Abstract
Glis2 is a member of the Gli-similar (Glis) subfamily of Krüppel-like zinc finger transcription factors. It functions as an activator and repressor of gene transcription. To identify potential co-activators or co-repressors that mediate these actions of Glis2, we performed yeast two-hybrid analysis using Glis2 as bait. C-terminal binding protein 1 (CtBP1) was identified as one of the proteins that interact with Glis2. This interaction was confirmed by mammalian two-hybrid analysis. CtBP1 did not interact with other members of the Glis subfamily suggesting that this interaction is specific for Glis2. Pulldown analysis with GST-CtBP1 demonstrated that CtBP1 physically interacts with Glis2. Analysis of CtBP1 and Glis2 deletion mutants identified several regions important for this interaction. CtBP1 repressed transcriptional activation induced by Glis2(1–171). Repression by Glis2 appears to involve the recruitment of both CtBP1 and histone deacetylase 3 (HDAC3). Confocal microscopic analysis demonstrated that Glis2 localized to nuclear speckles while in most cells CtBP1 was found diffusely in both cytoplasm and nucleus. However, when CtBP1 and Glis2 were co-expressed, CtBP1 was restricted to nuclear speckles and co-localized with Glis2. Our observations suggest that the co-repressor CtBP1 and HDAC3 are part of transcription silencing complex that mediates the transcriptional repression by Glis2.
INTRODUCTION
Krüppel-like zinc finger proteins, named after the Drosophila segmentation gene Krüppel, constitute a large superfamily of transcription factors (1). Typically, these proteins contain two or more Cys2-His2 type zinc fingers that are separated by a conserved consensus sequence, (T/S)GEKP(Y/F)X (2,3). Gli, Zic and Glis proteins are members of three closely related subfamilies of Krüppel-like zinc finger proteins (1,4–7). These proteins contain a highly conserved zinc finger domain consisting of five tandem Cys2-His2 zinc finger motifs. The Gli subfamily, which consists of the three mammalian proteins, Gli1, Gli2 and Gli3, and the Drosophila homolog Cubitus interruptus (Ci), is the best studied (1,7–9). Gli proteins play a critical role in embryonic development and have been implicated in several diseases, including cancer. The transcriptional activity of Gli and Ci proteins is controlled by the sonic hedgehog (Shh) signaling pathway.
The Gli-similar (Glis) subfamily contains three members Glis1–3 (4,6,10–13). Glis2, also referred to as NKL, is a 56 kDa transcriptional regulator that is expressed in several adult tissues, most abundantly in kidney. During embryonic development Glis2 is expressed in a temporal and spatial manner suggesting that Glis2 is involved in the regulation of gene transcription at specific stages of development. During metanephric development Glis2 mRNA is predominantly expressed in the ureteric bud, precursor of the collecting duct, and inductor of nephronic tubule formation (4). Glis2 regulates gene transcription by binding to Glis-response elements (GRE) containing the consensus sequence CCACCCA. The N-terminus of Glis2 contains a transactivation and a repressor function suggesting that Glis2 can act as a repressor as well as an activator of transcription (4).
Little is known about the mechanisms by which Glis2 regulates the transcription of target genes. Very probably transcriptional regulation by Glis2 is mediated through interaction with other nuclear proteins that function as co-repressors or co-activators. In an attempt to identify proteins that interact with and mediate the action of Glis2, we performed yeast two-hybrid analysis with Glis2 as bait. This analysis identified C-terminal binding protein 1 (CtBP1) as a putative Glis2-interacting protein. CtBP1 was initially identified as a protein that interacts with the C-terminus of the adenoviral transforming protein E1A (14,15). Vertebrates express two closely related members, CtBP1 and CtBP2, which are expressed in many tissues and play an important role in embryonic development (16). Subsequent studies have shown that CtBPs can interact with a large number of transcriptional repressors and other regulatory proteins, including BKLF, Knirps, MEF2-interacting transcription repressor (MITR), RIP140, HIC1 and Sox6 (14,15,17–20). CtBPs interact with many of these proteins through a PXDLS consensus motif or a degenerate version of this motif. However, not all interactions with CtBP appear to involve PXDLS-like motifs (19,21). CtBPs function as transcriptional co-repressors that together with other proteins, including various histone deacetylases (HDACs), are assembled in large gene silencing complexes (14,15).
In this study, we show that Glis2 is able to recruit CtBP1. We characterize the interaction of Glis2 and CtBP1 and demonstrate that CtBP1 physically interacts with Glis2 and represses Glis2(1–171)- and Glis2(30–148)-induced transcriptional activation. In addition to CtBP1, Glis2 is able to recruit histone deacetylase HDAC3 into the repressor complex. In the nucleus CtBP1 co-localizes with Glis2 in nuclear speckles, regions of accumulation of transcriptional and mRNA splicing factors. Our results suggest that CtBP1 is part of a co-repressor complex that mediates transcriptional repression by Glis2.
MATERIALS AND METHODS
Plasmids
The plasmids pGBKT7, pGADT7, pM, pVP16 and pEGFP/C1 were purchased from BD Biosciences (Palo Alto, CA). The reporter plasmid pFR-LUC, containing five copies of the Gal4 upstream-activating sequence (UAS) and referred to as (UAS)5-LUC, was obtained from Stratagene (La Jolla, CA). The Renilla luciferase reporter plasmid phRL-SV40 was purchased from Promega (Madison, WI). The full-length coding region of mouse CtBP1 was amplified by PCR using IMAGE clone 5005681 as template. The PCR products were cloned into the EcoRI and SalI sites of pGADT7. The various pM-Glis2 deletion mutants, encoding Gal4(DBD) fused to various regions of Glis2, and pVP16-CtBP1 mutants, encoding Gal4(AD) fused with various regions of CtBP1, were generated by inserting different fragments obtained by PCR amplification into pM or pVP16. The point mutations DL9AS (PLDLK→PLASK), DL80AS (LVDLS→LVASS), DL380AS (PLDLS→PLASS), DL487AS (VLDLS→VLASS), and the double and quadruple point mutations in Glis2 were generated by a Quickchange site-directed mutagenesis kit (Stratagene). pM-Glis1(Δ544) was described previously (6). pCMV-myc-CtBP1 expressing myc-CtBP1 fusion protein was obtained by inserting full-length CtBP1 obtained by PCR amplification into the EcoRI and SalI sites of pCMV-myc. The various pCMV-3XFlag-Glis2 constructs were generated by inserting different fragments of Glis2 obtained by PCR amplification into the EcoRI and XbaI sites of pCMV-3XFlag-7.1 (Sigma, St Louis, MO). To generate pGEX-CtBP1, full-length CtBP1 was amplified by PCR and inserted into the EcoRI and NotI sites of pGEX5X-3 (GE Healthcare, Piscataway, NJ). pEGFP-Glis2 was obtained by cloning full-length Glis2 into pEGFP/C1. The sequences of all plasmids were verified by restriction enzyme analysis and DNA sequencing analysis.
Yeast two-hybrid screening
The Gal4 yeast two-hybrid system was purchased from BD Biosciences. Library screening was conducted according to the manufacturer's instructions. Briefly, the bait construct pGBKT7-Glis2 was generated by cloning the full-length region into the EcoRI and SalI sites of pGBKT7. The pGBKT7-Glis2 was transformed into Saccharomyces cerevisiae strain AH109(MATa). Yeast two-hybrid library screening was carried out by the yeast-mating procedure using S.cerevisiae Y187(MATα) pretransformed with a pACT2, E11 mouse embryo, MATCHMAKER cDNA library (BD Biosciences). After mating, clones were selected on minimal Synthetic Dropout medium (-Trp-Leu-His) containing 5 mM 3-amino-1,2,4-triazole (3AT). After sequencing the positive clones, protein–protein interactions were independently confirmed by yeast two-hybrid analysis and β-galactosidase assay. pGBKT7-p53, encoding Gal4(DBD)-p53, and pGADT7-TD1-1, encoding the Gal4-activation domain fused to the SV40 large T antigen, were used as a positive control in yeast two-hybrid analysis.
Immuno-pulldown (IP) assay
CV-1 or CHO cells were transiently transfected with p3XFlag-CMV, p3XFlag-CMV-Glis2, pEGFP-Glis2, p3XFlag-CMV-HDAC3 or pCMV-myc-CtBP1, as indicated, using Fugene 6 transfection reagent (Roche, Indianapolis, IN). At 48 h after transfection, cells were harvested and lysed in RIPA buffer (Upstate, Charlottesville, VA) containing protease inhibitor cocktails I and II (Sigma). The cell lysates were centrifuged at 14 000 r.p.m., 15000 g at 4°C for 10 min. The supernatants were subsequently stored at −70°C. Flag-Glis2 protein complexes were isolated using anti-Flag M2 affinity resin according to the manufacturer's instructions (Sigma). The resin was washed three times with 0.6 ml phosphate-buffered saline (PBS)/0.05% Tween 20. The bound protein complexes were then solubilized in sample buffer and analyzed by western blot analysis using mouse anti-Flag M2 (Sigma), anti-myc (Invitrogen, Carlsbad, CA) or anti-GFP (Miltenyi Biotec, Auburn, CA) antibodies.
In vitro pulldown assay
Escherichia coli Tuner(DE3) cells (Novagen, Madison, WI) transformed with pGEX-CtBP1 or pGEX-5X-3 plasmid DNA were grown at 37°C to mid-log phase, the synthesis of GST proteins was then induced by the addition of isopropyl-β-d-thiogalactopyranoside; 0.1 mM final concentration) at 30°C. After 3 h of incubation, cells were collected, resuspended in BugBuster protein extraction reagent (Novagen) and incubated as described by the manufacturer's protocol. Cellular extracts were then centrifuged at 15 000 g, and the supernatants containing the soluble GST proteins were collected. Equal amounts of GST-CtBP1 proteins or GST protein (Sigma) were incubated with glutathione–Sepharose 4B beads and washed in PBS. [35S]methionine-labeled Glis2 was obtained using the TNT Quick Coupled Transcription/Translation system (Promega, Madison, WI). The GST- and GST-CtBP1 bound beads were then incubated with the [35S]methionine-labeled Glis2 in 0.2 ml binding buffer (20 mM Tris–HCl, pH 7.6, 100 mM KCl, 0.05% Nonidet P-40, 0.1 mM EDTA, 10% glycerol and 1 mM phenylmethlysulfonyl fluoride). After 1 h incubation at 4°C, beads were washed five times in binding buffer and then boiled in 15 µl of 2× SDS–PAGE loading buffer. Solubilized proteins were then separated by 4–20% SDS–PAGE and the radiolabeled proteins visualized by autoradiography.
Reporter gene assay
CHO, CV-1, COS-1 and 293HT cells were maintained in DMEM/F-12 supplemented with 10% fetal bovine serum. Cells were transfected with the reporter plasmids pFR-LUC and phRL-SV40, and the pM and pVP16 expression plasmids indicated, using Fugene 6 transfection reagent (Roche). Luciferase activity was measured using the Dual-Luciferase® Reporter Assay System from Promega (Madison, WI). All transfections were performed in triplicate. Experiments were carried out at least twice independently. To determine equal expression of VP16-fusion proteins cell lysates were examined by western blot analysis using an anti-VP16 antibody (BD Biosciences).
Subcellular localization
CHO, CV-1, COS-1 and 293HT cells were plated in glass-bottom culture dishes (MatTek Corp., Ashland, MA) and 24 h later transfected with pEGFP-Glis2 (0.2 µg) and/or pCMV-myc-CtBP1 using Fugene 6 reagent. After 48 h, cells were washed in PBS, fixed in 4% paraformaldehyde, washed with PBS. Cells were then permeabilized with 0.3% Triton X-100, washed again with PBS, and subsequently incubated with mouse anti-myc antibody. After 30 min incubation cells were washed five times with PBS and then incubated for an additional 30 min with secondary antibody (goat anti-mouse Alexa 594). Cells were washed with PBS and nuclei stained with 4′-6-diamidino-2-phenylindole (DAPI). Fluorescence was observed in a Zeiss LSM 510 NLO confocal microscope (Zeiss, Thornwood, NY).
RESULTS
Identification of CtBP1 as a Glis2-interacting protein
Previous studies have shown that Glis2 can function as a repressor and activator of transcription (4). As reported for other transcription factors, this transcriptional regulation is probably mediated through interaction with other nuclear proteins that function either as co-repressors or co-activators. To identify proteins that mediate or modulate the transcriptional activity of Glis2, we performed yeast two-hybrid analysis using full-length Glis2 as bait and an E11 mouse embryo cDNA library as prey. This yeast two-hybrid analysis yielded 30 positive clones, 6 of which were identified as CtBP1 after DNA sequencing.
To confirm this interaction, yeast strains AH109(MATα) and Y187(MATα) were transformed with pGBKT7-Glis2 or pGADT7-CtBP1, respectively, mated and the transformants selected at 30°C on -Leu-Trp plates and -Leu-Trp-His+3AT plates. Only yeast that contain bait and interacting prey, are able to grow on -Leu-Trp-His(+3AT) plates. As shown in Figure 1A, only yeast expressing both Gal4(DBD)-Glis2 and Gal4(AD)-CtBP1 fusion proteins were able to grow on -Leu-Trp-His(+3AT) plates. Yeast expressing pGBKT7-53 and pTD1-1 were used as a positive control and also grew on -His(+3AT) plates (Figure 1A). Colonies isolated from -Leu-Trp-His(+3AT) plates were grown in liquid culture and assayed for β-galactosidase activity. As shown in Figure 1B, only yeast colonies containing both Gal4(DBD)-Glis2 and Gal4(AD)-CtBP1 fusion proteins and the positive control expressed high β-galactosidase activity. These observations support the conclusion that Glis2 and CtBP1 interact with each other.
Figure 1.
Identification of CtBP1 as a Glis2-interacting protein by yeast two-hybrid analysis. (A) Yeast strains harboring the plasmids indicated were grown on -Leu-Trp or on -His-Leu-Trp plates supplemented with 5 mM 3AT. All yeast strains grew on -Leu-Trp plates; however, only yeast containing both pGBKT7-Glis2 and pGADT7-CtBP1 were able to grow on -His-Leu-Trp plates containing 5 mM 3AT. Yeast containing pGBKT7-p53 and pGADT7-TD1-1 were used as a positive control. (B) Yeast was grown in -His-Leu-Trp liquid medium supplemented with 5 mM 3AT and then assayed for β-galactosidase activity. Three different colonies from each original plate were analyzed. E, indicates empty pGBKT7 or pGADT7 vector was used.
Mammalian two-hybrid and immuno-pulldown analysis
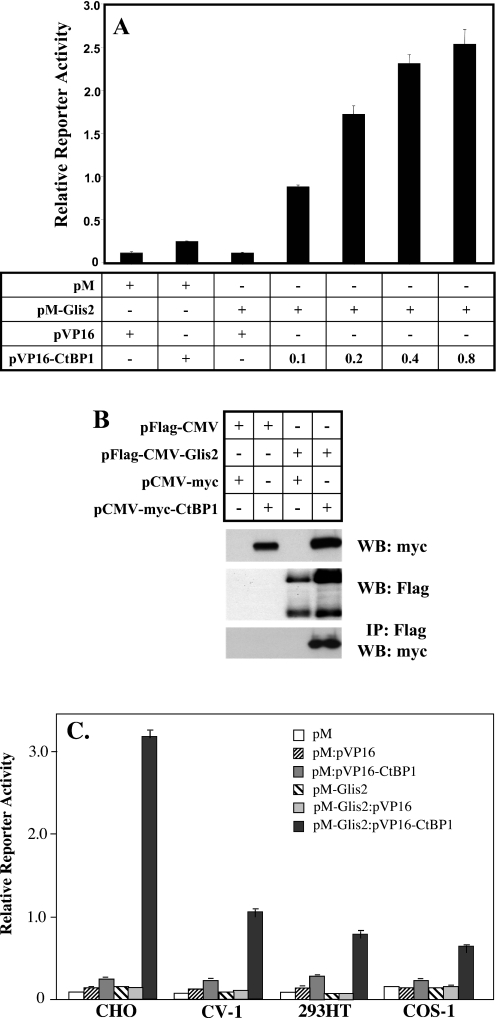
The interaction between Glis2 and CtBP1 was confirmed by mammalian two-hybrid analysis. CHO cells were co-transfected with (UAS)5-LUC reporter, pM or pM-Glis2, encoding the fusion protein Gal4(DBD)-Glis2, in the presence of increasing amounts of pVP16-CtBP1, as indicated (Figure 2A). Co-expression of VP16-CtBP1 increased reporter activity in a dose-dependent manner owing to the interaction of CtBP1 with Glis2 (Figure 2A). These observations are consistent with the results obtained by yeast two-hybrid analysis and support the conclusion that also in mammalian cells Glis2 and CtBP1 are able to interact with each other.
Figure 2.
Analysis of the interaction between Glis2 and the co-repressor CtBP1 by mammalian two-hybrid and immuno-pulldown analysis. (A) Mammalian two-hybrid analysis. CHO cells were co-transfected with (UAS)5-LUC, phRL-SV40, pM, pVP16, pM-Glis2, and increasing amounts (µg) of pVP16-CtBP1 as indicated. Cells were assayed for reporter activity 48 h after transfection. The relative LUC activity was calculated and plotted. (B) Immuno-pulldown analysis. CV-1 cells were co-transfected with p3XFlag-CMV-Glis2, pCMV-myc-CtBP1, p3XFlag-CMV, and pCMV-myc as indicated in the graph. Cell lysates were prepared and Flag-Glis2 protein complexes isolated using anti-Flag M2 agarose affinity resin as described in Materials and Methods. Proteins in the cellular lysates and immunoprecipitated (IP) proteins were subsequently examined by western blot (WB) analysis using anti-Flag M2 or anti-myc antibody. (C) Glis2 interacts with CtBP1 in different cell types. CHO, CV-1, 293HT and COS-1 cells were co-transfected with (UAS)5-LUC, phRL-SV40, pM, pVP16, pM-Glis2, pVP16 and pVP16-CtBP1 as indicated. Cells were assayed for reporter activity 48 h after transfection and the relative LUC activities calculated and plotted.
We next examined the interaction between Glis2 and CtBP1 by immuno-pulldown analysis. For this purpose, CV-1 cells were co-transfected with pCMV-myc-CtBP1 and p3XFlag-CMV(empty) or p3XFlag-CMV-Glis2 expression plasmids and 48 h later, cell extracts were prepared. One half of each protein extract was used for western blot analysis, the other half for the isolation of Flag-Glis2 protein complexes using anti-Flag M2 affinity resin. Western blot analysis shows that the anti-Flag antibody recognized two bands representing full-length Flag-Glis2 and a smaller, proteolytically processed form of Glis2. Proteolytic cleavage has also been reported for the closely related Ci and Gli proteins (8,9). The functional significance of the processing of Glis2 has yet to be determined. As shown in Figure 2B, Flag-Glis2 was able to pulldown CtBP1 whereas control cells containing p3XFlag-CMV(empty) did not. These observations are consistent with the concept that Glis2 is able to recruit CtBP1 and are in agreement with the results obtained by two-hybrid analysis.
In a number of instances protein–protein interactions are greatly dependent on the cell type used. We, therefore, examined the interaction of Glis2 and CtBP1 in several cell types. Mammalian two-hybrid analysis demonstrated that although the induction of reporter activity was highest in CHO cells, the interaction of Glis2 with CtBP1 was not limited to CHO cells but was also observed in several other cell lines, including CV-1, COS-1 and 293HT cells (Figure 2C). The lowest induction in reporter activity was observed in 293HT and COS-1 cells. The observed variation in reporter activity between cell types is interesting and could be due to many factors, including competition of other nuclear proteins with CtBP1 for Glis2 binding and differences in posttranslational modification(s) of either Glis2 or CtBP1.
Specificity of the interaction between Glis2 and CtBP1
Glis2 together with Glis1 and Glis3 constitute the Glis subfamily of Krüppel-like zinc finger proteins (10,13,22–24). The zinc finger domain of Glis2 exhibits 68% homology with those of Glis1 and Glis3 while these proteins exhibit low homology outside their zinc finger domain. To determine whether CtBP1 interacted with other Glis family members, we examined their possible interaction by mammalian two-hybrid and immuno-pulldown analysis. As shown in Figure 3A, mammalian two-hybrid analysis indicated that CtBP1 did not interact with Glis1 or Glis3 suggesting that the interaction is specific for Glis2. This was confirmed by immuno-pulldown analysis; only Glis2 was able to pulldown CtBP1 (Figure 3B).
Figure 3.
Interaction between Glis proteins and CtBP1 is specific for Glis2. (A) Mammalian two-hybrid analysis. CHO cells were transfected with (UAS)5-LUC, phRL-SV40, pM-Glis1, pM-Glis2, pM-Glis3, pVP16, pVP16-CtBP1 and pVP16-CtBP2. Reporter activity was assayed 48 h after transfection and the relative LUC activity calculated and plotted. (B) Immuno-pulldown analysis. CV-1 cells were transfected with p3XFlag-CMV-Glis1, p3XFlag-CMV-Glis2, p3XFlag-CMV-Glis3, pCMV-myc-CtBP1, p3XFlag-CMV and pCMV-myc as indicated. Cell lysates were prepared and Flag-Glis2 protein complexes isolated using anti-Flag M2 affinity resin as described in Materials and Methods. Proteins in the cellular lysates and immunoprecipitated (IP) proteins were subsequently examined by Western blot (WB) analysis using anti-Flag M2 or anti-myc antibody. Asterisk indicates the position of the different Flag-Glis fusion proteins (Glis1, 84.3 kDa; Glis2, 55.8 kDa; Glis3, 83.8 kDa).
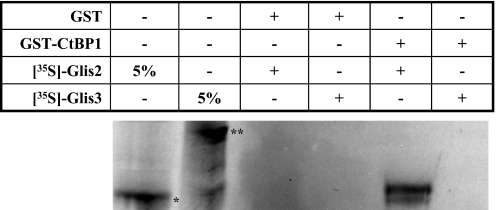
CtBP1 interacts with Glis2 in vitro
To determine whether CtBP1 physically interacts with Glis2, we performed in vitro pulldown analysis using GST-CtBP1 and [35S]-labeled Glis2. GST-CtBP1 bound to glutathione–Sepharose beads was incubated with [35S]-labeled Glis2 and GST-CtBP1 protein complexes examined by PAGE. As shown in Figure 4, GST-CtBP1 was able to bind radiolabeled Glis2 while in the control, GST did not pulldown Glis2. Moreover, GST-CtBP1 protein did not pulldown radiolabeled Glis3. Although we cannot totally rule out that the interaction between CtBP1 and Glis2 involves another protein, these results strongly suggest that CtBP1 and Glis2 physically interact with each other and that the interaction is specific. These observations support our findings obtained by mammalian two-hybrid and immuno-pulldown analyses (Figures 2 and 3).
Figure 4.
CtBP1 interacts directly with Glis2. GST and GST-CtBP1 fusion proteins were bound to glutathione–Sepharose 4B beads and then incubated with [35S]methionine-labeled Glis2 or Glis3. After 1 h incubation, beads were washed extensively and bound proteins solubilized and examined by PAGE. Radiolabeled proteins were visualized by autoradiography. Lane 1 and 2, 5% input of radiolabeled Glis2 (asterisk) and Glis3 (double asterisk), respectively; lanes 3 and 4, radiolabeled protein retained by GST; lane 5 and 6, protein retained by GST-CtBP1. Results show that only radiolabeled Glis2 was retained by GST-CtBP1.
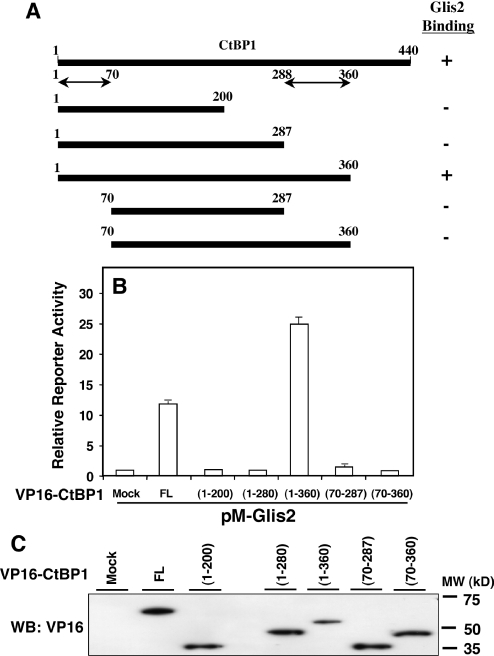
Effect of various deletions in CtBP1 on its interaction with Glis2
To obtain insight into the region(s) in CtBP1 important for its interaction with Glis2, we examined the effect of several deletions in CtBP1 on the interaction of Glis2 by mammalian two-hybrid analysis. For this purpose, CHO cells were co-transfected with (UAS)5-LUC, phRL-SV40, pM-Glis2 and pVP16 containing full-length CtBP1 or several CtBP1 deletion mutants. Figure 5 shows the effect of various deletions on the interaction of CtBP1 with Glis2. This analysis indicated that deletion of 70 amino acids at the N-terminus or deletion of 153 or 240 amino acids at the C-terminus each abolished the interaction with Glis2 completely. In contrast, deletion of 80 amino acids at the C-terminus had little effect. These results suggest that two regions within CtBP1, the first 70 amino acids at the N-terminus and the region from amino acid 288 to 360, are required for its interaction with Glis2.
Figure 5.
Effect of various deletions in CtBP1 on its interaction with Glis2. The reporter plasmids (UAS)5-LUC and phRL-SV40 were co-transfected with pM-Glis2 and various pVP16-CtBP1 deletions mutants (A) in CHO cells, as indicated in the graph. After 48 h, cells were assayed for luciferase reporter activities as described in Materials and Methods. The relative reporter activity was calculated and plotted (B). The level of each CtBP1 fusion protein in total cellular extract was examined by western blot analysis with an anti-VP16 antibody (C). Data are means ± SEM of triplicate samples. FL, full-length CtBP1.
Effect of various deletions in Glis2 on its interaction with CtBP1
To determine what region of Glis2 was necessary for its interaction with CtBP1, we examined the effect of various deletions on the ability of Flag-Glis2 to bind myc-CtBP1 in immuno-pulldown analysis. These results showed that the N-terminal regions from 1 to 143 and from 1 to 200 as well as the C-terminal regions from 170 to 520 and 324 to 520 of Glis2 were able to pulldown myc-CtBP1 (Figure 6A and B). In contrast, the middle regions 83–323 and 170–323, which contain the zinc finger domain, did not pulldown myc-CtBP1. These results indicate that CtBP1 can interact with both the N- and C-terminus of Glis2 whereas the zinc finger domain of Glis2 is not required. This conclusion was supported by mammalian two-hybrid analysis using pM-Glis2, several pM-Glis2 deletion mutants and VP16-CtBP1 (Figure 6C). Both pM-Glis2(1–323) and pM-Glis2(164–520), containing the N-terminus and C-terminus, respectively, enhanced pVP16-CtBP1-dependent transcriptional activation of the luciferase reporter whereas pM-Glis2(170–323) did not. However, the level of reporter activity generated by both Glis2(1–323) and Glis2(164–520) was reduced compared with that by full-length Glis2 suggesting that both regions are necessary for optimal interaction. These results show that both the N- and C-terminus play a role in the interaction of Glis2 with CtBP1 whereas the zinc finger domain is not required. These findings are in agreement with those obtained by immuno-pulldown analysis.
Figure 6.
(A) Effect of various deletions in Glis2 on its interaction with CtBP1. CV-1 cells were transiently transfected with pCMV-myc-CtBP1 and p3XFlag-CMV-7.1 containing various Glis2 deletion mutants. (B) At 48 h after transfection, protein lysates were prepared as described in Materials and Methods. One part was used in western blot (WB) analysis using anti-Flag M2 (upper panel) or anti-Myc antibodies (middle panel). The remaining was used in immuno-pulldown (IP) assay using anti-Flag M2 affinity resin. Bound proteins were then examined by western blot analysis using anti-Myc antibody (lower panel). Lane 1, Flag-Glis2(1–143); lane 2, Flag-Glis2(1–200); lane 3, Flag-Glis2(83–323); lane 4, Flag-Glis2(170–323); lane 5, Flag-Glis2(323–520); lane 6, Flag-Glis2(170–520); lane 7, Flag-Glis2(full-length). (C) Mammalian two-hybrid analysis. CHO cells were transfected with (UAS)5-LUC (0.1 µg), pM-Glis2 (FL), or pM-Glis2(1–325), pM-Glis2(170–323), pM-Glis2(164–520) deletion mutant constructs (0.1 µg), phRL-SV40 (5ng), and pVP16 or pVP16-CtBP1 (0.1 µg) as described in Materials and Methods. Reporter activity was determined and relative LUC activity plotted. Data are means ± SEM of triplicate samples.
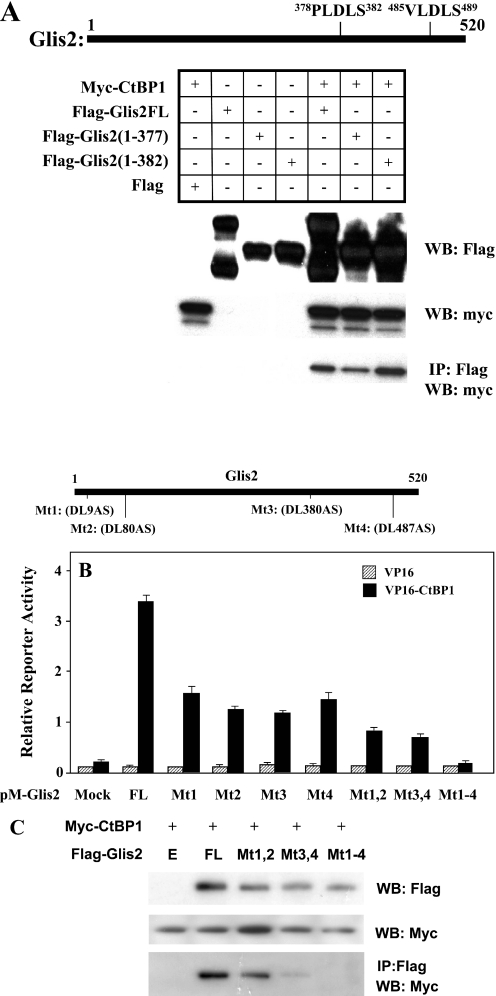
CtBP1 interacts with many proteins through the CtBP-binding signature sequence motif PXDLS or a very similar sequence. In many instances, deletion or mutation of a single critical PXDLS motif abrogates the interaction with CtBP1 and causes loss of the functional consequences of CtBP1 association. Examination of the Glis2 sequence identified one PXDLS consensus sequence at 378PLDLS (P3) and three degenerate PXDLS sequences at 7PLDLK (P1), 78LVDLS (P2) and 485VLDLS (P4). Immuno-pulldown analysis demonstrated that both Flag-Glis2(1–382), which lacks P4, and Flag-Glis2(1–377), which lacks both P3 and P4, were able to pulldown myc-CtBP1 in agreement with the conclusion that the P3 and P4 motifs of Glis2 are not essential for the recruitment of CtBP1 (Figure 7A). Moreover, two-hybrid analysis (Figure 6C) demonstrated that Glis2(1–323), lacking P3 and P4, and Glis2(164–520), lacking P1 and P2, are able to interact with CtBP1 indicating that these PXLDS-like motifs are not an absolute requirement for Glis2 to interact with CtBP1. To analyze further the role of these PXDLS-like motifs in the interaction of Glis2 with CtBP1, the effects of DL→AS mutations, introduced by site-directed mutagenesis, on the interaction was examined by mammalian two-hybrid analysis. This analysis showed that although each mutation (Mt1 to Mt4) caused a substantial reduction in the transcriptional activation of the LUC reporter, all Glis2 mutants were still able to significantly increase reporter activity (Figure 7B). The double mutations Mt1,2 and Mt3,4 caused a further reduction in transactivation while the quadruple mutation Mt1-4 totally abolished transactivation. Results obtained by immuno-pulldown analyses showed that Mt1,2 and Mt3,4 reduced the amount of CtBP1 pulled down while CtBP1 was not pulled down by Glis2 containing the quadruple mutation (Figure 7C) in agreement with the data shown in Figure 7B. These observations indicate that although single mutations do not block the recruitment of CtBP1 by Glis2, the quadruple mutation totally abolishes the interaction of Glis2 with CtBP1. These results suggest that these PXDLS-like motifs are necessary for optimal interaction of Glis2 with CtBP1.
Figure 7.
Role of PXDLS-binding motifs in the recruitment of CtBP1 by Glis2. (A) Immuno-pulldown analysis. CV-1 cells were co-transfected with p3XFlag-CMV-7.1, pCMV-myc-CtBP1 and several p3XFlag-CMV-Glis2 mutants as indicated. One half of the lysates were examined by western blot (WB) analysis. The other half was subjected to immunoprecipitation with anti-Flag M2 agarose affinity resin (IP: Flag) and bound proteins examined by western blot analysis with anti-myc antibody. (B) Effect of point mutations in the four PXDLS-like motifs (P1–P4) on the recruitment of CtBP1 by Glis2. CHO cells were transiently transfected with (UAS)5-LUC, phRL-SV40, pVP16 or pVP16-CtBP1, and various pM-Glis2 mutants (Mt1 to Mt4, Mt1,2, Mt3,4 and Mt1-4). At 48 h after transfection, cells were assayed for reporter activity. The relative LUC activity was calculated and plotted. (C) CV-1 cells were co-transfected with pCMV-myc-CtBP1 and several p3XFlag-CMV-Glis2 (FL) or several p3XFlag-CMV-Glis2 mutants (Mt1,2, Mt3,4 and Mt1–4) as indicated. One half of the lysates were examined by western blot analysis. The other half was subjected to immunoprecipitation with anti-Flag M2 agarose affinity resin (IP: Flag) and bound proteins examined by western blot analysis with anti-myc antibody.
CtBP1 inhibits Glis2-mediated transcriptional activation
Previous studies showed that Glis2 contains a transactivation domain at its C-terminus between amino acids 30 and 148 (4). In 293HT cells, Gal4(DBD)-Glis2(1–171) and Gal4(DBD)-Glis2(30–148) cause, respectively, a 3.5- and 90-fold increase in UAS-dependent transcriptional activation of the luciferase reporter (Figure 8A and B). To analyze the effect of CtBP1 on this transactivation, 293HT cells were co-transfected with pM-Glis2(1–171) or pM-Glis2(30–148), (UAS)5-LUC, and pCMV-myc-CtBP1. As shown in Figure 8A, CtBP1 was able to inhibit Glis2-mediated transcriptional activation in a dose-dependent manner. CtBP1 also inhibited the transcriptional activation induced by Glis2(30–148) (Figure 8B). These observations suggest that CtBP1 functions as a co-repressor for Glis2. Glis1ΔN544 induced UAS-dependent transcriptional activation of the luciferase reporter about 13-fold. This activation was not significantly inhibited by CtBP1 suggesting that the inhibition is specific for Glis2 and not an effect on the general transcriptional machinery.
Figure 8.
(A) CtBP1 suppressed Glis2(1–171)-mediated transcriptional activation. 293HT cells were co-transfected with (UAS)5-LUC, phRL-SV40, pM, pM-Glis2(1–171), and increasing amounts of pCMV-myc-CtBP1 (0–0.4 µg) as indicated. (B) CtBP1 inhibited Glis2(30–148) but not Glis1ΔN544 induced transactivation. 293HT cells were co-transfected with (UAS)5-LUC, phRL-SV40, pM, pM-Glis2(30–148) or pM-Glis1ΔN544, and pCMV-myc-CtBP1 (0.2 µg) as indicated. At 48 h after transfection, cells were assayed for LUC reporter activities. The relative LUC activity was calculated and plotted.
Glis2 is able to recruit both CtBP1 and HDAC3
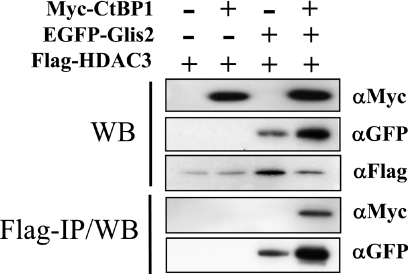
Repression by transcriptional regulators is mediated through the recruitment of co-repressor complexes that include proteins with histone deacetylase activity. In this study, we examined whether HDAC3 is part of the Glis2-CtBP1 repressor complex. CHO cells were co-transfected with p3XFlag-CMV, pEGFP-Glis2, p3XFlag-CMV-HDAC3 or pCMV-myc-CtBP1, as indicated, and the interaction between HDAC3, Glis2 and CtBP1 examined by immuno-pulldown analysis. In cells co-transfected with p3XFlag-CMV-HDAC3 and either pEGFP-Glis2 or pCMV-myc-CtBP1, HDAC3 was able to recruit Glis2 but not CtBP1 (Figure 9). However, HDAC3 was able to pulldown CtBP1 in cells co-expressing Glis2. These results indicate that the recruitment of CtBP1 by HDAC3 is dependent on the presence of Glis2. These observations suggest that Glis2 is able to recruit both HDAC3 and CtBP1 into the same protein complex and are in agreement with the concept that CtBP1 and HDAC3 are involved in mediating the transcriptional silencing by Glis2.
Figure 9.
Glis2 recruits CtBP1 and HDAC3. CHO cells were co-transfected with p3XFlag-CMV7.1, pEGFP-Glis2, p3XFlag-CMV-HDAC3 or pCMV-myc-CtBP1, as indicated. One part of the protein extracts was analyzed by western blot analysis (WB) for expression of EGFP-Glis2, Flag-HDAC-3 and myc-CtBP1. The other half was used for immuno-pulldown analysis as described in Materials and Methods. Proteins bound to anti-Flag M2 affinity resin were analyzed by western blot analysis using anti-GFP or anti-myc antibodies (Flag-IP/WB).
Glis2 and CtBP1 co-localize in the nucleus
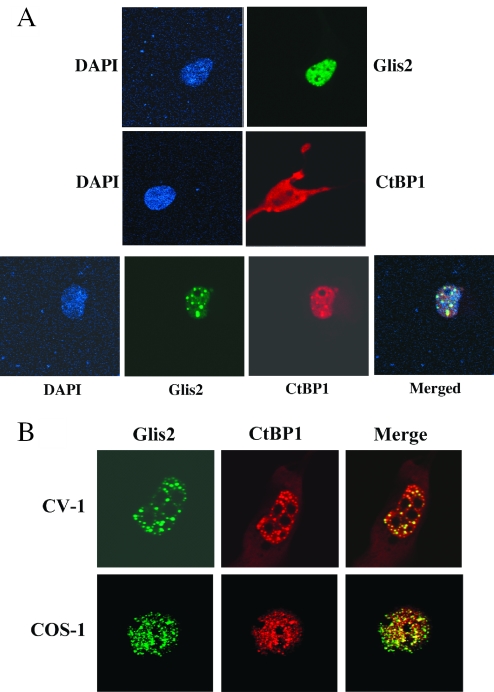
To study the subcellular localization of Glis2 and CtBP1, we expressed EGFP-Glis2 and myc-CtBP1 fusion proteins in CHO cells either alone or together and examined their localization by confocal microscopy (Figure 10A). Analysis of CHO cells transfected with pEGFP-Glis2 only showed that EGFP-Glis2 localized predominantly to the nucleus in 92% of the cells while in the other 8% Glis2 was found about equally in both cytoplasm and nucleus (Table 1). In the nucleus, Glis2 was distributed in a speckled pattern and was excluded from the nucleoli in agreement with previous observations (4). In CHO cells transfected with pCMV-myc-CtBP1 only, 83% of the transfected cells expressed myc-CtBP1 about equally in both nucleus and cytoplasm, in 9% of the cells it was present predominantly in the nucleus, and in the remaining 8% predominantly in the cytoplasm (Table 1). In most nuclei CtBP1 was expressed rather diffusely. In CHO cells co-expressing Glis2 and CtBP1, the distribution of Glis2 did not change significantly; however, CtBP1 became localized mostly to the nucleus in 71% of cells (Table 1). In addition, the pattern of distribution of CtBP1 in the nucleus changed from a diffused to a speckled pattern. The speckled pattern observed for CtBP1 overlapped with that of Glis2 indicating that CtBP1 and Glis2 co-localize to the same protein complexes in the nucleus. The co-localization of Glis2 and CtBP1 was also observed in other cell lines, including CV-1 and COS-1 cells (Figure 10B). In both cell types, Glis2 and CtBP1 localized to the nucleus and exhibited a speckled pattern that mostly overlapped.
Figure 10.
CtBP1 co-localizes with Glis2 in the nucleus. (A) COS-1 cells were transfected with a pEGFP-Glis2 (upper panels) or pCMV-myc-CtBP1 (middle panels) expression plasmids or co-transfected with both plasmids (lower panels). At 48 h later, expression of EGFP-Glis2 and myc-CtBP1 fusion proteins was visualized by fluorescence confocal microscopy as described in Materials and Methods. Myc-CtBP1 was detected by incubating cells with mouse anti-myc and subsequently with Alexa Fluor® 594 goat anti-mouse antibodies. Nucleus was identified by DAPI staining. (B) Co-localization of Glis2 and CtBP1 in CV-1 and COS-1 cells. Cells were co-transfected with pEGFP-Glis2 and pCMV-myc-CtBP1 and then processed as described under (A).
Table 1.
Glis2 expression enhances the nuclear localization of CtBP1
| Transfected-1 cells | EGFP-Glis2 | myc-CtBP1 | ||||
|---|---|---|---|---|---|---|
| %C | %N | %C + N | %C | %N | %C + N | |
| EGFP-Glis | 0 | 92.0 | 8.0 | — | — | — |
| myc-CtBP1 | — | — | — | 8.0 | 9.0 | 83.0 |
| EGFP-Glis2 + myc-CtBP1 | 0 | 94.0 | 6.0 | 2.0 | 71.0 | 27.0 |
Experiment was carried out as described in the legend of Figure 10A. Randomly, 100 cells were selected and the subcellular localization of CtBP1 or Glis2 determined. The percentage of cells containing Glis2 (EGFP fluorescence) or CtBP1 (Alexa Fluor® 594 fluorescence) predominantly in the nucleus (%N), cytoplasm (%C) or both (%N + C) was calculated. In the case of COS-1 cells co-transfected with both pEGFP-Glis2 and pCMV-myc-CtBP1, only cells were counted that contained both Glis2 and CtBP1.
DISCUSSION
Members of the Gli and Glis subfamilies contain multiple domains that regulate their localization, proteolytic processing and transcriptional activity (4,6,9,10,25,26). However, little is still know about the genes that are regulated by these transcription factors. The N-terminus of Glis2 contains a transactivation and a repressor function suggesting that Glis2 acts as a repressor as well as an activator of transcription (4). In a number of cell types expression of full-length Glis2 has been shown to repress GRE-mediated transcription. Whether Glis2 can function as a transcriptional activator and by what mechanism that regulates such activation has yet to be established. Regulation of gene expression by transcription factors is mediated through interactions with other nuclear proteins (27). This involves the recruitment of either co-activator and co-repressor complexes that contain histone acetylase or deacetylase activity, respectively. HDACs induce compactation of chromatin and repression of gene expression while histone acetylases induce decompactation of the chromatin and increased transcription of the target gene. The co-activator CBP has been shown to mediate the transcriptional activation by Ci, Gli3 and Gli2 (22,28,29). In many Krüppel-like zinc finger proteins transcriptional repression has been reported to involve specific repressor domains such as the Krüppel-associated box (KRAB) and SCAN domain (30,31). KRAB is an evolutionary conserved motif of 75 amino acids that can be found at the N-terminus of ∼30% of the Krüppel-like zinc finger proteins. This region has been shown to mediate repression by recruiting co-repressor complexes. For example, the transcriptional repression by the Krüppel-like zinc finger proteins KRAZ1 and KRAZ2 has been reported to involve recruitment of the co-repressor TIF1 to their KRAB repressor domain (32). However, the repressor domain identified in Glis2 does not show any homology with KRAB or SCAN and appear to represent a novel repressor function.
Thus far no proteins have been identified that mediate the transcriptional activity of Glis2. Therefore, yeast two-hybrid analysis was used to identify proteins that interact with Glis2 and mediate its transcriptional activity. This analysis identified CtBP1 as a putative Glis2-interacting protein. This was supported by mammalian two-hybrid and immuno-pulldown analysis while in vitro pulldown analysis using GST-CtBP1 indicated that CtBP1 and Glis2 physically interact. The interaction was shown to be specific for Glis2 and CtBP1 since Glis1 and Glis3 did not interact with CtBP1. The zinc finger region of Glis proteins is highly conserved while Glis proteins exhibit little sequence homology outside their zinc finger domain. Since the zinc finger domain of Glis2 is not involved in the interaction with CtBP1, it may not be surprising that the interaction of CtBP1 is specific for Glis2.
Analysis of Glis2 protein expression in several cell types demonstrated that in >90% of the cells Glis2 is restricted largely to the nucleus where it is expressed in a speckled pattern in agreement with previous observations (4). The lack of transcriptional activation is therefore not due to retention of Glis2 to the cytoplasm as reported for Gli but inherent to the repressor activity of Glis2. CtBP1 was found to be diffusely expressed in both cytoplasm and nucleus. However, when CtBP1 and Glis2 are co-expressed, CtBP1 is largely localized to the nucleus and restricted to nuclear speckles, regions of accumulation of transcriptional and mRNA splicing factors, where it co-localizes with Glis2. These observations are in agreement with the conclusion that Glis2 and CtBP1 interact and are part of the same nuclear protein complex.
CtBP1, which was initially identified through its ability to bind the C-terminus of the adenoviral transforming protein E1A, has been reported to interact with many different proteins believed to be involved in transcriptional silencing (14,15). These include the MITR, the tumor suppressor gene HIC1, HDACs, histone methyltransferases, the co-repressor RIP140 and the zinc finger protein Ikaros (10,18–20,33). Although the precise mechanisms by which CtBP1 mediates transcriptional repression has not been totally established, it appears to involve dimerization and recruitment of class I and class II HDACs, as well as of other proteins. But HDAC-independent mechanisms have also been identified (15). As mentioned above, full-length Glis2 acts as a transcriptional repressor in several cell types; however, Glis2(1–171) and Glis2(30–148) can induce transcription (Figure 8). We show that CtBP1 can effectively repress this transcriptional activation. The recruitment of CtBP1 by Glis2 together with the observed repression of Glis2-induced transcription suggests that CtBP1 functions as a co-repressor that mediates the transcriptional repression by Glis2. Since HDACs have been found to be part of CtBP1 gene silencing complexes, we examined the interaction between Glis2, CtBP1 and HDAC3. Our results show that HDAC3 was able to bind Glis2 in the absence of CtBP1. Interestingly, the recruitment of CtBP1 by HDAC3 was dependent on the presence of Glis2. These results suggest that Glis2 is able to recruit both HDAC3 and CtBP1 into the same protein complex and support the concept that the recruitment of both the co-repressor CtBP1 and HDAC3 are involved in the repression of transcription by Glis2. The observed interaction between Glis2 and HDAC3 in the absence of exogenous CtBP1 may suggest that Glis2 is able to repress transcription through a HDAC-dependent mechanism that may involve co-repressors other than CtBP1.
In many instances PXDLS or a degenerate PXDLS motif is the primary determinant for CtBP binding; however, certain CtBP partner proteins do not contain recognizable PXDLS motifs or these motifs do not appear to be important for the interaction (15,34). For example, the zinc finger protein Tramtrack69 contains a PPDLS motif that is not required for binding CtBP1 (21). HDAC5 contains a related motif (PVELR) that is dispensable for CtBP interaction (23). In these cases other contacts are made and other (nonpocket) regions of CtBP may be involved in the interaction. Deletion mutant analysis showed that the interaction of Glis2 with CtBP1 is complex. Both the amino- and carboxyl-terminus of Glis2 are able to interact with CtBP1 while both the amino-terminus and the region between A288 and T360 of CtBP1 are essential for its interaction with Glis2. The first 200 amino acids at the amino-terminus of CtBP1 have been previously implicated in the recognition of the PXDLS motif while deletion of the carboxyl-terminus greatly diminishes the interaction with Ikaros (33). We show that individual point mutations in the four potential PXDLS motifs of Glis2 reduced the interaction with CtBP1 but did not block the interaction. However, the Glis2 mutant in which all four motifs were mutated, did not interact with CtBP1. Although we cannot rule out effects of mutations on the proper folding of Glis2, our results suggest that the PXDLS-like motifs are required for optimal recruitment of CtBP1 by Glis2. In addition, the complex formation of Glis2 with CtBP1 appears to involve multiple domains in both proteins and probably involves the recruitment of other nuclear proteins that through their interaction with different regions of Glis2 and CtBP1, act in synergy to stabilize the complex.
In summary, in this study we identify CtBP1 as a protein that is recruited by Glis2. This association is supported by yeast and mammalian two-hybrid, immuno-pulldown analysis, and confocal microscopy. In vitro pulldown analysis suggests that Glis2 and CtBP1 interact directly with each other. We provide evidence indicating that Glis2 recruits both CtBP1 and HDAC3 into a larger protein complex. Moreover, we show that CtBP1 is able to repress Glis2(30–148)-mediated transcription activation. Our study suggests that the association of the co-repressor CtBP1 and HDAC3 with Glis2 is involved in the repression of transcription by Glis2.
Acknowledgments
The authors would like to thank Drs Christina Teng and James Zhang for their valuable comments on the manuscript. This research was supported by the Intramural Research Program of the NIEHS, NIH. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ruppert J.M., Kinzler K.W., Wong A.J., Bigner S.H., Kao F.T., Law M.L., Seuanez H.N., O'Brien S.J., Vogelstein B. The GLI-Kruppel family of human genes. Mol. Cell. Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang D.T., Pevsner J., Yang V.W. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner J., Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F., Nakanishi G., Kurebayashi S., Yoshino K., Perantoni A., Kim Y.S., Jetten A.M. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcriptional factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J. Biol. Chem. 2001;12:10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
- 5.Aruga J., Nagai T., Tokuyama T., Hayashizaki Y., Okazaki Y., Chapman V.M., Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J. Biol. Chem. 1996;271:1043–1047. doi: 10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y.S., Lewandoski M., Perantoni A.O., Kurebayashi S., Nakanishi G., Jetten A.M. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem. 2002;277:30901–30913. doi: 10.1074/jbc.M203563200. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler K.W., Ruppert J.M., Bigner S.H., Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz i Altaba A., Sanchez P., Dahmane N. GLI and hedgehog in cancer: tumours, embryos and stem cells. Nature. Rev. Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 9.Villavicencio E.H., Walterhouse D.O., Iannaccone P.M. The sonic hedgehog-patched-Gli pathway in human development and disease. Am. J. Hum. Genet. 2000;67:1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y.S., Nakanishi G., Lewandoski M., Jetten A.M. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31:5513–5525. doi: 10.1093/nar/gkg776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F., Jetten A.M. Genomic structure of the gene encoding the human GLI-related, Kruppel-like zinc finger protein GLIS2. Gene. 2001;280:49–57. doi: 10.1016/s0378-1119(01)00764-8. [DOI] [PubMed] [Google Scholar]
- 12.Nakashima M., Tanese N., Ito M., Auerbach W., Bai C., Furukawa T., Toyono T., Akamine A., Joyner A.L. A novel gene, GliH1, with homology to the Gli zinc finger domain not required for mouse development. Mech. Dev. 2002;119:21. doi: 10.1016/s0925-4773(02)00291-5. [DOI] [PubMed] [Google Scholar]
- 13.Lamar E., Kintner C., Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- 14.Turner J., Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 16.Hildebrand J.D., Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu J.R., Arnosti D.N. Functional similarity of Knirps CtBP-dependent and CtBP-independent transcriptional repressor activities. Nucleic Acids Res. 2003;31:4654–4662. doi: 10.1093/nar/gkg491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castet A., Boulahtouf A., Versini G., Bonnet S., Augereau P., Vignon F., Khochbin S., Jalaguier S., Cavailles V. Multiple domains of the receptor-interacting protein 140 contribute to transcription inhibition. Nucleic Acids Res. 2004;32:1957–1966. doi: 10.1093/nar/gkh524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C.L., McKinsey T.A., Lu J.R., Olson E.N. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 20.Deltour S., Pinte S., Guerardel C., Wasylyk B., Leprince D. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol. Cell. Biol. 2002;22:4890–4901. doi: 10.1128/MCB.22.13.4890-4901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen Y., Nguyen D., Li Y., Lai Z.C. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics. 2000;156:195–203. doi: 10.1093/genetics/156.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai P., Shinagawa T., Nomura T., Harada J., Kaul S.C., Wadhwa R., Khan M.M., Akimaru H., Sasaki H., Colmenares C., et al. Ski is involved in transcriptional regulation by the repressor and full-length forms of Gli3. Genes Dev. 2002;16:2843–2848. doi: 10.1101/gad.1017302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y., Zhang S., Rehn M., Itaranta P., Tuukkanen J., Heljasvaara R., Peltoketo H., Pihlajaniemi T., Vainio S. Induced repatterning of type XVIII collagen expression in ureter bud from kidney to lung type: association with sonic hedgehog and ectopic surfactant protein C. Development. 2001;128:1573–1585. doi: 10.1242/dev.128.9.1573. [DOI] [PubMed] [Google Scholar]
- 24.McKinsey T.A., Zhang C.L., Olson E.N. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 26.Walterhouse D.O., Yoon J.W., Iannaccone P.M. Developmental pathways: Sonic hedgehog-Patched-GLI. Environ. Health Perspect. 1999;107:167–171. doi: 10.1289/ehp.99107167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acevedo M.L., Kraus W.L. Transcriptional activation by nuclear receptors. Essays Biochem. 2004;40:73–88. doi: 10.1042/bse0400073. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki H., Nishizaki Y., Hui C., Nakafuku M., Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 29.Akimaru H., Chen Y., Dai P., Hou D.X., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 30.Williams A.J., Khachigian L.M., Shows T., Collins T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J. Biol. Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- 31.Agata Y., Matsuda E., Shimizu A. Two novel Kruppel-associated box-containing zinc-finger proteins, KRAZ1 and KRAZ2, repress transcription through functional interaction with the corepressor KAP-1 (TIF1beta/KRIP-1) J. Biol. Chem. 1999;274:16412–16422. doi: 10.1074/jbc.274.23.16412. [DOI] [PubMed] [Google Scholar]
- 32.Le Douarin B., You J., Nielsen A.L., Chambon P., Losson R. TIF1alpha: a possible link between KRAB zinc finger proteins and nuclear receptors. J. Steroid Biochem. Mol. Biol. 1998;65:43–50. doi: 10.1016/s0960-0760(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 33.Koipally J., Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 2000;275:19594–19602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 34.Turner J., Crossley M. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays. 2001;23:683–690. doi: 10.1002/bies.1097. [DOI] [PubMed] [Google Scholar]