX | 0.93 | 0.59 | 0.84 | 0.70 | |||
| IPMA-IFA | 0.93 | 0.64 | 0.88 | 0.63 | |||
Sensitivity (Se) = number of positive samples in both the test and the reference test/total number of positive samples in the reference test. Specificity (Sp) = number of negative samples in both the test and the reference test/total number of negative samples in the reference test.
US ELISA for samples from the United States; EU ELISA for samples from Germany and the United Kingdom.
DISCUSSION
The PRRSV N-protein was used to establish an ELISA allowing detection and differentiation of antibodies against European and North American strains of PRRSV. We had shown before that recombinant N-protein can be used as an ELISA antigen for detection of PRRSV antibodies (10). Comparison of the amino acid sequences of North American and European strains showed intratype sequence homologies of approximately 97% (18, 26), whereas there was only about 60% homology when the two types were compared to each other (17, 18). These data suggested that a test based on the respective N-proteins of the two virus types might be sufficiently sensitive and specific for serological detection and differentiation of North American and European PRRSV antibodies.
For cloning and expression of recombinant N-protein, the prokaryotic vector pQE31 containing an IPTG-inducible promoter and designed for synthesis of foreign proteins fused to a hexahistidine tag was selected. Histidine-tagged recombinant N-protein containing additional vector-derived amino acid residues (Fig. 1) at both ends was expressed very efficiently. Isolation of soluble protein under nondenaturing conditions was possible, but the yields obtained under denaturing conditions were about 10 times higher. Native as well as denatured protein was affinity purified (Fig. 2), and the suitability for detection and differentiation of PRRSV antibodies was evaluated. Both forms worked similarly well for detection, but differentiation was more specific with denatured proteins. Four distinct antigenic sites (A, B, C, and D) have been identified for N-protein of PRRSV (19). Antigenic sites A to C are predicted to represent linear epitopes, whereas site D is thought to be conformation dependent. As the amino acid compositions of epitopes B and D are identical for the two virus types, these sites can be considered to be responsible for the observed cross-reactivity. Hence, the conformational epitope D likely accounts for the higher cross-reactivity observed when native N-protein is used. In contrast, epitopes A and C differ between the two types and thus could be responsible for type-specific reactivity of the antibodies. The observed reactivity of serum US364/d42 with rEU-N-protein in Western blot analysis (Fig. 3) but not in the ELISA could be due to renaturation of the D epitope during transfer of the protein from the gel to the nitrocellulose membrane. Antigen purified and stored under denaturing conditions also resulted in long-term stability of the N-protein, in contrast to native antigen, which rapidly lost its activity after purification (data not shown).
Validation of the tandem PRRS ELISA was carried out with a total of 1,571 pig sera. According to our classification, 942 sera were considered seronegative for PRRSV, whereas 172 American sera were considered seropositive for US PRRSV and 456 European sera were considered seropositive for EU PRRSV.
Each of the sera was tested simultaneously in the two ELISAs contained in the tandem PRRS ELISA (Fig. 4). Based on the selected cutoff value of 5% reactivity, both ELISAs showed a sensitivity and specificity of higher than 0.92. These values are considered to be excellent (13) for a diagnostic test. As shown in Fig. 6, calculation of r values for double-positive samples allowed a clear differentiation of the US PRRSV-positive sera from the EU PRRSV-positive sera. Only one of the American sera and six of the European sera reacted more strongly with the heterologous antigen than with the homologous antigen. Interestingly, all of the European sera which reacted more strongly with the heterologous antigen (r value of >0) originated from the United Kingdom and were collected in 1995. At that time, the US PRRSV-based live attenuated vaccine was already widely used in North America, whereas in the European Union it was admitted only in 1996. Whether the nonconsistent result is due to a particular virus strain with unexpected antigenic properties or to illegal use of PRRSV vaccine from the United States remains an open question. We also identified four sera originating from Germany (1991) which were considered positive based on the IDEXX ELISA but which reacted only in the US ELISA, although very weakly. Again, the occurrence of virus strains for which the respective antibodies are not differentiated in the tandem PRRS ELISA cannot be excluded, although the weak reactivity argues rather for an unspecific reaction. It is assumed that US PRRSV was not present in Germany in the early 1990s, although it has occasionally been speculated that US PRRSV has been introduced into Central Europe by a means other than through live vaccines (I. Psikal et al., Proc. 3rd Int. Symp. PRRS Aujeszky's Dis., abstr. P4-P-06, 1999). False-negative results could also be due to the fact that the tandem PRRS ELISA is based on a single viral structural protein as the antigen, whereas IPMA-IFA and allegedly also the IDEXX ELISA apply complete virus as the antigen. Some animals may mainly produce antibodies to other viral proteins or to the conformational antigenic site D and, consequently, would not be recognized in the tandem PRRS ELISA. Finally, the possibility that false-negative results are due to PRRSV strains with N-proteins differing in their antigenic structure from those used here cannot be excluded. When sera experimentally raised against eight different strains of US PRRSV were examined, differences in the ability to detect PRRSV antibodies between the tandem PRRS ELISA and the two reference tests were observed only for the US PRRSV strain VR-2332 (data not shown). The route of virus inoculation used in this experiment is unknown, but we assume that it was by injection. In contrast, sera from pig US364, which was infected oronasally with the same vaccine strain, scored positive as early as day 14 in both the tandem PRRS and the IDEXX ELISA (Fig. 7). Possibly, the route of inoculation modulates the immune response in a way such that early antibodies are not detected by the tandem PRRS ELISA in some cases.
Based on the two reference tests used, false-positive results occurred in the tandem ELISA at frequencies of 4.8% of the sera tested in the US ELISA and 3.3% of the sera tested in the EU ELISA. Seven these sera originated from European PRRSV-positive herds and therefore are very likely to be positive. The negative result in the reference tests would then presumably be due to lower sensitivity of these tests. However, the remaining sera reacting only in the tandem ELISA are derived from New Zealand and Switzerland, where PRRSV has never been reported so far. Therefore, the reactivity observed in these cases is likely to be unspecific.
Comparison of the three tests (Table 1) showed that the tandem PRRS ELISA was superior to IPMA-IFA in both subsets (EU and US PRRS ELISAs) when the IDEXX ELISA was taken as a reference. The IPMA-IFA was highly sensitive but had a very poor specificity. This was even more pronounced when IDEXX ELISA and IPMA-IFA were compared to the US and EU ELISAs as reference tests. Interestingly, in this setup the IDEXX ELISA showed a very poor specificity, particularly in detection of European sera. Finally, comparison of the tandem PRRS ELISA with the IDEXX ELISA using the IPMA-IFA as a reference resulted in almost equal performance concerning sensitivity but clearly superior specificity of the EU ELISA. The high sensitivity of the tandem ELISA is additionally supported by the results obtained for the sera from experimentally infected pigs EU8787 and US364 (Fig. 7). The tandem PRRS ELISA detected antibodies against EU PRRSV significantly earlier after infection than did both reference tests.
Analyses of intratest and intertest repeatabilities showed acceptable results (14).
In order to reduce the risk of false classification, we decided to define an intermediate, questionable range of from 5 to 10% reactivity (Fig. 4 and 5) for future routine diagnostic purposes instead of a single cutoff. In view of the serious economic consequences after positive diagnosis of PRRS, further confirmation tests must be performed in the case of questionable or positive results. Employing this intermediate range to the sets of data shown in our study, specificity values of 0.96 in the US ELISA and 0.99 in the EU ELISA were obtained as analyzed by the TwoGraph-ROC (Fig. 5).
The tandem PRRS ELISA is designed to fulfill the requirements of a diagnostic assay in Switzerland, which is still considered to be free from PRRS. The ability to differentiate between European and North American PRRSV isolates is important for tracing the origin of any PRRS antibody-positive case. When differentiation is not an issue, the test could be modified by combining rUS-N-protein and rEU-N-protein in one single well, allowing more economic screening.
In conclusion, the tandem PRRS ELISA is a simple, reliable, low-cost, and robust test for the detection of antibodies against PRRSV. To our knowledge it is the first test for distinction of antibodies directed against either North American or European strains of PRRSV which is based on a recombinant antigen. Thus, it represents a valuable addition for PRRS serology which will be useful for routine diagnostics, epidemiological surveys, and follow-up investigations of outbreaks.
Acknowledgments
This work was supported by Swiss Federal Veterinary Office grant 1.99.05.
REFERENCES
- 1.Albina, E. 1997. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet. Microbiol. 55:309-316. [DOI] [PubMed] [Google Scholar]
- 2.Albina, E., Y. Leforban, T. Baron, J. P. Plana Duran, and P. Vannier. 1992. An enzyme linked immunosorbent assay (ELISA) for the detection of antibodies to the porcine reproductive and respiratory syndrome (PRRS) virus. Ann. Rech. Vet. 23:167-176. [PubMed] [Google Scholar]
- 3.Benfield, D. A., E. Nelson, J. E. Collins, L. Harris, S. M. Goyal, D. Robison, W. T. Christianson, R. B. Morrison, D. Gorcyca, and D. Chladek. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Invest. 4:127-133. [DOI] [PubMed] [Google Scholar]
- 4.Botner, A., B. Strandbygaard, K. J. Sorensen, P. Have, K. G. Madsen, E. S. Madsen, and S. Alexandersen. 1997. Appearance of acute PRRS-like symptoms in sow herds after vaccination with a modified live PRRS vaccine. Vet. Rec. 141:497-499. [DOI] [PubMed] [Google Scholar]
- 5.Canon, N., L. Audige, H. Denac, M. Hofmann, and C. Griot. 1998. Evidence of freedom from porcine reproductive and respiratory syndrome virus infection in Switzerland. Vet. Rec. 142:142-143. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 7.Cho, H. J., D. Deregt, and H. S. Joo. 1996. An ELISA for porcine reproductive and respiratory syndrome: production of antigen of high quality. Can. J. Vet. Res. 60:89-93. [PMC free article] [PubMed] [Google Scholar]
- 8.Dea, S., C. A. Gagnon, H. Mardassi, B. Pirzadeh, and D. Rogan. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145:659-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dea, S., L. Wilson, D. Therrien, and E. Cornaglia. 2000. Competitive ELISA for detection of antibodies to porcine reproductive and respiratory syndrome virus using recombinant E. coli-expressed nucleocapsid protein as antigen. J. Virol. Methods 87:109-122. [DOI] [PubMed] [Google Scholar]
- 10.Denac, H., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J. Virol. Methods 65:169-181. [DOI] [PubMed] [Google Scholar]
- 11.Dewey, C., G. Charbonneau, S. Carman, A. Hamel, G. Nayar, R. Friendship, K. Eernisse, and S. Swenson. 2000. Lelystad-like strain of porcine reproductive and respiratory syndrome virus (PRRSV) identified in Canadian swine. Can. Vet. J. 41:493-494. [PMC free article] [PubMed] [Google Scholar]
- 12.Egli, C., B. Thur, L. Liu, and M. A. Hofmann. 2001. Quantitative TaqMan(R) RT-PCR for the detection and differentiation of European and North American strains of porcine reproductive and respiratory syndrome virus. J. Virol. Methods 98:63-75. [DOI] [PubMed] [Google Scholar]
- 13.Greiner, M., D. Pfeiffer, and R. D. Smith. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 45:23-41. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson, R. H. 1998. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Technol. 17:469-526. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Mardassi, H., S. Mounir, and S. Dea. 1995. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Quebec reference strain. Arch. Virol. 140:1405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng, X. J., P. S. Paul, P. G. Halbur, and M. A. Lum. 1995. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch. Virol. 140:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meulenberg, J. J., A. P. van Nieuwstadt, A. Essen-Zandbergen, J. N. Bos-de Ruijter, J. P. Langeveld, and R. H. Meloen. 1998. Localization and fine mapping of antigenic sites on the nucleocapsid protein N of porcine reproductive and respiratory syndrome virus with monoclonal antibodies. Virology 252:106-114. [DOI] [PubMed] [Google Scholar]
- 20.Mittelholzer, C., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. Generation of cytopathogenic subgenomic RNA of classical swine fever virus in persistently infected porcine cell lines. Virus Res. 51:125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motha, J., K. Stärk, and J. Thompson. 1997. New Zealand is free from PRRS, TGE and PRCV. Surveillance 24:10-11. [Google Scholar]
- 22.Oleksiewicz, M. B., A. Botner, K. G. Madsen, and T. Storgaard. 1998. Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Vet. Microbiol. 64:7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 24.Sorensen, K. J., A. Botner, E. S. Madsen, B. Strandbygaard, and J. Nielsen. 1997. Evaluation of a blocking Elisa for screening of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus. Vet. Microbiol. 56:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Sorensen, K. J., B. Strandbygaard, A. Botner, E. S. Madsen, J. Nielsen, and P. Have. 1998. Blocking ELISA's for the distinction between antibodies against European and American strains of porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 60:169-177. [DOI] [PubMed] [Google Scholar]
- 26.Suarez, P., R. Zardoya, M. J. Martin, C. Prieto, J. Dopazo, A. Solana, and J. M. Castro. 1996. Phylogenetic relationships of European strains of porcine reproductive and respiratory syndrome virus (PRRSV) inferred from DNA sequences of putative ORF-5 and ORF-7 genes. Virus Res. 42:159-165. [DOI] [PubMed] [Google Scholar]
- 27.Takikawa, N., S. Kobayashi, S. Ide, Y. Yamane, Y. Tanaka, and H. Yamagishi. 1996. Detection of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus in swine sera by enzyme-linked immunosorbent assay. J. Vet. Med. Sci. 58:355-357. [DOI] [PubMed] [Google Scholar]
- 28.Wensvoort, G., E. P. de Kluyver, J. M. Pol, F. Wagenaar, R. J. Moormann, M. M. Hulst, R. Bloemraad, A. den Besten, T. Zetstra, and C. Terpstra. 1992. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: a review of mystery swine disease research at Lelystad. Vet. Microbiol. 33:185-193. [DOI] [PubMed] [Google Scholar]
- 29.Wensvoort, G., C. Terpstra, J. M. Pol, E. A. ter Laak, M. Bloemraad, E. P. de Kluyver, C. Kragten, L. van Buiten, A. den Besten, and F. Wagenaar. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 30.Witte, S. B., C. Chard-Bergstrom, T. A. Loughin, and S. Kapil. 2000. Development of a recombinant nucleoprotein-based enzyme-linked immunosorbent assay for quantification of antibodies against porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab Immunol. 7:700-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon, I. J., H. S. Joo, W. T. Christianson, H. S. Kim, J. E. Collins, R. B. Morrison, and G. D. Dial. 1992. An indirect fluorescent antibody test for the detection of antibody to swine infertility and respiratory syndrome virus in swine sera. J. Vet. Diagn. Invest. 4:144-147. [DOI] [PubMed] [Google Scholar]