Abstract
HOX proteins are important transcriptional regulators in mammalian embryonic development and are dysregulated in human cancers. However, there are few known direct HOX target genes and their mechanisms of regulation are incompletely understood. To isolate and characterize gene segments through which HOX proteins regulate transcription we used cesium chloride centrifugation-based chromatin purification and immunoprecipitation (ChIP). From NIH 3T3-derived HOXA13-FLAG expressing cells, 33% of randomly selected, ChIP clones were reproducibly enriched. Hox-enriched fragments (HEFs) were more AT-rich compared with cloned fragments that failed reproducible ChIP. All HEFs augmented transcription of a heterologous promoter upon coexpression with HOXA13. One HEF was from intron 2 of Enpp2, a gene highly upregulated in these cells and has been implicated in cell motility. Using Enpp2 as a candidate direct target, we identified three additional HEFs upstream of the transcription start site. HOXA13 upregulated transcription from an Enpp2 promoter construct containing these sites, and each site was necessary for full HOXA13-induced expression. Lastly, given that HOX proteins have been demonstrated to interact with histone deacetylases and/or CBP, we explored whether histone acetylation changed at Enpp2 upon HOXA13-induced activation. No change in the general histone acetylation state was observed. Our results support models in which occupation of multiple HOX binding sites is associated with highly activated genes.
INTRODUCTION
Hox transcription factors are essential for normal growth and patterning and regulate regional specification along the anteroposterior axis of the developing embryo and in the developing limb bud (1,2). Also, alterations in Hox gene expression and chromosomal translocations involving Hox genes have been reported in many human cancers (3). HOX proteins bind DNA and regulate the expression of downstream targets (4–6). In vitro the majority of the HOX proteins bind preferentially to the simple core sequence TAAT with the exception of the posterior, Abdominal-B-like genes, which show preference for the simple core sequence TTAT/C (7–9). Enhancement of binding specificity can be accomplished through HOX protein cofactor interaction, and HOX transcriptional activity can be regulated through these interactions (10). HOX proteins have also been shown to associate with chromatin modifying enzymes, including CBP and HDACs (11–15), but the in vivo functions of these interactions are unclear.
Despite the critical importance of HOX proteins in mammalian development and our knowledge of their in vitro binding site preferences, individual gene function in mammals is complicated by functional redundancy, tissue-specific effects and dosage effects. Additionally, a scarcity of in vivo DNA binding sites and direct downstream targets has held up progress in elucidating mammalian HOX biochemical function.
Chromatin immunoprecipitation (ChIP) is one method used to identify transcription factor binding sites in vivo. This method uses formaldehyde to covalently crosslink proteins to their native DNA binding sites followed by transcription factor specific immunoprecipitation and DNA isolation (16–18). For HOX proteins, this method has been used to verify binding of HOX proteins within the promoters of selected downstream targets (19,20) and to identify new HOX binding sites (21), but unbiased genomic approaches to in vivo direct HOX targets have not been reported.
In this paper, we report our initial results with a genomic approach, using ChIP, to isolate and characterize genomic fragments that are bound by HOXA13. We took advantage of our recent report showing that stable expression of HOX proteins in NIH 3T3-derived embryonic fibroblasts causes reproducible expression changes of multiple genes, many of which are biologically significant and/or previously reported putative HOX targets (22). We used this system to stably express FLAG-tagged HOX proteins followed by ChIP with anti-FLAG antibody. We used these cells to construct a library of immunoprecipitated genomic sequences from HOXA13-FLAG and HOXD13-FLAG expressing cells. As proof of principle we confirmed the association of HOXA13-FLAG with a subset of these targets, assessed expression changes of genes located close to these genomic binding locations and explored the ability of these sequences to confer transcriptional activity in transient transfection assays upon coexpression of HOXA13. With one newly identified direct target we explored the density of HOXA13-FLAG binding to its promoter region, the effects of HOXA13 coexpression in target promoter driven reporter assays and the histone acetylation status of ChIP-enriched gene fragments.
MATERIALS AND METHODS
Retroviral vectors and cell culture
All retroviral experiments were performed under BL2 containment and were approved by the University of Michigan Institutional Biosafety Committee. Exon 1 from Hoxa13 was obtained by PCR amplification and subcloned from pCMV-HOXA13, a wild-type cDNA clone (23), and placed together with PCR-amplified C-terminal-FLAG tagged Hoxa13 exon 2 into the BamHI site of pGem5zf+ upstream of an IRES-EGFP cassette (22). An AgeI/XhoI fragment from this clone containing Hoxa13-FLAG-IRES-EGFP was cloned into pRet2 (24). PCR was used to amplify and insert a FLAG-tag in-frame at the C-terminus of Hoxd13, which was then subcloned using AgeI/EcoRI in pRet2-IRES-EGFP (22). Cells were cultured in high glucose DMEM (GIBCO-BRL) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM l-glutamine (GIBCO-BRL). Phoenix-A amphotropic packaging cells (Gary Nolan, Stanford University) were transfected with retroviral plasmid vectors using calcium phosphate Pro-Fection kit (Promega). Retrovirus-containing medium was collected at 24, 48 and 72 h post-transfection, filtered through a 0.45 µM filter (Nalgene) and incubated for 24 h with GP+E86 ecotropic packaging cells (25). Infected GP+E86 cells were sorted by GFP-based FACS to obtain a heterogeneous population of virus producing cells, which were expanded for experiments.
Immunocytochemistry
GP+E86 expressing cell lines were plated on Growth Coverslips (Fisher Scientific) 24 h prior to fixation. Protein expression of Hoxa13-FLAG and Hoxd13-FLAG was analyzed as described previously (23), except cells were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 5 min at room temperature, incubated with a 1:200 dilution of either rabbit anti-HOXA13 (23) or rabbit anti-HOXD13 (MAP-peptide with HOXD13 amino acids 176–198) primary antibody followed by a 1:500 dilution of donkey anti-rabbit Rhodamine conjugated secondary antibody (Santa Cruz Biotechnology). Slides were then mounted with VECTASHIELD containing DAPI (Vector Labs) and photographs were taken using a Zeiss Axiophot microscope (40× objective).
Protein isolation for western blotting
Cells (estimated 70% confluent) were collected by treatment with trypsin-EDTA, pelleted and solubilized in 60 mM Tris, pH 6.8, 2% SDS, 10 mM EDTA, 10 mM EGTA, 10% glycerol and 1× EDTA-free protease inhibitor cocktail (Roche Applied Sciences). Protein concentrations were determined using the BioRad Protein Assay. Equivalent quantities of protein were separated by electrophoresis using 12% SDS–polyacrylamide gels and subsequently electro-transferred to nitrocellulose. Western analysis used primary antibodies at a 1:10 000 dilution of either rabbit anti-HOXA13 (23) or rabbit anti-HOXD13. Secondary antibodies were used at a 1:15 000 dilution of donkey anti-rabbit horseradish peroxide-conjugated secondary antibody (Amersham Biosciences). All antibodies were incubated with blots in PBST with 5% Carnation nonfat dry milk. Protein expression was visualized using Supersignal chemilluminescent substrate (Pierce Biotechnology).
Protein isolation for immunoprecipitation
Cells (estimated 70% confluent) were collected by treatment with trypsin-EDTA, pelleted and lysed by the addition of 1 ml lysis buffer [1% Triton X-100, 0.1% SDS, 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 1× EDTA-free protease inhibitor cocktail (Roche Applied Sciences)] on ice, and mechanically disrupted by aspiration and extrusion through a 26G needle. Immunoprecipitation was preformed using anti-FLAG M2 affinity gel following the manufacturer's protocol (Sigma–Aldrich). Eluted proteins were separated by 12% SDS–polyacrylamide gel electrophoresis. Proteins were electro-transferred to nitrocellulose for western analysis using methods described above.
RNA isolation and semi-quantitative RT–PCR
RNA was isolated using Trizol (Invitrogen) and semi-quantitative one-step RT–PCR was performed using primers to Enpp2, Fhl1, M32486, Ppic and Ngef as described previously (22).
ChIP
A modified version of previously reported ChIP was used (26–28). Cells (1 × 108) were fixed in 10 mM dimethyl adipimidate (DMA) in PBS for 30 min at room temperature, rinsed in PBS and subsequently fixed with 1% formaldehyde in PBS for 10 min at room temperature. Fixation was terminated by the addition of glycine to 125 mM for 5 min at room temperature. Cells were collected at 1000 r.p.m. and lysed on ice for 10 min in ChIP lysis buffer [50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.25% Triton X-100, 1% glycerol and 1× protease inhibitor cocktail (Roche Applied Sciences)]. Nuclei were collected at 4000 r.p.m. for 10 min and then resuspended in sonication buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 0.5 mM EGTA and 1× protease inhibitors). Sonication was performed with a Branson 185 Cell Disrupter on ice to an average length of 500 bp as determined by agarose gel electrophoresis (data not shown). Sonicated samples were centrifuged at 10 000 r.p.m. to remove debris and adjusted to 1.42 g/ml with cesium chloride. Samples were centrifuged at 40 000 r.p.m. in a Ti70.1 rotor for 48 h. 1 ml fractions were collected from the bottom of the gradient. Fractions containing crosslinked chromatin were combined and dialyzed versus TE buffer (data not shown). The dialyzed samples were adjusted to the equivalent of 1.5 × 107 cells/ml with immunoprecipitation buffer to a final concentration of 10 mM Tris, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 0.1% Triton X-100 and 1× protease inhibitors. To obtain a PCR positive control, 500 µl of each sample was incubated at 65°C overnight to reverse the crosslinks, then 10 µg RNAse A and proteinase K were added and incubated at 55°C for 3 h. Samples were extracted twice with Tris-saturated phenol followed by extraction with chloroform and then ethanol precipitated with 5 µg glycogen carrier. Resulting DNA was dissolved in 50 µl sterile water and quantitated using a BioRad SmartSpec 3000. The remaining chromatin samples were adjusted to contain equivalent quantity of total input DNA with ChIP immunoprecipitation buffer and to each ml 20 µl (50% slurry) of washed anti-FLAG M2 agarose (Sigma–Aldrich) was added and rotated overnight at 4°C. For each sample, agarose beads were collected at 1000 r.p.m. and washed six times in ChIP wash buffer (50 mM HEPES, pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% deoxycholate and 1× protease inhibitors) and then one time with TE (pH 8.0) each for 5 min on ice. Immunoprecipitated complexes were eluted with 500 µl 200 mM glycine (pH 2.5) for 5 min on ice. Each eluate was neutralized with 1/10 vol of 1 M Tris, pH 8.0 and incubated at 65°C overnight to reverse crosslinks. To each sample 10 ug each RNAse A and proteinase K were added and the samples were incubated for 3 h at 55°C. Samples were extracted twice with Tris-saturated phenol followed by extraction with chloroform and then ethanol precipitated with 5 µg glycogen carrier. The precipitated material was resuspended in 25 µl sterile water.
Variations of this protocol used with PCR detection include exclusion of DMA fixation, exclusion of CsCl chromatin purification and/or elution by 1% SDS/0.1 M sodium carbonate. For a typical PCR ChIP experiment 1 × 106 cells were fixed for 10 min with FA, lysed with ChIP lysis buffer, sonicated to shear the chromatin to an average length of 500 bp, immunoprecipitated directly with anti-FLAG M2 agarose (Sigma-Aldrich) or with anti-acetyl histone H3 or anti-acetyl histone H4 followed by collection with protein A agarose (Upstate Biotechnology), washed in ChIP wash buffer, eluted with 1% SDS/0.1 M sodium carbonate and DNA was collected as described above.
ChIP fragment cloning
Cloning of DNA fragments collected by ChIP was performed following methods described previously (29). Immunoprecipitated material was treated with T4 DNA polymerase and cloned into PCR-blunt vector (Invitrogen) in 2 µl total volume. Each ligation was transformed into XL-10 Blue (Stratagene) chemically competent cells. The entire transformation was plated onto kanamycin treated Luria broth agar plates. Colonies with inserts were identified by PCR using primers (5′-GTGCTGCAAGGCGATTAAGTTGG-3′ and 5′-CCAGGCTTTACACTTTATGCTTCC-3′) spanning the cloning site in the vector. Inserts >50 bp were chosen for sequencing at the University of Michigan DNA Sequencing Core Lab.
ChIP PCR
PCR mixtures contained equivalent volume immunoprecipitated DNA, 10 mM primer pair, 1.5 mM MgCl2, 0.2 mM each dNTP, 1× PCR buffer (Promega) and 1.25 U Taq DNA polymerase (Promega) in a total volume 25 µl. Primer pairs were designed for each of 21 fragments of interest and designed to amplify fragments of 64–416 bp with an annealing temperature of 53°C. Following 34–40 cycles of PCR amplification, the PCR products were resolved on a 2% agarose gel and visualized using ethidium bromide staining. Primers for ChIP: HEF1 for 5′-GACAGAGAAGTTGGGAGTG-3′; HEF1 rev 5′-TAACGAGTCTGTGAAAGTCTA-3′; HEF2 for 5′-ATACAACATATTTGTAAATTCTAC-3′; HEF2 rev 5′-TAAATGGCATGGGTACATCTA-3′; HEF3 for 5′-CAATGTGGCCTGCCCTTG-3′; HEF3 rev 5′-TGAAGGCTCAGAAGCTGAC-3′; HEF4 for 5′-ACTGGACAGTGAGGTTTCC-3′; HEF4 rev 5′-ACTGCACTGTTCTCGAAGC-3′; HEF5 for 5′-GACATCTTTGTGGATTGTTCA-3′; HEF5 rev 5′-CACATATGTACATAAGGCATC-3′; HEF6 for 5′-CAAGGAATTGGTACAGACTG-3′; HEF6 rev 5′-GCATCACCCTTAAAGTAACATT-3′; HEF7 for 5′-TACCCCATTATCTTTGCTGG-3′; HEF7 rev 5′-ACCTCAGCGTTCCAAGTTC-3′; Enpp2 promoter A-for 5′-CTGGCAGCCCCAGTATTTGAT-3′; Enpp2 promoter A rev 5′-CTCTGTCTTTACCATTTGTTTATTT-3′; Enpp2 promoter B for 5′-CATCTCGAGATGGAATATATAGTAG-3′; Enpp2 promoter B rev 5′-GCATAAAACCCGATAGCACAAAAG-3′; Enpp2 promoter C for 5′-ATTGTCTACATTACCAGGCTCAG-3′; Enpp2 promoter C rev 5′-TGTCTCCACCCCCAACAGGG-3′; Enpp2 promoter D for 5′-GATGAACGGTTACTCACAGATC-3′; Enpp2 promoter D rev 5′-AGGCAAGCTTCAATACAAATGCC-3′; Enpp2 transcription start for 5′-TTGCCTTAAGCCTCTTCTGC-3′; Enpp2 transcription start rev 5′-CCTGGTATGCCCGAAACAG-3′.
Vectors, transient transfections and transcriptional activity assay
To isolate the chicken β-actin minimal promoter pTriEx1.1 (Novagen) was digested with SacII/NsiI, which was cloned into pGL3-basic modified to contain a SacII and NsiI site between HindIII and BglII (pGL3-β-actin). HEFs cloned into PCR-blunt were digested with EcoRI and subcloned into EcoRI digested pBluescript II SK+ (Stratagene). pBluescript cloned HEFs were digested with SacI/XbaI or SacI/KpnI and cloned upstream into pGL3-β-actin digested with either SacI/XbaI or SacI/KpnI to obtain both orientations. The promoter region of Enpp2 was amplified using PCR: P1 = 5′-CTTGGAAAACCTCTGCAGGGTG-3′; P2 = 5′-ATTGTCTACATTACCAGGCTCAG-3′ and cloned in the forward orientation into pGL3-basic (Promega). Site-directed mutagenesis was performed (Stratagene) using oligos: mutA 5′-GAAGTAGCTGACAATGTATCCCCATAGATACAGGGTCATTC-3′; mutB 5′-GAAAAACCTTACATGGGGTCCCCATCTTTTAGAAGCCTTGC-3′; mutC 5′-TATTTGAGATAATGAGAGTCCCCTGTGTTTTACAGTTGATTC-3′. Cos7 cells were transfected with luciferase expression vector, pβactin-βgal (a gift from Didi Robins), and either pCMV5 or pCMV-HOXA13 (23). A total of 125 000 COS7 cells were plated per well in 12-well dishes, in triplicate per condition, 24 h prior to transfection. Fugene-6 transfection reagent (Roche) was used according to the manufacturer's directions. An aliquot of 800 ng total DNA was used per transfection and 2.4 µl total transfection reagent per well. Twenty-four hours post-transfection cell lysates were prepared by rinsing cells two times in PBS followed by incubation for 15 min at room temperature in 200 µl Passive Lysis Buffer (Promega). Each well was scraped and collected into microfuge tubes, vortexed and the debris was pelleted by centrifugation at 4°C. Supernatant solutions were subsequently used in β-galactosidase and luciferase activity assay systems following the manufacturer's protocols (Clontech and Promega, respectively) using a Monolight 3010 luminometer (PharMingen).
Quantitative RT–PCR
First strand cDNA was synthesized from 1 µg total RNA, isolated as described above, using Superscript First Strand cDNA synthesis kit following the manufacturer's protocol (Invitrogen). cDNA equivalent to 10 ng RNA was used in Taqman Gene Expression Assays (Applied Biosystems) following the manufacturer's protocol. Each sample was performed in triplicate. For each reaction 15 µl cDNA in sterile water, 4 µl 5× SurePRIME and GO reaction mixture (Qbiogene Inc.), and 1 µl Taqman Gene expression assay for Ube2v2 (Mm00786385_s1), Mcm4 (Mm00725863_s1) or Hprt (Mm00446968_M1) was used. PCR was performed using an ABI Prism 7700 sequence detection system and software (Applied Biosystems). The results were analyzed using the comparative CT method (Applied Biosystems).
RESULTS
Creation and characterization of cell lines
In a recent report, we described the use of a retroviral system for creating stable cell lines expressing members of the Hox family of genes and demonstrated reproducible gene expression changes (22). We used this approach to create stable cell lines expressing C-terminal FLAG-tagged HOXA13/EGFP and C-terminal FLAG-tagged HOXD13/EGFP with retroviral vectors modified with bicistronic C-terminal FLAG Hox-IRES-EGFP expression cassettes as described in Materials and Methods. The control HOX (−) cell line used was previously described and exhibited no HOXA13 or HOXD13 RNA or protein (22).
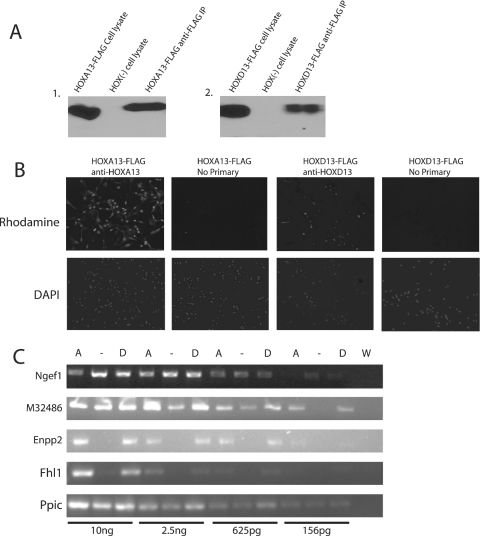
To confirm expression and nuclear localization of HOXA13-FLAG or HOXD13-FLAG proteins, we used western analysis and immunocytochemistry. The HOXA13-FLAG and HOXD13-FLAG cells expressed nuclear localized FLAG-tagged HOXA13 or HOXD13 (Figure 1A and B). To confirm that FLAG-tagged HOX proteins also reproducibly alter gene expression we performed semi-quantitative RT–PCR (Figure 1C). Gene expression changes for Ngef1, M32486, Enpp2 and Fhl1 were consistent with those we reported for the non-tagged HOX proteins (22). Anti-FLAG M2 agarose immunoprecipitation experiments showed that the FLAG-tagged HOX proteins could be immunoprecipitated from total cell lysates (Figure 1A). Thus, these cells produce FLAG-tagged, nuclear localized HOX proteins and exhibit expected changes in gene expression.
Figure 1.
Characterization of HOXA13-FLAG/EGFP or HOXD13-FLAG/EGFP expressing cells. (A) Western blot using HOX-specific antibodies demonstrating HOXA13 expression and anti-FLAG immunoprecipitation from HOXA13-FLAG cell line and absent HOXA13 expression in the HOX (−) cell line (1); HOXD13-FLAG expression and anti-FLAG immunoprecipitation from HOXD13-FLAG cell line but absent HOXD13 expression in the HOX (−) cell line (2). (B) Immunocytochemistry using Hox specific antibodies and DAPI staining demonstrate expression and nuclear localization of HOXA13 or HOXD13 in their respective cell lines. (C) Input RNA using serial dilutions ranging from 156 pg to 10 ng was used in semi-quantitative RT–PCR to look for expression changes of four reported targets. Fhl1 (+6-fold), Enpp2 (+18.8-fold) and M32486 (+2.3) are upregulated in the HOXA13-FLAG (A) and HOXD13-FLAG (D) cell lines and Ngef (−2.4) is downregulated compared to HOX (−). Water (W) was used as a PCR control. Ppic was a loading control and was shown to be unchanged in the HOXA13-FLAG and HOXD13-FLAG cell lines.
Generation of Hox-enriched fragments (HEFs)
To isolate DNA segments associated with HOXA13 and HOXD13 in vivo we used ChIP after crosslinking of protein to DNA. Formaldehyde (FA) is a protein–DNA crosslinker that covalently links amino acids with a primary amine group to DNA through cytosine (30). HOX DNA binding sites are adenine- and thymidine-rich, and we hypothesized that FA alone may not be sufficient to crosslink HOX proteins to DNA. Second, if HOX proteins in some cases are not directly bound to DNA but are part of multiprotein complexes (31,32), FA crosslinking alone may not allow for reproducible recovery of fragments. Homeodomain proteins have been shown in some contexts, including our cells (22), to retain function in the absence of in vitro monomeric DNA binding capability (9,33,34). For these reasons, DMA was included as a pre-FA-crosslink step. DMA promotes formation of covalent protein–protein crosslinks (35) thereby crosslinking HOX proteins to putative DNA-binding protein cofactors that could then be subsequently crosslinked to DNA through FA. Cesium chloride (CsCl)-based chromatin isolation was used to minimize recovery of repeated sequences and free DNA and to enrich for protein-crosslinked DNA (26).
The HOX (−), HOXA13-FLAG and HOXD13-FLAG cells were subjected to crosslinking, lysis, sonication, chromatin isolation by CsCl centrifugation and immunoprecipitation as described in Materials and Methods. For a typical experiment 100 million cells produced ∼340 µg DNA in the crosslinked chromatin prior to immunoprecipitation. After immunoprecipitation direct assessment of the amount of DNA obtained was not performed. Eluted DNA fragments were cloned without PCR amplification and colonies from the resulting transformations were screened for inserts using vector-specific primers. A total of 100 colonies (50 of 934 HOXA13-FLAG and 50 of 826 HOXD13-FLAG derived clones) were randomly screened and, of those screened, 87 had inserts ranging from 43 to 1152 bp (data not shown). A total of 25 of 724 colonies were screened from the control HOX (−) cells and the size variation was consistent with those from the HOX group 13 expressing cells (data not shown). Clones that were determined to be 100% repeated sequence by RepeatMasker (http://www.repeatmasker.org) were not included for further analysis (19 clones). While carryover of some repeats was expected as reported by other studies, it is difficult to evaluate whether these fragments are specifically associated with HOX proteins (36). An additional 13 sequences were of unknown origin, meaning that their identity could not be found in the NCBI m33 Feb 05 database. Thus a total of 32 sequences were excluded from further analysis.
The remaining 55 ChIP fragments were prioritized based on their genomic locations. Twenty-one fragments fulfilled prioritization requirements detailed below. Of these twenty-one, seven were found to be within 500 kb of a gene whose expression was shown to be significantly altered in HOXA13 expressing cells (22); nine individual clones contained at least 50 bp of sequence that was conserved a minimum of 75% to their homologous region in the Homo sapiens genome; the closest gene to nine fragments was a gene known to be important in limb development or a transcription factor; and eight were located within 5 kb of the putative transcription start site of any gene. Ten fragments fell into more than one of these categories and were placed at the top of the priority list (Table 1). Of the remaining 34 clones, 11 were located in intragenic regions outside of the first 5 kb of transcribed sequence. A total of 23 were in intergenic regions over 5 kb away from the transcription start site of the closest gene, within regions not conserved in human syntenic sequence, and over 500 kb away from any gene significantly altered in expression in HOXA13 expressing cells.
Table 1.
Prioritization of 21 cloned ChIP fragments
| Name | Near a regulated genea | Distance to regulated gene | Conserved Hs/Mm | Closest important gene | Near the promoter of any gene | Chromosome | Ensemble Start (NCBI m33 Feb 05) | Size | % AT | Number of replicates enriched |
|---|---|---|---|---|---|---|---|---|---|---|
| BC024683 | Intron 2 (BC024683) | 3 | 88826566 | 98 | 44.3 | 1 of 5 | ||||
| C79747 | 300 kb | Intron 2 (PPP2ce) | 3 | 69879357 | 174 | 50.6 | 0 of 5 | |||
| HEF1 | Ube2v2 | 28 kb | Yes | Mcm4 | 16 | 15397757 | 589 | 58 | 5 of 5 | |
| HEF2 | Enpp2 | Intragenic | Enpp2 | Intron 2 (Enpp2) | 15 | 55091172 | 140 | 73.6 | 5 of 5 | |
| P4ha2 | 218 kb | Acsl6 | 11 | 53985541 | 68 | 47 | 0 of 5 | |||
| HEF3 | Ttrap | 175 kb | Intron 1 (6330500D04) | 13 | 24035157 | 211 | 57.8 | 5 of 5 | ||
| Yes | Ppp1r12b | 1 | 134718064 | 269 | 50.2 | 1 of 5 | ||||
| HEF4 | Yes | Intron 1 (LOC215415) | 1 | 37422455 | 373 | 48 | 5 of 5 | |||
| NEF1 | Yes | Intron 1 (LOC384064) | 4 | 132506680 | 77 | 51.7 | 1 of 5 | |||
| HEF5 | Rbms1 | Intron 1 (Rbms1) | 2 | 60887764 | 250 | 68.8 | 4 of 5 | |||
| Trpm7 | 360 kb | 2 | 126185532 | 243 | 51.9 | 2 of 5 | ||||
| Upstream (LOC233053) | 7 | 23326578 | 552 | 58.4 | 1 of 5 | |||||
| Ngfb | 425 kb | 3 | 102877953 | 307 | 52.8 | 0 of 5 | ||||
| Yes | 10 | 20442119 | 412 | 52.4 | 2 of 5 | |||||
| NEF2 | Yes | 10 | 30931007 | 315 | 56.2 | 2 of 5 | ||||
| Yes | 17 | 26568731 | 153 | 49 | 0 of 5 | |||||
| Yes | 6 | 89186862 | 135 | 38.5 | 0 of 5 | |||||
| Yes | 4 | 102043918 | 570 | 52.5 | 1 of 5 | |||||
| HEF6 | Unc84b | 15 | 80062394 | 160 | 56.3 | 4 of 5 | ||||
| Fhl2 | 1 | 43504079 | 283 | 52.3 | 1 of 5 | |||||
| HEF7 | Foxn2 | 17 | 86974455 | 918 | 49.5 | 4 of 5 |
aWilliams et al. (22).
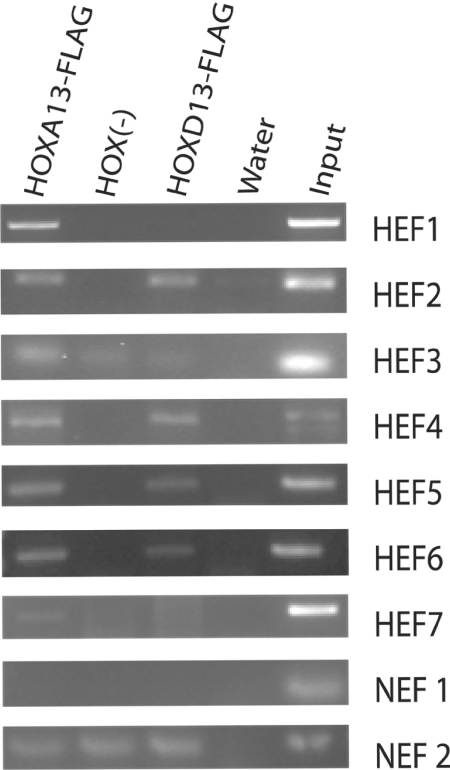
Seven of twenty-one ChIP-cloned fragments are reproducibly enriched
The 21 cloned fragments from the prioritization were tested for reproducibility of enrichment using ChIP and 7 (33%) fragments, hereafter referred to as HEFs, were consistently enriched in the HOXA13-FLAG cells compared with the HOX (−) cells (Figure 2). Enrichment of these HEFs from the HOXD13-FLAG cell line was variable. The reproducibility of HEF enrichment as well as the consistent non-enrichment of other fragments cloned from these cells support our conclusion that HOXA13-FLAG can be recovered in association with specific DNA regions within the genome. We used two non-enriched fragments (NEF1 and NEF2) as negative controls in later experiments.
Figure 2.
Representative PCR enrichment of HEFs. ChIP was performed in HOXA13-FLAG, HOX (−) and HOXD13-FLAG cell lines using anti-FLAG agarose. PCR detection was performed using primers specific to each HEF, NEF1 and NEF2. NEF1 resulted in no detectable product for each cellular sample and NEF2 resulted in product with no detectable difference between the HOXA13-FLAG or HOXD13-FLAG cell lines and the HOX (−) cells. Water was used as a negative PCR control and input ChIP DNA from the HOX (−) cells was used as the positive PCR control.
Sequence comparisons between the 7 HEFs and the 14 NEFs showed few differences. A significant increase in the %A–T in the HEFs (average 58.9%) versus NEFs (average 50.5%) (student's t-test P = 0.029) was observed. While five of the top six fragments with the highest %A–T were reproducibly enriched, HEF4 and HEF7 lie below the average at 48 and 49.5%, respectively (Table 1). This shows that while the %A–T may be a general indicator of HOXA13 association it alone cannot determine whether a fragment will be reproducibly associated with HOXA13 in chromatin. Moreover, 4 HEFs also contained 1 or 2 matches to 7 bp HOX group 13 binding sites (9,20); however of the other 14 fragments, 3 also had 1 or 2 of these sites, a number not statistically different (P = 0.111).
HEFs are enriched independent of DMA crosslinking and CsCl chromatin purification
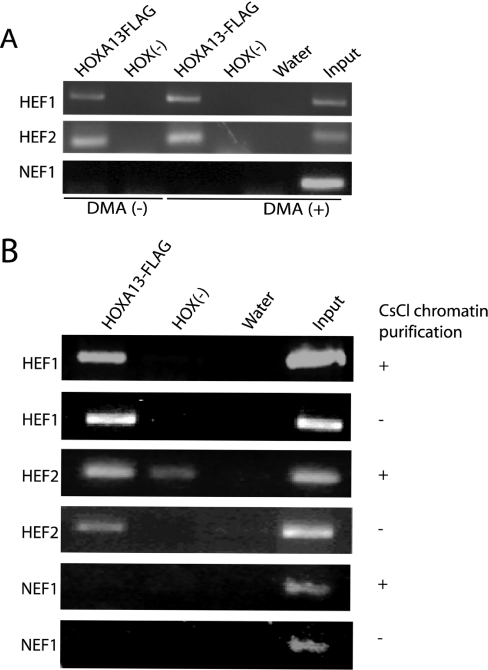
To test the necessity for DMA to obtain enrichment of HEFs, we performed ChIP on HOXA13-FLAG cells and HOX (−) cells each with or without DMA fixation using HEFs1–7 and NEF1 (Figure 3A and data not shown). Enrichment in HOXA13-FLAG cells relative to HOX (−) cells for each HEF was still apparent without DMA treatment; however, for HEF1 upon addition of DMA the enrichment was visibly better (∼1.9-fold). This demonstrates that DMA addition may help recover a selective population of fragments, but may not be necessary.
Figure 3.
HEF enrichment without protein–protein crosslinking and chromatin purification. (A) ChIP was performed in HOXA13-FLAG and HOX (−) cells using anti-FLAG agarose. Elimination of crosslinking with DMA preceding formaldehyde crosslinking is represented in indicated lanes [DMA(−)]. HEF1 and HEF2 are enriched upon addition of DMA as well as without DMA in the HOXA13-FLAG versus HOX (−) cells. HEF1 demonstrated a visibly higher signal with DMA versus DMA (−) in the HOXA13-FLAG cells (1.9-fold by BioRad Quantity One analysis) while HEF2 recovery was equal between the samples. (B) ChIP was performed in HOXA13-FLAG and HOX (−) cell lines using anti-FLAG agarose. CsCl purification of chromatin was eliminated in the indicated samples (−). HEF1 and HEF2 are both enriched in the HOXA13-FLAG cells versus the HOX (−) cells both with and without chromatin purification. There was consistently no product in the HOX (−) sample for HEF1; however, there is a product present in the CsCl purified HEF2 sample.
Chromatin purification via CsCl centrifugation was used to minimize the number of repeated sequence and free DNA fragments cloned into the library and to enrich for crosslinked chromatin as the source for ChIP input. The availability of HEFs allowed us to test whether the CsCl purification was necessary to obtain enrichment of selected fragments using PCR. ChIP was performed on HOXA13-FLAG cells and HOX (−) cells side-by-side with and without chromatin purification. HEF enrichment was still robust and, for HEF2, more apparent without CsCl chromatin purification (Figure 3B and data not shown). Importantly, NEF1 was similarly not enriched under each condition (Figure 3B). The consistent HEF enrichment using several manipulations of ChIP supports our conclusion that the fragments recovered reproducibly from these cells are associated with HOXA13-FLAG in cellular chromatin.
Ube2v2 is upregulated in HOXA13 expressing cells
To correlate gene regulation with HEF location, we examined whether endogenous genes near HEF1 and HEF2 show altered expression in the HOXA13-FLAG cells versus the HOX (−) cells. HEF2 lies within the second intron of Enpp2 whose expression is highly upregulated in these cells as demonstrated by semi-quantitative RT–PCR (Figure 1C). The next closest genes to HEF2 are 2600005O03Rik (210 kb away) and Nov (150 kb away). While the Riken gene was not represented on Affymetrix array MG_U74Av2, the expression of Nov was not altered in previous microarray data (22). Thus, Enpp2 is the only gene close to HEF2 for which HOXA13 expression is known to change steady-state RNA levels.
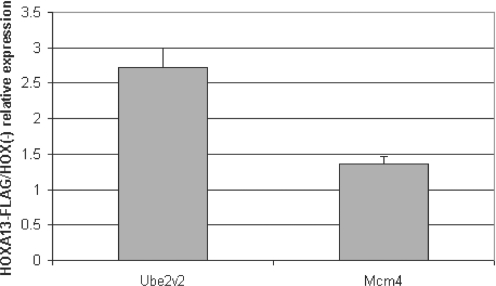
For HEF1, we used real-time PCR. HEF1 is located 30 kb upstream of Ube2v2 and 500 bp downstream of Mcm4 on mouse chromosome 16. The steady-state RNA abundance of Ube2v2 was increased (2.5-fold) in the HOXA13-FLAG cells, while the expression of Mcm4 was unchanged, demonstrating a gene-specific effect of HOXA13-FLAG expression in these cells (Figure 4). The proximity of HEF1 to Ube2v2 and the upregulation of Ube2v2 in the HOXA13-FLAG cells suggest that HOXA13 binding to HEF1 may modulate Ube2v2 expression in vivo.
Figure 4.
Ube2v2 is upregulated 2.5-fold in HOXA13-FLAG cells. Input RNA isolated from HOXA13-FLAG and HOX (−) cell lines were used in real time PCR assays for Ube2v2 and Mcm4. Hprt was used as a loading control to normalize the values between cell lines. Ube2v2 expression was 2.5-fold upregulated in the HOXA13-FLAG cell versus the HOX (−) cells. Mcm4 expression was not changed between cell lines.
HEFs are capable of modifying reporter activity in vitro
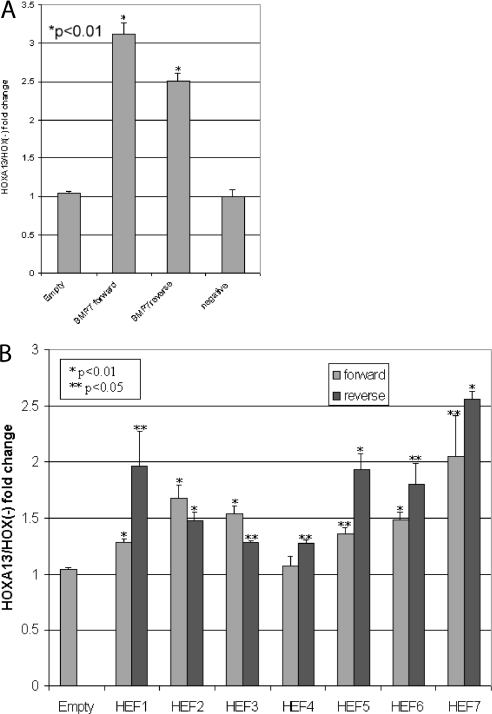
To test the transcriptional activity function, each HEF (1 copy) was cloned into a luciferase reporter plasmid with a minimal promoter and tested for transcriptional regulation by HOXA13. Recently, HOXA13 was reported to enhance (1.8- to 2.0-fold) reporter expression via a fragment in the promoter region of BMP7 (20). We cloned this fragment into our reporter vector and used it as a positive control for HOXA13 function (Figure 5A).
Figure 5.
HOXA13 enhances transcription from HEFs. (A) The enhancer region of Bmp7 (20) was cloned upstream of the chicken β-actin minimal promoter driving luciferase and cotransfected in COS7 cells with CMV-HOXA13 or pCMV as a positive control for HOXA13 function. Addition of CMV-HOXA13 resulted in a 2.5- to 3.1-fold increase in activity over empty vector. NEF1 was also cloned upstream of the chicken β-actin minimal promoter driving luciferase and used in the same assay resulted in no detectable difference in normalized reporter activity upon addition of CMV-HOXA13 over empty vector. A β-galactosidase expression vector was cotransfected as a transfection control and all samples were normalized to its activity. (B) HEFs were cloned in both a forward and reverse orientation upstream of the chicken β-actin minimal promoter driving luciferase and cotransfected in COS7 cells with CMV-HOXA13 or pCMV. Luciferase reporter activity upon addition of HOXA13 resulted in a significant (*P < 0.01; **P < 0.05) increase in relative luciferase activity when compared with identical transfections with control pCMV vector. A β-galactosidase expression vector was used as a transfection control and all samples were normalized to lacZ activity.
Coexpression of HOXA13 with the HEF-reporter plasmids revealed an orientation-independent increase in luciferase activity when compared with control transfections lacking HOXA13 (Figure 5B). The HOXA13-induced HEF transcriptional enhancement ranged from 1.3- to 2.6-fold and transcription of the BMP7 positive control was increased by 2.5 in the forward orientation and 3.2-fold in the reverse orientation over empty vector control. As a negative control, NEF1 was cloned into the reporter vector and, when coexpressed, HOXA13 was unable to change reporter activity demonstrating a fragment specific effect (Figure 5A). The recovery of these fragments using ChIP and their ability to drive reporter expression in the presence of HOXA13 indicates that HOXA13 is able to mediate enhancement of gene expression in vivo via these fragments.
HOXA13 is bound to the Enpp2 promoter in vivo
In transient experiments, HOXA13 was able to enhance reporter expression driven by HEF1 2-fold, consistent with the endogenous gene expression change upon HOXA13 expression in the stable cell lines. However, HEF2 maps to intron 2 of Enpp2 whose endogenous expression in HOXA13-FLAG cells was found to be at least 18-fold upregulated as determined by gel signal analysis (BioRad Quantity One 4.2.1) (Figure 1C) (22). HOXA13 was able to enhance reporter expression driven by HEF2 by 1.5- to 1.7-fold, which does not completely account for the expression level difference seen between the cell lines. In Drosophila homeotic proteins eve and ftz were shown to bind at uniformly high levels through the genomic locus of their activated target genes (37). It is possible that the same phenomenon is seen in the mammalian genome and together multiple binding site occupancy may result in a larger expression change. We hypothesized that multiple, additional binding sites for HOXA13 within the Enpp2 gene, and possibly other genes whose expression was determined to be highly upregulated in these cells, could explain the higher level of expression.
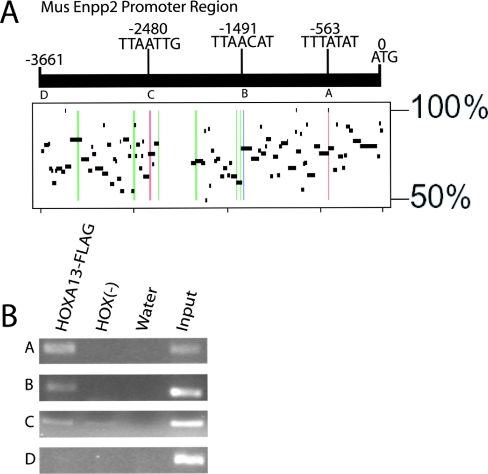
To identify potential HOXA13 binding sites within Enpp2, we searched 3.6 kb upstream of the start methionine (ATG) of the mouse Enpp2 gene for putative binding sites similar in sequence to those previously reported (20). We incorporated nucleotide variations to reflect core binding site differences found in vitro (9). This region of the Enpp2 promoter is highly conserved to the homologous human sequence, which has been shown to contain transcription start sites at −64 bp and −61 bp (38). Three putative HOXA13 binding sites were identified at −563 bp, −1491 bp and −2480 bp (Figure 6A). PipMaker (http://pipmaker.bx.psu.edu/pipmaker/) was used to determine the conservation of this region to the human Enpp2 upstream region. All three potential HOXA13 binding sites fall within human/mouse conserved regions with identity >50% over 50 bp; however, only the 7 bp putative HOX binding site at −1491 bp is 100% conserved over the entire 7 bp (Figure 6A). Within these conserved regions there are several additional candidate HOXA13 binding sites present only in the human sequence; thus the location of candidate sites are not always conserved between species.
Figure 6.
Candidate HOXA13 binding sites in the Enpp2 upstream region are enriched in HOXA13-FLAG expressing cells. (A) Candidate in vivo binding sites for HOXA13 were identified upstream of the mouse Enpp2 translational start methionine (ATG) using in vitro core sequence variations (9,20) and are labeled with their position relative to the ATG. The plot resulting from an analysis using Advanced Pipmaker (http://pipmaker.bx.psu.edu/pipmaker/) shows sequence conservation to the region upstream of the human Enpp2 start codon. The candidate HOXA13 binding sites' locations in the mouse sequence are shown as vertical colored lines. The site conserved between mouse and human is labeled in blue (B). Sites that were found in the mouse sequence but were not fully conserved to human are labeled in red (A and C). Candidate sites that were found in the human sequence but are not fully conserved in mouse are labeled in green. PCR primers were designed around each mouse candidate sites (A–C) as well as one additional sequence within the mouse Enpp2 promoter region without a putative HOXA13 binding motif (D). (B) Chromatin was prepared from HOXA13-FLAG expressing and HOX (−) cells and subjected to anti-FLAG ChIP. The DNA recovered from the ChIP experiments was used in PCR for the sites upstream of the Enpp2 mouse promoter (A–D). Reproducible enrichment (n = 4) of sites ‘A’ and ‘B’ and to a lesser extent ‘C’ was observed in HOXA13-FLAG expressing cells. Site ‘D’ was not detectably enriched between cell lines.
PCR primers were designed around the three candidate mouse sites (A–C) as well as one additional upstream segment devoid of any putative HOX binding motifs (D) as a control. PCR with these primers was used to look for enrichment in HOXA13-FLAG cells versus HOX (−) cells in an anti-FLAG ChIP experiment (Figure 6B). PCR surrounding the potential binding sites A and B and to a lesser extent C consistently demonstrated ChIP enrichment in the HOXA13-FLAG cells. PCR surrounding site D consistently showed no enrichment. To our knowledge this is the first study to demonstrate binding by a mammalian HOX protein across a genetic region at multiple sites encompassing the promoter as well as the transcribed region of an endogenous target gene.
Enpp2 promoter activity is augmented by HOXA13 expression in vitro in transient transfections
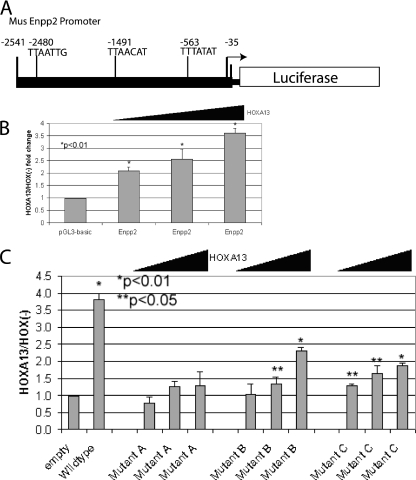
Since ChIP enrichment could be correlated to increased Enpp2 expression we sought to test the ability of HOXA13 to regulate transcription from the Enpp2 promoter in a transient transcription assay. We cloned a segment of the promoter, −35 bp to −2541 bp relative to the start methionine, upstream of luciferase in the pGL3-Basic vector (Promega) (Figure 7A). We cotransfected the reporter construct, a β-Galactosidase expression vector to normalize for transfection efficiency, and increasing concentrations of HOXA13 expression vector and then tested for reporter activity 24 h after transfection. Luciferase activity increased in a HOXA13 dose-dependent manner (Figure 7B).
Figure 7.
HOXA13 augments transcriptional activity from the mouse Enpp2 promoter. (A) PCR was used to amplify the region −35 to −2541 upstream of the mouse Enpp2 start methionine and cloned into the pGL3-basic vector in a forward orientation. (B) Cotransfection of the Enpp2 promoter-driven luciferase reporter vector with increasing concentrations of HOXA13 relative to empty vector results in a dose dependent increase in reporter activity. A β-galactosidase expression vector was cotransfected as a transfection control and all samples were normalized to its activity. (C) Site-directed mutagenesis was used to change the AT-rich core of each putative HOX binding site within the Enpp2 promoter in the context of the pGL3 vector. These vectors were cotransfected with increasing concentrations of HOXA13 plasmid. A marked reduction of relative luciferase activity at each HOXA13 concentration tested was seen for each individually mutated site A, B or C when compared with the wild-type promoter. Significant increases in luciferase activity were observed upon addition of HOXA13 (*P < 0.01; **P < 0.05) with mutant B and mutant C but not mutant A.
To test the necessity of each putative HOXA13 binding site within the Enpp2 promoter region on reporter expression we used site-directed mutagenesis to alter the core AT-rich binding sites of each putative site (Figure 7C). When sites B and C were individually mutated partial activation of the reporter was observed. However, when site A was mutated no significant activation was achieved. Thus, site A appears to be the most indispensable for activation. However, each site was necessary for full activation by HOXA13. Therefore, HOX-activation of this target gene may be realized through a net-additive effect of occupancy of multiple sites.
Enpp2 promoter histone acetylation state is not altered in HOXA13-FLAG cells
HOX proteins are likely to achieve their regulator function through interactions with coactivators and corepressors. One such class of coregulators is histone-acetylation state modifying enzymes (39). Members of the HOX family of proteins have been shown to interact with histone deacetylases (HDACs) (12,15) as well as the histone acetyl transferase (HAT), CBP (11–13). The effect of HOX protein/HDAC or HOX protein/CBP interactions on the acetylation state of histones at in vivo HOX binding sites is unknown.
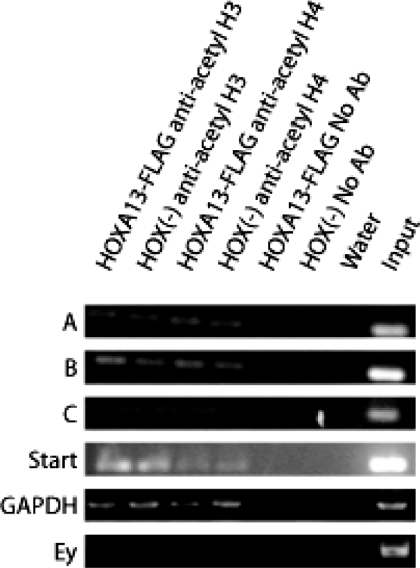
To test whether HOXA13-FLAG occupancy at the Enpp2 promoter has an effect on the overall acetylation status of local histones we performed ChIP using anti-acetyl (K9 and K14) histone H3 and anti-acetyl (K5, K8, K12 and K16) histone H4 (Upstate Biotechnology) which includes residues acetylated by CBP (histone H3 K14; histone H4 K5) (40,41). PCR for fragments A, B, C, D, HEF2 and primers surrounding the Enpp2 transcription start site were used. There was no enrichment for acetyl-histone H3 or H4 within the loci bound by HOXA13 or at the transcription start site in the HOXA13-FLAG cell line versus the HOX (−) cell line (Figure 8 and data not shown). A region of GAPDH was used as a positive control and was equally recovered between the two cell types. A region of the mouse Ey-globin locus (Ey) was used as a negative control and was not recovered from both cell types as expected. Our results indicate that HOXA13 does not affect the general acetylation status of histones when bound at the Enpp2 genetic locus as detectable by this method; however, this experiment does not eliminate the possibility that HOXA13 may be affecting the acetylation of a single histone residue not detectable using these general acetyl histone antibodies or otherwise involving other chemical histone modifications.
Figure 8.
Histone acetylation state is not altered at HOXA13 genomic binding sites within the Enpp2 promoter. ChIP was performed on the HOXA13-FLAG cells versus the HOX (−) cells using anti-acetyl Histone H3 or anti-acetyl Histone H4 antibodies or no antibody. Recovery of each site was detectable using both antibodies versus no antibody but there was no detectable difference between cell lines for recovery of sites A-C or the transcription start site.
DISCUSSION
Using a ChIP-based cloning approach, we identified novel HOXA13 genomic binding regions. We found that 33% of the randomly selected ChIP clones were reproducibly enriched in independent ChIP assays. To our knowledge, these HEFs represent the first reported mammalian HOX binding sites outside of the immediate upstream regions of candidate genes. The HEFs ranged in location from 3 to 80 kb from their closest gene lending evidence to the hypothesis that HOX proteins can bind and may function at considerable distances from the promoters of genes.
Emphasis on defining a HOX binding sequence has led to the use of in vitro methods to define the DNA binding sequence for HOX proteins (42). However, the existence of a putative site within the genome does not necessarily indicate that a HOX protein would be bound in vivo or even functional when bound (1,43–45). Thus, using sequence information alone overestimates the library of putative sites within regions of target genes and the prospect of long-range (megabase) interactions, as demonstrated for some enhancers (46), further complicates the identification of relevant binding sites in silico. Additionally, variable flanking DNA sequences (47) may confer modest binding affinity differences for individual sites. Occupancy of sites will also vary between tissues and may be dependent on HOX protein expression level, chromatin conformation and presumably the presence of cofactors. Therefore, assessing whether an in silico site is occupied and functional in vivo requires experimental exploration as we have shown.
Chromatin immunopurification protocol modifications
To obtain the purest population of authentic HOXA13 binding fragments, we included DMA crosslinking and chromatin purification prior to immunoprecipitation when the library was cloned. These steps were included in previous reports and significantly increased enrichment of fragments (26,35). Using this technique we showed that 33% of clones tested were reproducibly enriched in up to five independent ChIP experiments, consistent with what has been reported with ChIP followed by cloning (29,48–50). In our hands, DMA crosslinking and CsCl-based chromatin purification are not necessary to obtain enrichment. However, the proportion of ChIP-derived clones that could be obtained from exclusion of these steps was not tested and could be <33%.
HEFs and gene expression
Three (HEF1, HEF2 and HEF3) of the seven HEFs were located within 500 kb of a gene that was changed in expression in HOXA13 stable cells. HEF1 was located 28 kb upstream of Ube2v2, HEF3 was located 175 kb upstream of Ttrap, and HEF2 was located within the Enpp2 gene. However, we only sequenced ∼5% of the total clones and it is reasonable to expect that within the library there are many additional clones, mapping within or immediately upstream of transcriptionally altered genes. Additionally, it is possible that we did not detect expression changes in genes near each of the other HEFs for several reasons. (i) Chromatin structure may inhibit the ability of HOXA13 to exert function on nearby genes. (ii) There may be absence of necessary cofactors in certain genomic regions as suggested by the widespread binding model (4). (iii) The level of gene expression change may have been below the level of detection or significance for the microarray assays.
HEF2 was located in intron 2 of Enpp2. The vast increase in the expression of Enpp2 in these cells is consistent with the data in Drosophila of multiple binding sites within a single genetic locus that might account for the large expression change (37). We were able to use ChIP to demonstrate reproducible enrichment of fragments located in the immediate upstream region of the Enpp2 promoter and at least four HOXA13 associated fragments are present within this region tested (Figure 6). This was not an exhaustive study and further analysis of the entire Enpp2 genetic locus, perhaps with ChIP followed by microarray hybridization (51), would reveal several additional fragments. The genomic extent of HOXA13 occupancy over the Enpp2 locus would be interesting to determine and might shed light on the mechanism that HOX proteins use to regulate their downstream targets.
Dose-dependent HOXA13 response
Similar to other HOX family members, HOXA13 and HOXD13 have been shown to demonstrate a gene dosage-dependent effect in their regions of expression, including the developing limb bud, digestive tract and urogenital tract (52–56). The promoter region of Enpp2 demonstrates a dose-dependent increase in reporter gene activity upon HOXA13 addition (Figure 7B). Our data support a model that quantitative regulation of HOX direct downstream targets is one method in which dose dependency can be realized (57). This could be accomplished through the use of multiple binding sites. While we have shown only one gene with multiple sites, our data are also consistent with that from Drosophila showing the accumulation of multiple, functional binding sites within homeotic target genes (37,58).
HOXA13 does not affect local histone acetylation status at the Enpp2 promoter
Two models to explain HOX protein binding site specificity are the activity regulation model (4,59,60) and the binding site selection model (61). For either model HOX proteins are likely to achieve their transcriptional regulation through association with coactivators and corepressors. We found no differences in the acetylation status of histones at the HOXA13 bound fragments of the Enpp2 genetic locus even though it is also clear that histones are acetylated at transcriptionally active genes. Recruitment of HOXA13 to a target locus may not necessarily influence the proportion of acetylated histones at a detectable level using this method. This does not eliminate the possibility that Enpp2 regulation in the HOXA13 expressing cells is associated with other histone modifications. For example, site-specific methylation of K9 and K4 in histone H3 has been strongly implicated in roles in gene repression and gene activation, respectively (62). The vast upregulation of Enpp2 in the HOXA13 expressing cells implicates histone modification at this locus and the methylation state is a compelling candidate to examine even though HOX proteins have not been found to be associated with histone methyltransferases.
Biological relevance
The direct regulation of Enpp2 expression by HOXA13 is intriguing; however, the tissue-specific role this regulation plays in vivo is unclear and needs further investigation. Enpp2 was first identified as ‘autocrine motility factor’ for melanoma cells and later shown to be a member of the ecto-nucleotide pyrophosphatase/phosphodiesterase family of proteins (63). Recently, Enpp2 was found to be a regulator of adhesive properties of the extra-cellular matrix (64) and to catalyze the production of lysophosphatidic acid, a lipid mediator with potent roles in diverse cell types for cell proliferation, migration and survival (65,66). Many HOX downstream genes that have been identified are known to be involved in these processes (67,68). This suggests that induction of Enpp2 expression may allow HOXA13 to function in cell survival or cell motility pathways.
HOXA13 specifically has been shown to be (i) expressed in migrating premyoblasts (69), (ii) necessary for autopod outgrowth and development (53), (iii) involved in organization of mesenchymal and endothelial cell layers of the umbilical arteries (70) and (iv) critical for apoptosis in the urethra without which results in hypospadias (71). Thus, it is intriguing to speculate a role for Enpp2 in these tissues.
Using Enpp2 as a direct target of HOXA13 will allow us to thoroughly explore the domains of the HOXA13 protein that are necessary for its transcriptional effects, determine whether addition of known cofactors will augment the ability of HOXA13 to report expression from the promoter, explore the ability of other HOX family members to regulate expression from this promoter and explore the chromatin structure at a known direct target containing multiple, necessary HOXA13 binding sites.
Acknowledgments
The authors thank Tom Williams for the creation of the HOX (−) cell line and training in cell line creation. The authors also thank Amanda Vespers for training in real-time PCR and Sally Camper for reagents. The authors also thank Mara Steinkamp for advice and Didi Robins for luciferase expression reagents. The authors also thank Greg Dressler for a critical review of the manuscript. This work was supported by NIH grant HD37486 to J.W.I. C.D.M. was supported in part by a NIH Training Grant Fellowship in Cellular Biotechnology (T32 GM08353). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Veraksa A., Del Campo M., McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Mol. Genet. Metab. 2000;69:85–100. doi: 10.1006/mgme.2000.2963. [DOI] [PubMed] [Google Scholar]
- 2.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 3.Grier D.G., Thompson A., Kwasniewska A., McGonigle G.J., Halliday H.L., Lappin T.R. The pathophysiology of HOX genes and their role in cancer. J. Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 4.Biggin M.D., McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 5.Graba Y., Aragnol D., Pradel J. Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays. 1997;19:379–388. doi: 10.1002/bies.950190505. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi S., Scott M.P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- 7.Benson G.V., Nguyen T.H., Maas R.L. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol. Cell. Biol. 1995;15:1591–1601. doi: 10.1128/mcb.15.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen W.F., Rozenfeld S., Lawrence H.J., Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 9.Caronia G., Goodman F.R., McKeown C.M., Scambler P.J., Zappavigna V. An I47L substitution in the HOXD13 homeodomain causes a novel human limb malformation by producing a selective loss of function. Development. 2003;130:1701–1712. doi: 10.1242/dev.00396. [DOI] [PubMed] [Google Scholar]
- 10.Mann R.S., Affolter M. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 11.Chariot A., van Lint C., Chapelier M., Gielen J., Merville M.P., Bours V. CBP and histone deacetylase inhibition enhance the transactivation potential of the HOXB7 homeodomain-containing protein. Oncogene. 1999;18:4007–4014. doi: 10.1038/sj.onc.1202776. [DOI] [PubMed] [Google Scholar]
- 12.Saleh M., Rambaldi I., Yang X.J., Featherstone M.S. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen W., Chrobak D., Krishnan K., Lawrence H.J., Largman C. HOXB6 protein is bound to CREB-binding protein and represses globin expression in a DNA binding-dependent, PBX interaction-independent process. J. Biol. Chem. 2004;279:39895–39904. doi: 10.1074/jbc.M404132200. [DOI] [PubMed] [Google Scholar]
- 14.Shen W.F., Krishnan K., Lawrence H.J., Largman C. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 2001;21:7509–7522. doi: 10.1128/MCB.21.21.7509-7522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y., Goldenberg I., Bei L., Andrejic J., Eklund E.A. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J. Biol. Chem. 2003;278:47792–47802. doi: 10.1074/jbc.M305885200. [DOI] [PubMed] [Google Scholar]
- 16.Ren B., Dynlacht B.D. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 2004;376:304–315. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- 17.Weinmann A.S., Farnham P.J. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 18.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 19.Lei H., Wang H., Juan A.H., Ruddle F.H. The identification of Hoxc8 target genes. Proc. Natl Acad. Sci. USA. 2005;102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knosp W.M., Scott V., Bachinger H.P., Stadler H.S. HOXA13 regulates the expression of bone morphogenetic proteins 2 and 7 to control distal limb morphogenesis. Development. 2004;131:4581–4592. doi: 10.1242/dev.01327. [DOI] [PubMed] [Google Scholar]
- 21.Tomotsune D., Shoji H., Wakamatsu Y., Kondoh H., Takahashi N. A mouse homologue of the Drosophila tumour-suppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature. 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- 22.Williams T.M., Williams M.E., Kuick R., Misek D.E., McDonagh K., Hanash S., Innis J.W. Candidate downstream regulated genes of HOX group 13 transcription factors with and without monomeric DNA binding capability. Dev. Biol. 2005;279:462–480. doi: 10.1016/j.ydbio.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Post L.C., Margulies E.H., Kuo A., Innis J.W. Severe limb defects in Hypodactyly mice result from the expression of a novel, mutant HOXA13 protein. Dev. Biol. 2000;217:290–300. doi: 10.1006/dbio.1999.9550. [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Friedman M.S., Bian H., Crofford L.J., Roessler B., McDonagh K.T. Highly efficient genetic transduction of primary human synoviocytes with concentrated retroviral supernatant. Arthritis Res. 2002;4:215–219. doi: 10.1186/ar409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz D., Hesdorffer C., Ward M., Goff S., Bank A. Retroviral gene transfer using safe and efficient packaging cell lines. Ann. NY Acad. Sci. 1990;612:407–414. doi: 10.1111/j.1749-6632.1990.tb24328.x. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi Y., Rayman J.B., Dynlacht B.D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 27.Orlando V., Strutt H., Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 28.Fujita N., Jaye D.L., Kajita M., Geigerman C., Moreno C.S., Wade P.A. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 29.Weinmann A.S., Bartley S.M., Zhang T., Zhang M.Q., Farnham P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGhee J.D., von Hippel P.H. Formaldehyde as a probe of DNA structure. II. Reaction with endocyclic imino groups of DNA bases. Biochemistry. 1975;14:1297–1303. doi: 10.1021/bi00677a030. [DOI] [PubMed] [Google Scholar]
- 31.Shen W.F., Rozenfeld S., Kwong A., Kom ves L.G., Lawrence H.J., Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shanmugam K., Green N.C., Rambaldi I., Saragovi H.U., Featherstone M.S. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 1999;19:7577–7588. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vershon A.K., Jin Y., Johnson A.D. A homeo domain protein lacking specific side chains of helix 3 can still bind DNA and direct transcriptional repression. Genes Dev. 1995;9:182–192. doi: 10.1101/gad.9.2.182. [DOI] [PubMed] [Google Scholar]
- 34.Fitzpatrick V.D., Percival-Smith A., Ingles C.J., Krause H.M. Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature. 1992;356:610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- 35.Kurdistani S.K., Grunstein M. In vivo protein–protein and protein-DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- 36.Martens J.H., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter J., Dever C.A., Biggin M.D. Two homeo domain proteins bind with similar specificity to a wide range of DNA sites in Drosophila embryos. Genes Dev. 1994;8:1678–1692. doi: 10.1101/gad.8.14.1678. [DOI] [PubMed] [Google Scholar]
- 38.Kawagoe H., Stracke M.L., Nakamura H., Sano K. Expression and transcriptional regulation of the PD-Ialpha/autotaxin gene in neuroblastoma. Cancer Res. 1997;57:2516–2521. [PubMed] [Google Scholar]
- 39.Lopez-Rodas G., Brosch G., Georgieva E.I., Sendra R., Franco L., Loidl P. Histone deacetylase. A key enzyme for the binding of regulatory proteins to chromatin. FEBS Lett. 1993;317:175–180. doi: 10.1016/0014-5793(93)81271-z. [DOI] [PubMed] [Google Scholar]
- 40.Schiltz R.L., Mizzen C.A., Vassilev A., Cook R.G., Allis C.D., Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 41.McManus K.J., Hendzel M.J. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol. Cell. Biol. 2003;23:7611–7627. doi: 10.1128/MCB.23.21.7611-7627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991;30:11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 43.Scott M.P., Weiner A.J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Natl Acad. Sci. USA. 1984;81:4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaRonde-LeBlanc N.A., Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gehring W.J., Qian Y.Q., Billeter M., Furukubo-Tokunaga K., Schier A.F., Resendez-Perez D., Affolter M., Otting G., Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 46.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 47.Zeng W., Andrew D.J., Mathies L.D., Horner M.A., Scott M.P. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- 48.Wang H., Tang W., Zhu C., Perry S.E. A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J. 2002;32:831–843. doi: 10.1046/j.1365-313x.2002.01455.x. [DOI] [PubMed] [Google Scholar]
- 49.Stevens T.A., Iacovoni J.S., Edelman D.B., Meech R. Identification of novel binding elements and gene targets for the homeodomain protein BARX2. J. Biol. Chem. 2004;279:14520–14530. doi: 10.1074/jbc.M310259200. [DOI] [PubMed] [Google Scholar]
- 50.Seki K., Hata A. Indian hedgehog gene is a target of the bone morphogenetic protein signaling pathway. J. Biol. Chem. 2004;279:18544–18549. doi: 10.1074/jbc.M311592200. [DOI] [PubMed] [Google Scholar]
- 51.Kirmizis A., Farnham P.J. Genomic approaches that aid in the identification of transcription factor target genes. Exp. Biol. Med. (Maywood) 2004;229:705–721. doi: 10.1177/153537020422900803. [DOI] [PubMed] [Google Scholar]
- 52.Galant R., Carroll S.B. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 53.Fromental-Ramain C., Warot X., Messadecq N., LeMeur M., Dolle P., Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- 54.Davis A.P., Witte D.P., Hsieh-Li H.M., Potter S.S., Capecchi M.R. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 55.Horan G.S., Ramirez-Solis R., Featherstone M.S., Wolgemuth D.J., Bradley A., Behringer R.R. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995;9:1667–1677. doi: 10.1101/gad.9.13.1667. [DOI] [PubMed] [Google Scholar]
- 56.Zakany J., Fromental-Ramain C., Warot X., Duboule D. Regulation of number and size of digits by posterior Hox genes: a dose-dependent mechanism with potential evolutionary implications. Proc. Natl Acad. Sci. USA. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cribbs D.L., Benassayag C., Randazzo F.M., Kaufman T.C. Levels of homeotic protein function can determine developmental identity: evidence from low-level expression of the Drosophila homeotic gene proboscipedia under Hsp70 control. EMBO J. 1995;14:767–778. doi: 10.1002/j.1460-2075.1995.tb07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galant R., Walsh C.M., Carroll S.B. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development. 2002;129:3115–3126. doi: 10.1242/dev.129.13.3115. [DOI] [PubMed] [Google Scholar]
- 59.Li X., Murre C., McGinnis W. Activity regulation of a Hox protein and a role for the homeodomain in inhibiting transcriptional activation. EMBO J. 1999;18:198–211. doi: 10.1093/emboj/18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Veraksa A., McGinnis W. A sequence motif distinct from Hox binding sites controls the specificity of a Hox response element. Development. 1999;126:5581–5589. doi: 10.1242/dev.126.24.5581. [DOI] [PubMed] [Google Scholar]
- 61.Mann R.S., Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu. Rev. Cell Dev. Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- 62.Lachner M., Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell. Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 63.Stracke M.L., Clair T., Liotta L.A. Autotaxin, tumor motility-stimulating exophosphodiesterase. Adv. Enzyme Regul. 1997;37:135–144. doi: 10.1016/s0065-2571(96)00017-9. [DOI] [PubMed] [Google Scholar]
- 64.Fox M.A., Colello R.J., Macklin W.B., Fuss B. Phosphodiesterase-Ialpha/autotaxin: a counteradhesive protein expressed by oligodendrocytes during onset of myelination. Mol. Cell Neurosci. 2003;23:507–519. doi: 10.1016/s1044-7431(03)00073-3. [DOI] [PubMed] [Google Scholar]
- 65.Tokumura A., Majima E., Kariya Y., Tominaga K., Kogure K., Yasuda K., Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 66.Moolenaar W.H., van Meeteren L.A., Giepmans B.N. The ins and outs of lysophosphatidic acid signaling. Bioessays. 2004;26:870–881. doi: 10.1002/bies.20081. [DOI] [PubMed] [Google Scholar]
- 67.Hombria J.C., Lovegrove B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 68.Lohmann I., McGinnis N., Bodmer M., McGinnis W. The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell. 2002;110:457–466. doi: 10.1016/s0092-8674(02)00871-1. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto M., Gotoh Y., Tamura K., Tanaka M., Kawakami A., Ide H., Kuroiwa A. Coordinated expression of Hoxa-11 and Hoxa-13 during limb muscle patterning. Development. 1998;125:1325–1335. doi: 10.1242/dev.125.7.1325. [DOI] [PubMed] [Google Scholar]
- 70.Stadler H.S., Higgins K.M., Capecchi M.R. Loss of Eph-receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development. 2001;128:4177–4188. doi: 10.1242/dev.128.21.4177. [DOI] [PubMed] [Google Scholar]
- 71.Morgan E.A., Nguyen S.B., Scott V., Stadler H.S. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development. 2003;130:3095–3109. doi: 10.1242/dev.00530. [DOI] [PubMed] [Google Scholar]