Abstract
Repetitive elements represent a large portion of the human genome and contain much of the CpG methylation found in normal human postnatal somatic tissues. Loss of DNA methylation in these sequences might account for most of the global hypomethylation that characterizes a large percentage of human cancers that have been studied. There is widespread interest in correlating the genomic 5-methylcytosine content with clinical outcome, dietary history, lifestyle, etc. However, a high-throughput, accurate and easily accessible technique that can be applied even to paraffin-embedded tissue DNA is not yet available. Here, we report the development of quantitative MethyLight assays to determine the levels of methylated and unmethylated repeats, namely, Alu and LINE-1 sequences and the centromeric satellite alpha (Satα) and juxtacentromeric satellite 2 (Sat2) DNA sequences. Methylation levels of Alu, Sat2 and LINE-1 repeats were significantly associated with global DNA methylation, as measured by high performance liquid chromatography, and the combined measurements of Alu and Sat2 methylation were highly correlative with global DNA methylation measurements. These MethyLight assays rely only on real-time PCR and provide surrogate markers for global DNA methylation analysis. We also describe a novel design strategy for the development of methylation-independent MethyLight control reactions based on Alu sequences depleted of CpG dinucleotides by evolutionary deamination on one strand. We show that one such Alu-based reaction provides a greatly improved detection of DNA for normalization in MethyLight applications and is less susceptible to normalization errors caused by cancer-associated aneuploidy and copy number changes.
INTRODUCTION
DNA methylation in mammalian cells is required for normal embryonic development, X-chromosome inactivation and genomic imprinting, and involves the addition of a methyl group to the C-5 position of cytosine, predominantly in a 5′-CpG-3′ sequence context [reviewed in (1)]. This is accomplished by the activities of one or more DNA methyltransferases (DNMTs), which use S-adenosylmethionine (AdoMet) as a cofactor. CpG dinucleotides are underrepresented in the human genome by a factor of about 5, due to the spontaneous deamination of 5-methylcytosine residues, resulting in C-to-T transition mutations at CpG dinucleotides (2). However, there are regions of the genome termed CpG islands that have retained their expected CpG content (3,4). Most CpG islands overlap the 5′ end of gene regions, including promoters, and are typically unmethylated in normal somatic tissues (4,5). However, only 40% of promoter regions are associated with CpG islands (4,5). The unique and repeated sequences in the remainder of the genome are often highly methylated at their CpG sites in somatic tissues (6).
CpG dinucleotides are often aberrantly methylated in human cancers to give regional hypermethylation at some CpG islands despite an overall reduction in 5-methylcytosine in the DNA (global DNA hypomethylation) (7–9). The frequent hypomethylation of repetitive elements in diverse human cancers is thought to largely account for the global hypomethylation commonly seen in human cancers (6).
Repetitive elements comprise ∼45% of the human genome (10,11) and consist of interspersed repeats derived from non-autonomous or autonomous transposable elements (12–14) and tandem repeats of simple sequences (satellite DNA) or complex sequences. The most plentiful short interspersed nucleotide element (SINE) in human DNA is the Alu repeat, an ∼282 bp non-LTR (Long Terminal Repeat) DNA sequence, which comprises 10% of the human genome and is present in ∼1 million copies per haploid genome (12). Other abundant non-LTR sequences are long interspersed nucleotide elements (LINEs) of up to 6 kb that comprise ∼20% of the human genome [reviewed in (6,13)]. LINE-1 elements are present at over 500 000 copies in the human genome; however, only 3000–4000 are full length and 30–100 are active retrotransposons (6,13).
LINE-1 elements are usually methylated in somatic tissues, and LINE-1 hypomethylation is a common characteristic of human cancers (15–18). Moreover, Alu sequences are also normally methylated in somatic tissues (19–21) and are thought to become hypomethylated in human cancer cells. However, not all Alus are hypomethylated in human cancers. Alu sequences located upstream of the CDKN2A promoter were found to be hypermethylated in cancer cell lines (22), and an Alu sequence located in intron 6 of TP53 showed extensive methylation in normal and cancer cells (22,23).
While LINEs and SINEs are interspersed throughout the genome, satellite DNA is largely confined to the centromeres or centromere-adjacent (juxtacentromeric) heterochromatin and to the large region of heterochromatin on the long arm of the Y chromosome. Satellite α (Satα) repeats are composed of 170 bp DNA sequences and represent the main DNA component of every human centromere (24). Satellite 2 (Sat2) DNA sequences are found predominantly in juxtacentromeric heterochromatin of certain human chromosomes and are most abundant in the long juxtacentromeric heterochromatin region of chromosome (Chr) 1. Sat2 sequences are composed of variants of two tandem repeats of ATTCCATTCG followed by one or two copies of ATG (25). Both Chr1 Satα and Chr1 Sat2 sequences, as well as Satα repeats present throughout all the centromeres, are highly methylated in normal postnatal tissues, hypomethylated in sperm and often hypomethylated in various cancers (26–29). In addition, Sat2 sequences on Chr1 and Chr16 are also hypomethylated in the ICF (immunodeficiency, centromeric region instability and facial abnormalities) syndrome, which usually involves mutations in DNMT3B (30,31).
Previous studies describing repetitive element DNA methylation have been mostly based on Southern blot analyses, which require large amounts of high-molecular-weight genomic DNA (7,27,29,32,33). Accurate global genomic 5-methylcytosine content is often determined by high performance liquid chromatography (HPLC) (7,27,29,32,33), which, although highly quantitative and reproducible, also requires large amounts of high-quality genomic DNA and is not suitable for high-throughput analyses. In a recent report, Alu and LINE-1 methylation levels were obtained by COBRA [COmbined Bisulfite Restriction Analysis, first described in (34)] and pyrosequencing of bisulfite-converted DNA (18). Although these quantitative methods represent major advancements in determining repetitive element DNA methylation levels, both require post-PCR manipulation, are labor-intensive and, therefore, may not be suitable for high-throughput analyses.
In this study, we advanced MethyLight assay technology, a quantitative, TaqMan-based real-time PCR system to analyze DNA methylation profiles (35), by extending it to the analysis of highly repeated DNA sequences. We designed and applied MethyLight assays to examine the methylation levels of Alu, LINE-1, and Chr1 centromeric Satα and juxtacentromeric Sat2 repeat sequences. We evaluated repetitive element MethyLight measurements on a panel of normal and tumor DNA samples for which accurate HPLC-based global DNA methylation measurements were available. These data suggest that methylation of either interspersed or tandem repeats can be used as a surrogate marker for estimating global DNA methylation levels. The combination (mean) of Alu and Sat2 repeat methylation measurements yielded a particularly close correlation with global genomic 5-methylcytosine content measurements obtained by HPLC.
Additionally, we exploited the high Alu copy number to design an Alu-based MethyLight control reaction to sensitively determine input DNA levels for normalization in MethyLight assays.
MATERIALS AND METHODS
Design of the Alu-based MethyLight control reaction
The Alu-based control reaction was designed in silico based on a deaminated Alu consensus DNA sequence, in which we assumed that all CpGs on one strand underwent evolutionary deamination. The strategy is indicated with deamination of the top strand in Figure 2 for simplicity; however, either the top or the bottom strands can be chosen. In designing the control reaction, we deaminated the CpGs on the bottom (opposite) strand of the consensus sequence to TpGs in silico, and as a result, CpG dinucleotides on the top strand of the consensus sequence became CpA dinucleotides. These CpA dinucleotides are converted to TpA dinucleotides upon bisulfite conversion and PCR, thereby generating a methylation-independent unique sequence. Using this deaminated and bisulfite-converted DNA sequence, we selected the PCR primer and probe sequences.
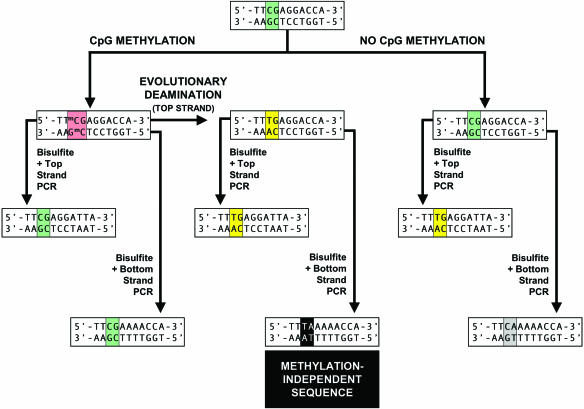
Figure 2.
Strategy for designing an Alu-based MethyLight control reaction against the Alu consensus DNA sequence in Figure 1. Since CpGs in Alu repeats can either be methylated (red) or unmethylated (green), one cannot distinguish if a CpG dinucleotide was subjected to evolutionary deamination of a methylated CpG (yellow) or was an unmethylated CpG deaminated due to reaction with bisulfite, as both events result in a TpG after bisulfite-specific PCR. However, if the bisulfite PCR primers are designed for amplification of the strand opposite to that which was evolutionarily deaminated, a TpA sequence results (see Methylation-Independent Sequence, shaded in black), which is distinct from the PCR products from an unmethylated CpG (CpA or TpG) after bisulfite conversion.
Bisulfite conversion and DNA recovery
DNA, in an 18 µl volume, was denatured at 100°C for 10 min, then centrifuged briefly, and chilled on ice. NaOH was added to a final concentration of 0.3 M in a 20 µl volume, and the sample was incubated at 42°C for 20 min. Sodium bisulfite solutions were prepared on the day of use by adding 1.9 g sodium metabisulfite (Sigma) to 3.2 ml of 0.44 M NaOH and heating at 50°C to dissolve the bisulfite. After addition of hydroquinone (0.5 ml of a 1 M solution), 120 µl of this solution was mixed with each DNA sample. The reaction was allowed to proceed at 50°C for 16 h in the dark.
Following bisulfite conversion, the DNA was recovered using the Qiagen Viral RNA Mini Kit (Qiagen) according to the manufacturer's specifications with the following changes: after loading the column with the supplied lysis buffer and 100% EtOH, the filtrate was re-loaded to increase DNA recovery. After washing with two supplied wash buffers, the DNA was eluted in 80 µl (2 × 40 µl elutions). To desulfonate the sample, 50 µl of 0.2 M NaOH was then added for 15 min, followed by neutralization with 10 µl of 1 M HCl. The supplied lysis buffer and EtOH was added to the desulfonated sample, and the bisulfite-converted DNA was then purified a second time. The eluted DNA sample was stored at −20°C.
M.SssI methylation assay
Peripheral blood leukocyte (PBL) DNA (Promega) was used as a substrate for M.SssI treatment. PBL DNA (0.05 µg/µl) was incubated with M.SssI at a concentration of 1 U/µg DNA (0.05 U/µl) and 0.16 mM AdoMet overnight at 37°C. Then extra AdoMet (to 0.20 mM) and M.SssI (to 0.065 U/µl) were added followed by a second overnight incubation at 37°C. The sample was stored at 4°C, and 18 µl (0.9 µg DNA) aliquots were used for bisulfite conversion and recovery as described above.
Whole-genome amplification (WGA)
To generate unmethylated human DNA as control samples for testing the MethyLight reactions, sperm and PBL DNAs (10 ng each) were amplified using a WGA kit (Molecular Staging) as described by the manufacturer. The DNA was then recovered by phenol–chloroform extraction and ethanol-precipitation, dissolved in water and stored at −20°C. An aliquot (1–2 µg) was then treated with bisulfite and recovered as described above.
MethyLight reactions
The PCR primers and probes are listed in Table 1. Probes were either labeled with a black hole quencher (BHQ-1, Biosearch Technologies), or a minor groove binder non-fluorescent quencher (MGBNFQ, Applied Biosystems). MethyLight PCR was performed in a 30 µl reaction volume with 200 µM dNTPs, 0.3 µM forward and reverse PCR primers, 0.1 µM probe, 3.5 mM MgCl2, 0.01% Tween-20, 0.05% gelatin and 0.1 U of Taq polymerase using the following PCR program: 95°C for 10 min, then 50 cycles of 95°C for 15 s followed by 60°C for 1 min. The samples in 96-well plates were analyzed on an Opticon DNA Engine Continuous Fluorescence Detector (MJ Research/Bio-Rad). A standard curve for the Alu repeat control reaction was generated from 1:25 serial dilutions of bisulfite-converted, M.SssI-treated DNA for the methylated reactions and 1:25 serial dilutions of bisulfite converted DNA after WGA for the unmethylated MethyLight reactions.
Table 1.
Description of repetitive element MethyLight reaction information
| Reaction ID | GenBank number | Amplicon start | Amplicon end | Forward primer sequence 5′ to 3′ | Reverse primer sequence 5′ to 3′ | Probe sequence 5′ to 3′a |
|---|---|---|---|---|---|---|
| ALU-C4 | Consensus Seq., Figure 1 | 1 | 98 | GGTTAGGTATAGTGGTTTATATTTGTAATTTTAGTA | ATTAACTAAACTAATCTTAAACTCCTAACCTCA | CCTACCTTAACCTCCC-MGBb |
| ALU-M1c | Y07755 | 5059 | 5164 | ATTATGTTAGTTAGGATGGTTTCGATTTT | CAATCGACCGAACGCGA | CCCAACACTTTAAAAAACCGAAATAAATAAATCACGA |
| ALU-M2 | Consensus Seq., Figure 1 | 7 | 95 | GCGCGGTGGTTTACGTTT | AACCGAACTAATCTCGAACTCCTAAC | AAATAATCCGCCCGCCTCGACCT |
| ALU-M3 | Consensus Seq., Figure 1 | 133 | 210 | ATTAGTCGGGCGTGGTGG | CCCGAATTCAAACGATTCTCC | CGCGTTTGTAATTTTAGTTATTCGGGAGGTTG |
| ALU-M5d | AC007256 | 156130 | 156221 | GGTATGATGGCGTATGTTTGTAATTT | CGACTCACCACAACTTCCACC | AAACGATTCTCCTACCTCAACCTCCCGAA |
| ALU-U3 | Consensus Seq., Figure 1 | 91 | 192 | TGGTTAATATGGTGAAATTTTGTTTTTATT | TCCTACCTCAACCTCCCAAATAACT | CAAACACACACCACCACACCCAACTAATTT |
| LINE-1-M1 | X52235 | 251 | 331 | GGACGTATTTGGAAAATCGGG | AATCTCGCGATACGCCGTT | TCGAATATTGCGTTTTCGGATCGGTTT |
| LINE-1-U3 | X52235 | 110 | 210 | TTTATTAGGGAGTGTTAGATAGTGGGTG | CCTTACACTTCCCAAATAAAACAATACC | CACCCTACTTCAACTCATACACAATACACACACCC |
| SATα-M1 | M38468 | 139 | 260 | TGATGGAGTATTTTTAAAATATACGTTTTGTAGT | AATTCTAAAAATATTCCTCTTCAATTACGTAAA | TATCCCGTTTCCAACGAA-MGBb |
| SATα-U1 | M38468 | 138 | 261 | TTGATGGAGTATTTTTAAAATATATGTTTTGTAGT | AAATTCTAAAAATATTCCTCTTCAATTACATAAA | TTTATCCCATTTCCAACAAA-MGBb |
| SAT2-M1 | X72623 | 1074 | 1153 | TCGAATGGAATTAATATTTAACGGAAAA | CCATTCGAATCCATTCGATAATTCT | CGATTCCATTCGATAATTCCGTTT-MGBb |
aAll probes contain a 6FAM fluorophore and a BHQ-1 probe unless otherwise noted.
bMGB refers to a Minor Groove Binder non-fluorescent quencher in the 3′ terminus of the probe (MGBNFQ).
cThis MethyLight reaction was designed toward an Alu sequence in the S100A2 gene.
dThis MethyLight reaction was designed toward an Alu sequence in the CASP8 gene.
Methylation calculations
The MethyLight data specific for methylated repetitive elements were expressed as percent of methylated reference (PMR) values and were calculated similarly to a recent report (36), but with the following changes. DNA treated with M.SssI served as a methylated reference, and the Alu-based control reaction (ALU-C4 in Table 1) was used as a control reaction to measure the levels of input DNA to normalize the signal for each methylation reaction. The levels of unmethylated repetitive elements were expressed as percent of unmethylated reference (PUR) values and were calculated similarly to PMR values except that bisulfite-converted, unmethylated human DNA obtained by WGA, as described above, was used as an unmethylated reference for PUR determinations of each repetitive element.
In analyzing the panel of DNAs described in Figure 4, each MethyLight reaction was performed 3–6 times, except for ALU-M3, which was only analyzed in duplicate. The PMR or PUR values represent the mean values, and the error bars represent standard error of the mean. Standard error of the mean values were not included for the ALU-M3 reaction because we obtained only two PMR measurements for this reaction. In correlating MethyLight measurements to HPLC-based global 5-methylcytosine levels (Figure 5), each MethyLight reaction was performed in triplicate, and the data shown are the mean PMR or PUR values of the three measurements. The data were plotted as PMR or PUR mean values for each repetitive element versus HPLC-based global 5-methylcytosine measurements for each sample. The composite methylation measurements of Alu and Sat2 (Figure 5H) were determined by obtaining the mean between the triplicate ALU-M2 and SAT2-M1 PMR values, and then plotting the composite mean PMR value versus the HPLC-based global 5-methylcytosine measurement for each sample. Linear regression analyses were performed using GraphPad InStat version 3.0a for Macintosh (GraphPad Software, San Diego, CA).
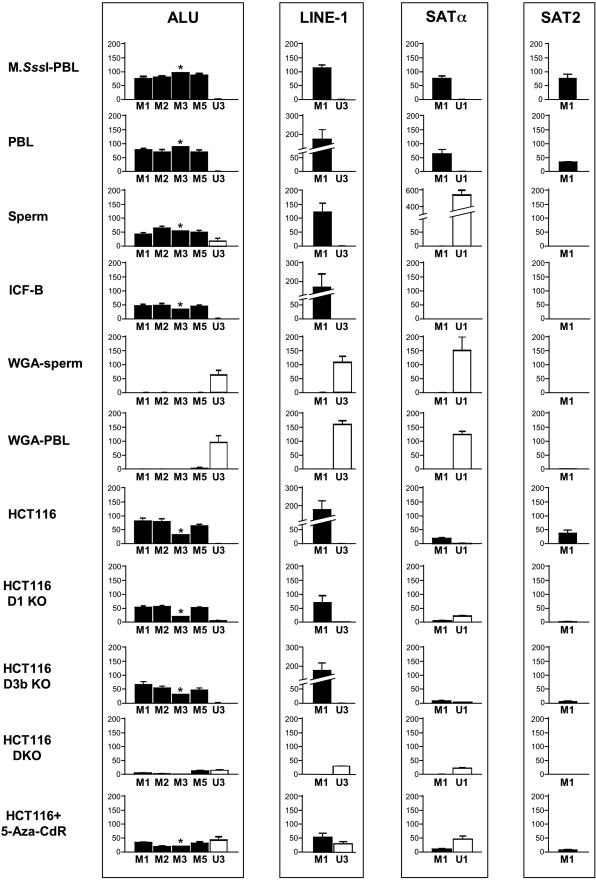
Figure 4.
Evaluation of MethyLight reactions toward the methylated and unmethylated versions of Alu, LINE-1, Satα and Sat2 sequences on a panel of DNA samples. Levels of methylation (in black) are expressed as PMR using DNA treated with M.SssI as a methylated reference. Levels of unmethylated DNA (in white) are expressed as PUR in which a WGA-DNA sample was used as an unmethylated reference. Each value represents the mean of 3–6 methylation measurements, except for the ALU-M3 reaction, which is the average of two measurements. Error bars indicate the standard error of the mean and have been omitted for the ALU-M3 reaction, as indicated by an asterisk, since we have only two PMR measurements for this reaction. We detected PMR or PUR values of <0.01 due to cross-reactivity of the methylated or unmethylated primers to either unmethylated or methylated template DNA, respectively, except for the ALU-M5 reaction, which gave slightly higher PMR values on the WGA-PBL DNA sample.
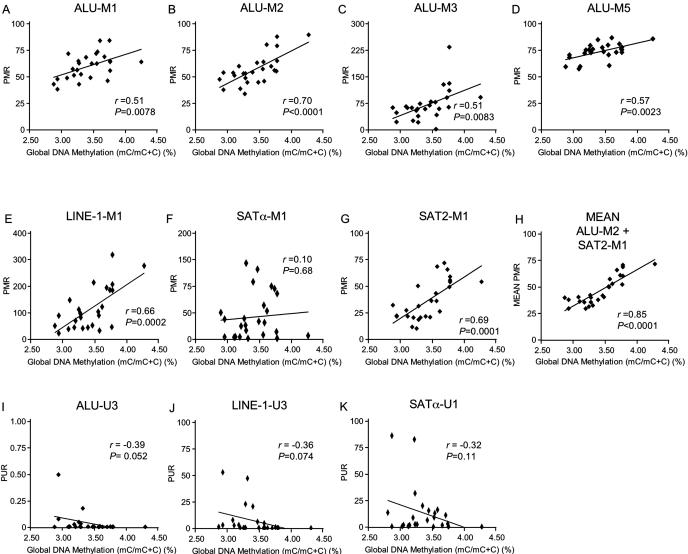
Figure 5.
Correlation of MethyLight-based measurements of each repetitive element with HPLC-based global DNA methylation measurements for the samples described in Table 2. PMR values for the methylated Alu sequences (A–D), LINE-1 (E), Satα (F), Sat2 (G), are correlated with HPLC measurements. The correlation between the composite mean PMR values of ALU-M2 and SAT2-M1 reactions and global methylation measurements is shown in (H). The correlation of PUR values for the unmethylated Alu, LINE-1 and Satα sequences with HPLC-based 5-methylcytosine measurements are shown in (I–K). The HPLC data represent the mean of 2–3 measurements and the MethyLight data represent the mean of three measurements. The MethyLight and HPLC-based methylation data were correlated using linear regression analysis for each repetitive element.
HPLC measurements of global genomic 5-methylcytosine content
The overall DNA 5-methylcytosine content was determined by HPLC on heat-denatured DNA digested to nucleosides. The global 5-methylcytosine content for each sample is listed in Table 2 and represents the mean value of 2–3 measurements (29). The average replicate percentage standard deviation for replicates was 2% (37). Linear regression was used to determine a correlation between MethyLight-based PMR values and HPLC-based global methylation measurements for each repetitive element. Global 5-methylcytosine content in humans has been shown to be tissue-specific with a range of 3.43–4.26% of cytosine residues methylated in normal tissues (32,37). DNA samples were classified as hypomethylated if the global methylation was between 3.20 and 3.40% and substantially hypomethylated if the amount of global methylation was ≤3.20%. These designations were only included as an indication of global 5-methylcytosine content in various tissues and in cancer cells.
Table 2.
HPLC-based measurements of global 5-methylcytosine content in normal and cancer DNA samples
| Sample | Laird ID | Normal/cancer | Global methylation: mC/(mC + C) (%) |
|---|---|---|---|
| WT4 | 6027 | Cancer | 2.88 |
| OvCa E | 6020 | Cancer | 2.94 |
| WT31 | 6036 | Cancer | 2.94 |
| WT9 | 6030 | Cancer | 3.09 |
| OvCa I | 6021 | Cancer | 3.11 |
| WT14B | 6031 | Cancer | 3.18 |
| WT6 | 6028 | Cancer | 3.20 |
| WT29 | 6035 | Cancer | 3.25 |
| OvCa L | 6023 | Cancer | 3.27 |
| WT7A | 6029 | Cancer | 3.28 |
| WT24 | 6033 | Cancer | 3.28 |
| OvCa D | 6019 | Cancer | 3.31 |
| WT22 | 6032 | Cancer | 3.39 |
| WT 27 | 6034 | Cancer | 3.46 |
| OvCa B | 6018 | Cancer | 3.48 |
| Liver A | 6042 | Normal | 3.55 |
| OvCa J | 6022 | Cancer | 3.56 |
| OvCa Q | 6026 | Cancer | 3.57 |
| OvCa M | 6024 | Cancer | 3.61 |
| Lung C | 6046 | Normal | 3.68 |
| OvCa O | 6025 | Cancer | 3.73 |
| OvCa A | 6017 | Cancer | 3.76 |
| Kidney B | 6043 | Normal | 3.77 |
| Spleen C | 6045 | Normal | 3.77 |
| Thyroid B | 6044 | Normal | 3.77 |
| Cerebellum A | 6041 | Normal | 4.26 |
Southern blot-based method of determining Chr1 Sat2 hypomethylation
Chr1 Sat2 hypomethylation was determined by Southern blot analysis, comparison of band patterns in autoradiograms and quantitation of Phosphorimager results as described previously (33). The MethyLight PMR values were compared with the hypomethylation scores for each sample using ANOVA.
RESULTS
Development of an Alu-based, methylation-independent MethyLight control reaction
MethyLight is a quantitative, TaqMan-based, real-time PCR assay to measure DNA methylation profiles, using bisulfite-converted DNA as a substrate (35,38). MethyLight is compatible with DNA samples derived from fresh tissue, cell lines, as well as formalin-fixed, paraffin-embedded tissues or bodily fluids such as plasma or serum, where the amount of DNA available is usually limiting. Each MethyLight-based methylation data point, expressed as a PMR value [first described in (39,40)], involves the use of a CpG-independent, bisulfite-specific control reaction to measure input DNA levels. The control reaction should be highly sensitive to accurately measure small amounts of DNA and should not detectably vary in its ability to be amplified from different human DNA samples, including cancer tissues.
Control reactions amplifying the low- or single-copy genes MYOD1, ACTB and COL2A1 have been routinely used in previous reports as a measure of input bisulfite-treated DNA levels (33,35,36,38). However, these single-copy genes may not always be reliable in human cancers, where chromosomal deletions, duplications and gene amplifications are frequent events. We therefore designed an Alu-based MethyLight control reaction to evaluate input DNA levels that would be both more sensitive in analyzing low amounts of input DNA, and at the same time would be less subject to local cancer-associated genetic alterations, compared with the single-copy control genes that we have traditionally used. The high copy number of Alu repeats, which are dispersed throughout the genome, makes it unlikely that copy number shifts at specific genomic loci would substantially influence their PCR product yield and also allows for sensitive detection of minute amounts of DNA. In addition, the presence of rare single-nucleotide polymophisms (SNPs) should not interfere with the PCR amplification of the Alu control reaction, but may hinder the PCR amplification of single- or low-copy sequences. Recently, Stroun et al. (41) used an Alu-based real-time PCR to analyze the amount of free DNA in plasma/serum of cancer patients and healthy controls.
Alu repeats are highly heterogeneous due to the depletion of CpG dinucleotides by spontaneous evolutionary deamination, in which C-to-T transition mutations are generated. In order to develop a methylation-independent, Alu-based control reaction, we first generated an Alu consensus sequence based on a panel of young and old individual Alu repeats subfamilies in which we identified all CpG dinucleotides as well as those that became evolutionarily deaminated to TpG (or CpA on the strand opposite of an evolutionary deamination event) in older Alu sequences (Figure 1). However, we could not identify a subregion in the consensus sequence that was devoid of CpGs. We therefore took advantage of the evolutionary deamination process to design a control reaction toward Alu sequences in which all cytosines in a CpG context have been deaminated on one of the two DNA strands. These deaminated Alu sequences should be CpG methylation independent. We reasoned that such strand-specifically deaminated Alu sequences should exist in the genome by chance, even though they were not present among the selected Alu sequences listed in Figure 1.
Figure 1.
An Alu consensus DNA sequence determined from the sequences of young and old individual Alu repeats. Old Alu sequences (Alu-J, SluSp, AluSx, AluSq and AluSc) and young Alu sequences (AluY, AluSb2, AluYb8, AluYa5 and AluYa8) were compared in order to generate an Alu consensus sequence for the purpose of designing an Alu-based MethyLight control reaction. Since the goal was to design a methylation-independent reaction of as many individual Alu repeats as possible, all CpG dinucleotides became a part of the consensus sequence. Old Alu sequences are the predominant form in human cells, and this is also reflected in the Alu consensus sequence. The continuous and dashed lines underneath the consensus sequence panels indicate the MethyLight PCR amplicon locations within the consensus sequence for the Alu control reaction (ALU-C4), two reactions toward the methylated consensus sequence (ALU-M2 and ALU-M3) and one reaction toward the unmethylated Alu consensus sequence (ALU-U3).
The design of this control reaction is complicated because MethyLight reactions are specific for bisulfite-converted DNA. After bisulfite conversion, the two DNA strands are non-complimentary (Figure 2), and the PCR primers are designed toward either the top or the bottom DNA strands. Methylated CpGs are refractory to bisulfite and remain as CpG on both DNA strands, whereas an unmethylated CpG dinucleotide is deaminated to a TpG after bisulfite conversion. However, a TpG dinucleotide also results from the evolutionary deamination of a methylated CpG. Sequences deaminated during evolution can be distinguished from those resulting from bisulfite conversion if the bisulfite-PCR primers are specific for the DNA strand opposite to the evolutionarily deaminated DNA strand (Figure 2, Methylation-Independent Sequence). This CpA containing strand will be converted to a distinct TpA sequence after bisulfite conversion.
Using this strategy, we simulated evolutionary deamination of the Alu consensus sequence in silico by first replacing every CpG dinucleotide with a CpA dinucleotide (representing the evolutionary deamination of the opposite strand of the consensus sequence), and then selecting primers and a probe for the MethyLight control reaction specific for the bisulfite-converted form of this DNA sequence. The locations of the PCR amplicon and the primer/probe sequences are shown in Table 1, and the location of the PCR amplicon within the Alu consensus sequence is shown in Figure 1. To satisfy the PCR melting temperature requirements for the Alu control reaction, the probe contains a minor groove binding non-fluorescent quencher (MGBNFQ). The use of MGB probes in real-time PCR-based, DNA methylation analyses was also recently reported for the purpose of improving PCR specificity (42).
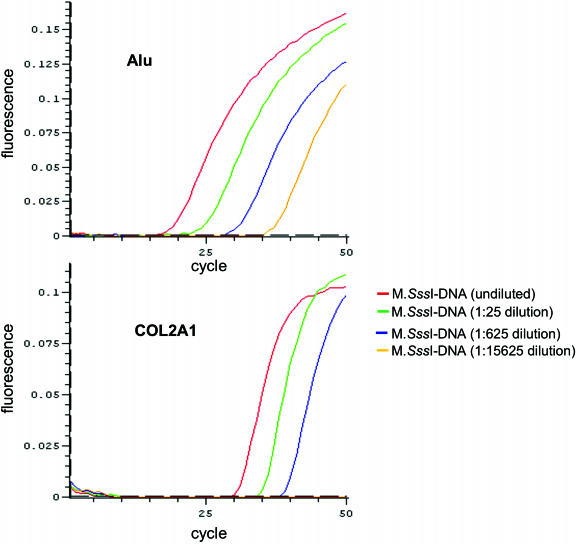
To assess whether the Alu-based MethyLight control reaction could detect a high number of evolutionarily deaminated Alu repeats, we compared the threshold cycle [C(t) value] of this reaction to the the C(t) value of the single-copy COL2A1 control reaction using real-time PCR on 1:25 serial dilutions of bisulfite-converted, M.SssI-treated DNA (Figure 3). The Alu reaction fluorescence was detected ∼15 cycles earlier than the COL2A1 reaction on undiluted, bisulfite-converted M.SssI-DNA. The Alu reaction, after a 1:15 000 dilution, still could detect an appreciable amount of input DNA compared with the COL2A1 reaction, which at the same dilution failed to amplify M.SssI-DNA (Figure 3). To address the variability of the Alu control reaction, we compared the cycle threshold [C(t)] values of the Alu and COL2A1 control reactions on M.SssI-treated PBL and HCT116 colon cancer cell line DNAs. The mean C(t) values for Alu and COL2A1 reactions on M.SssI DNA were 15.14 ± 0.21 and 29.71 ± 0.25, respectively, and the mean C(t) values on HCT116 DNA were 15.57 ± 0.04 and 30.86 ± 0.25, respectively, These data demonstrate not only the reproducibility of the Alu control reaction but also its high sensitivity. The greatly increased sensitivity for detecting input DNA in the methylation-independent reaction is especially useful when analyzing samples with limited amounts of DNA. However, this Alu-based control reaction does not increase the sensitivity of MethyLight-based methylation detection, which is a function of the methylation-specific MethyLight reactions. However, the availability of a highly sensitive control reaction allows us to determine the methylation detection sensitivity threshold for difficult samples.
Figure 3.
Evaluation of the performance of the Alu-based control reaction compared with a single-copy control reaction. Serial 1:25 dilutions of bisulfite-converted, M.SssI-treated DNA were used to compare the Alu and COL2A1 control reactions by real-time PCR. The fluorescence is plotted versus the PCR cycle number for both reactions and each sample dilution is indicated.
Development and evaluation of repetitive element MethyLight reactions
We next developed MethyLight reactions to target methylated Alu and LINE-1 elements, as well as the centromeric Satα and juxtacentromeric Sat2 repeats (Figure 4). We also designed MethyLight reactions specific for the unmethylated versions of Alu, LINE-1 and Satα sequences (Figure 4); however, we were unable to successfully develop a MethyLight reaction specific for unmethylated Sat2 repeats.
Our primers for the unmethylated reactions were designed by replacing CpG with TpG in the primers and probe. This design approach does not distinguish between unmethylated and evolutionarily deaminated CpGs in these repetitive elements. However, we assume that the fraction and genomic location of deaminated CpG dinucleotides are fairly constant in the human population, given the relatively recent evolutionary divergence of the human population, compared with the origin of Alu repeats, which predate the divergence of primates. Nevertheless, we acknowledge that a low level of inaccuracy in the measurements of unmethylated reactions may stem from this inability to discriminate between evolutionary and bisulfite-induced deamination. This problem does not arise for the methylation-specific reactions described below.
We designed four reactions toward methylated Alu sequences: the M1 reaction is designed toward an Alu repeat within the S100A2 gene; the M2 and M3 reactions are directed toward the consensus sequence (the locations of the ALU-M2 and ALU-M3 PCR amplicons are indicated in Figure 1); and the M5 reaction is specific toward an Alu repeat located upstream of the CASP8 gene. The S100A2 Alu sequence is similar to AluSx and AluSq subfamilies, and the CASP8 Alu sequence is most similar to the AluSp subfamily. We also designed one reaction toward the unmethylated Alu consensus sequence (ALU-U3). The methylated and unmethylated reactions specific for the LINE-1 sequences were based on a LINE-1 consensus sequence (GenBank accession number X52235). The Satα and Sat2 reactions were designed toward sequences specifically on chromosome 1 (GenBank accession numbers M38468 and X72623, respectively); however, satellite-specific sequences on other chromosomes may also be detected. Therefore, we classified these reactions generically as Satα and Sat2. Similar to the Alu control reaction, the probes for the Satα and Sat2 reactions also contain a 3′ MGBNFQ moiety to satisfy the TaqMan probe melting temperature requirements.
We tested the methylation specificities of the methylated and unmethylated Alu, LINE-1, Satα and Sat2 reactions on a panel of bisulfite-converted DNA samples (Figure 4). PBL DNA treated in vitro with the CpG methylase M.SssI (M.SssI-DNA) served as a methylated DNA template, and untreated PBL and sperm DNAs were also included. To prepare unmethylated DNA as a negative control for methylation, we used a strategy that takes advantage of the WGA reaction used to amplify minute amounts of DNA. An approach similar to this has recently been described (43). WGA is based on extending random hexamers annealed to genomic DNA with the highly processive phi 29 DNA polymerase that contains both 5′–3′ and 3′–5′ exonuclease (44) as well as strand displacement activities (45,46). As genomic DNA is amplified by this polymerase, the DNA methylation will be lost. We amplified sperm and PBL DNAs by WGA, followed by bisulfite conversion.
We also included DNA from human cell lines that have been previously characterized with regards to repetitive element methylation. ICF lymphoblastoid cell lines were included because they have extensive hypomethylation of chromosome 1 and 16 Sat2 sequences (47–49). In addition, we analyzed DNA from HCT116 human colon cancer cells, HCT116 cells containing DNMT1−/− (D1KO), DNMT3B−/− (D3bKO), DNMT1−/− and DNMT3B−/− cells (DKO) (50,51) and HCT116 cells after treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-Aza-CdR). Global DNA methylation is largely retained in the DNMT1−/− and DNMT3B−/− HCT116 cells, while DNA from the DKO HCT116 cells is almost completely hypomethylated (50,51). Alu sequences are detectably hypomethylated only in the DKO cells, while Sat2 sequences were hypomethylated in the single and the DKO cells (51). DNA from HCT116 cells after 5-Aza-CdR-mediated DNA methylation inhibition should be hypomethylated relative to the untreated HCT116 cells.
We evaluated the methylation specificity of each repetitive element MethyLight reaction on the panel of 11 DNA samples (Figure 4). The methylation values for the reactions directed toward methylated repetitive elements were expressed as PMR in which M.SssI-DNA was used as a methylated reference (39,40). For the unmethylated reactions, the amount of unmethylated DNA is expressed as a PUR in which a WGA-sperm sample was used as an unmethylated reference.
Our results indicated that Alu, LINE-1, Satα and Sat2 repeats were highly methylated in M.SssI-PBL DNA as well as PBL-DNA (Figure 4). Sperm DNA also showed substantial methylation of Alu and LINE-1 sequences. However, Satα and Sat2 sequences were hypomethylated in sperm relative to PBL-DNA (Figure 4), consistent with previous reports (26,33). These results are also in agreement with previous reports of hypomethylation of Chr1 Satα and Chr1 Sat2 sequences in sperm (47–49). Substantial Alu and LINE-1 methylation was also detected in ICF cells, while Satα and Sat2 methylation was not detected. While Alu methylation and Sat2 hypomethylation in ICF cells is consistent with previously reported studies (22,47,48), we could not detect unmethylated Satα DNA in the ICF sample. This may reflect the presence of either partially unmethylated or methylated Satα DNA in ICF cells, as the unmethylated MethyLight reactions were designed to recognize fully unmethylated target DNA sequences.
All four unmethylated reactions showed hypomethylation of the repetitive elements in both WGA-DNA samples, indicating that these samples are appropriate unmethylated DNA controls (Figure 4). However, centromeric and telomeric regions of the genome are underrepresented in the WGA assay (46). Although we detected a decrease in Satα and Sat2 input levels compared with the Alu and LINE-1 sequences in WGA-treated DNAs, there was still ample signal to accurately measure PMR and PUR values (data not shown).
We detected high levels of Alu and LINE-1 methylation in HCT116 cells, while both Satα and Sat2 sequences were hypomethylated relative to M.SssI-DNA. The hypomethylation of satellite repeats in this cancer cell line is consistent with the very frequent hypomethylation of these sequences in human cancers (6). Alu and LINE-1 methylation was similar or slightly reduced in the D1KO and D3bKO cells. Alu and LINE-1 repeats, as well as both satellite repeats were strongly hypomethylated in the HCT116 DKO cells, similar to previous findings by Southern blot analysis (50,51). The HCT116 cells also showed the expected increase in Alu, LINE-1 Satα and Sat2 hypomethylation after treatment with 5-Aza-CdR.
Our methylated-specific and unmethylated-specific reactions are designed to recognize fully methylated and fully unmethylated repetive elements, respectively. However, due to polymorphisms among the repetitive elements, and due to variable levels of methylation of these repeats in human DNA samples, it is difficult to determine exactly how many repeat units in the genome are recognized by each of our reactions. Nevertheless, we can obtain a good comparative measure of the number of copies recognized by each reaction by comparing the C(t) values of each reaction on its cognate optimal target DNA (M.SssI-treated DNA for methylated reactions and WGA DNA for unmethylated reactions). For example, the ALU-M2 and SAT2-M1 reactions both had C(t) values more than 10 cycles lower than the LINE-1-M1 and SATα-M1 reactions. Assuming an up to 2-fold amplification with each PCR cycle, this suggests that the ALU-M2 and SAT2-M1 reactions recognize up to 1000-fold more repeat elements than either the LINE-1-M1 or SATα-M1 reactions. We anticipate therefore that the ALU-M2 and SAT2-M1 reactions would be superior surrogate measures of global genomic 5-methylcytosine content than either the LINE-1-M1 or SATα-M1 reactions.
Correlation between repetitive element methylation and global DNA methylation levels
Using the MethyLight assay, each repetitive element reaction was tested on a panel of DNAs from normal tissues (liver, lung, kidney, spleen, thyroid and cerebellum) as well as ovarian carcinoma (OvCa) and Wilms tumor (WT) samples, all of which had been tested for global DNA methylation levels by HPLC (Table 2). Global DNA methylation levels in humans have been shown to be tissue-specific, with a range of 3.43–4.26% of cytosine residues methylated in normal tissues (32,37). DNA samples were classified as hypomethylated if the global methylation was between 3.20 and 3.40% and substantially hypomethylated if the amount of global methylation was ≤3.20%. However, these levels of hypomethylation were only used to better describe DNA hypomethylation in human cells and had no influence on the data analysis in this study.
We compared the PMR values (from reactions aimed at methylated sequences) or PUR values (from reactions toward unmethylated sequences) with HPLC-based global DNA methylation measurements for each sample. While the PMR values for all four methylated Alu reactions were significantly associated with global genomic 5-methylcytosine content (Figure 5A–D), linear regression analysis showed that the ALU-M2 methylation reaction was most closely associated with global DNA methylation as determined by HPLC (correlation coefficient, r = 0.70, P < 0.0001, Figure 5B). ALU-M3, another Alu reaction based on the consensus DNA sequence, was also correlative with global methylation measurements (r = 0.51, P = 0.0083, Figure 5C). However, the ALU-M2 fluorescence was greater than the ALU-M3 reaction (data not shown), suggesting that the ALU-M2 reaction is superior not only in correlating Alu methylation to global measurements, but also in PCR quality. Methylation levels of LINE-1 and Sat2 sequences were also significantly associated with global 5-methylcytosine content (Figure 5E and G) but there was not a significant association of Satα methylation with global DNA methylation (Figure 5F).
The ALU-M2 and SAT2-M1 reactions gave the best correlation with global 5-methylcytosine content, as we had anticipated from their relatively low C(t) values. Since these two reactions both recognize a relatively large number of repeat units, but represent quite distinct types of repetitive elements, we considered that their combined measurement could potentially provide a superior assessment of global DNA methylation across various human genomic DNA samples. Both measurements are expressed as PMR values, so we compiled a composite measurement by calculating the mean PMR for the two reactions for each sample. This composite measure yielded a remarkable improvement in the correlation with HPLC measurements of 5-methylcytosine content (r = 0.85, P < 0.0001) (Figure 5H). We therefore recommend that this composite measure be used for MethyLight-based estimates of genomic 5-methylcytosine content.
The reactions directed toward unmethylated versions of Alu and LINE-1 sequences were of borderline significance when compared with global DNA methylation measurements (Figure 5I and J). Unmethylated Satα repeats were also not significantly correlated with HPLC-based global DNA methylation measurements (Figure 5K). However, as expected, there was a clear trend of increased PUR measurements with decreasing global DNA methylation levels for all unmethylated repetitive element reactions.
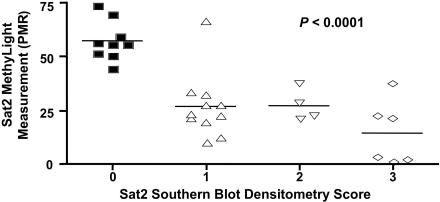
Correlation between Sat2 MethyLight measurements and Sat2 Southern blot-based hypomethylation scores
We showed here a statistically significant relationship between Sat2 methylation and global DNA methylation levels (Figure 5G). Sat2 methylation had been previously determined for these samples using Southern blot analysis in which Sat2 hypomethylation was graded on a scale of 0 (methylated) to 3 (extensively hypomethylated) using quantitation of Phosphorimager data and evaluation of band patterns from autoradiograms (33). We compared the MethyLight PMR measurements with the corresponding DNA hypomethylation scores from Southern blots for the same normal tissues, as well as ovarian carcinoma and Wilms tumor samples (Figure 6). Two ICF cell line DNAs were also included, as these samples were previously shown to harbor Sat2 hypomethylation (47–49). We found a statistically significant correlation between the Sat2 MethyLight PMR measurements and the Southern blot-based hypomethylation score (P < 0.0001), suggesting that the MethyLight-based assay for Sat2 methylation measurements are highly consistent with the Southern blot-based assay to determine Sat2 hypomethylation.
Figure 6.
MethyLight data (PMR) versus Southern blot-based Chr1 Sat2 hypomethylation densitometry scores. A score of 0, no hypomethylation; 1, small amounts of hypomethylation; 2, moderate hypomethylation; 3, strong hypomethylation on 7 normal tissues, two ICF cell lines, one control cell line and 20 cancer tissue samples (Wilms tumors and ovarian carcinomas). The data points are indicated by the squares (hypomethylation score = 0, upward triangles (hypomethylation score = 1), downward triangles (hypomethylation score = 2) and diamonds (hypomethylation score = 3). Mean PMR values are indicated by the horizontal bars. The significance of the association of both types of data after ANOVA analysis is shown.
DISCUSSION
Repetitive elements comprise ∼45% of the human genome, with the 1 million Alu sequences alone occupying ∼10% of the genome, and LINE-1 elements also representing a substantial portion of the genome. Since these interspersed repetitive elements as well as tandem repeated centromeric and juxtacentromeric repeats contain numerous CpG dinucleotides, the methylation status of these sequences is relevant to understanding global DNA methylation. It is generally thought that repetitive elements are heavily methylated in normal somatic tissues, but are methylated to a lesser extent in malignant tissues, driving the global genomic hypomethylation commonly found in human cancers.
In order to evaluate the repetitive element methylation in normal and malignant tissues, we designed MethyLight reactions specific for methylated Alu, LINE-1, and the Satα and Sat2 repeats, as well as unmethylated versions of Alu, LINE-1 and Satα repeats. These reactions were first evaluated on a panel of DNAs to test their methylation specificities. These samples included HCT116 colon cancer cells as well as HCT116 cells harboring gene-targeted disruptions of DNMT1, DNMT3B or both DNMT genes (50,51). In support of previous findings (50,51), DNMT1−/− and DNMT3B−/− cells retained Alu methylation, but were hypomethylated in the HCT116 DKO cells. Sat2 hypomethylation had been described for both the single and double knockout HCT116 cells when compared with wild-type HCT116 cells (51). Our MethyLight data directed toward this repeat showed similar findings.
More evidence of the specificity of the MethyLight-based Sat2 repetitive element reaction was shown with the analysis of cells from ICF patients. ICF cells exhibit hypomethylation of Sat2 repeats on chromosomes 1 and 16; however, global methylation is generally retained and Satα hypomethylation is found in only some patients (47–49). Here, we also show that Sat2 sequences were hypomethylated in ICF cells, while substantial levels of Alu and LINE-1 methylation remained. While DNMT3B is required for normal Sat2 methylation in vivo, both DNMT1 and DNMT3B physically interact (52) and cooperatively maintain genomic methylation (51).
The MethyLight measurements for each repetitive element were also compared with global 5-methylcytosine measurements by HPLC for a panel of normal and tumor tissue samples. Of all the MethyLight reactions tested, the methylated ALU-M2 reaction was most closely correlated with genomic 5-methylcytosine content, although all four reactions targeting methylated Alu sequences exhibited a significant association with global DNA methylation. MethyLight-based Sat2 and LINE-1 methylation levels were also significantly correlated to global DNA methylation. Therefore, methylation of diverse repetitive elements may serve as surrogate markers for genomic 5-methylcytosine content. Indeed, a composite measure combining ALU-M2 and SAT2-M1 PMR values yielded much better correlation with genomic 5-methylcytosine content (r = 0.85) than either measure alone. We recommend that this composite measure be used for MethyLight-based estimates of genomic 5-methylcytosine content.
A recent study from Yang et al. (18) describes the use of COBRA and pyrosequencing assays to determine Alu and LINE-1 methylation. Their PCR strategy can capture the composite methylation information of ∼15 000 Alu sequences, which represents ∼0.1% of the genome. The authors suggest that this measurement could serve as a surrogate for global DNA methylation measurement, but they do not confirm this with a validation using 5-methylcytosine determinations in the same samples. Instead they show that 5-aza-CdR treatment causes a reduction in their Alu measurement, which would be expected, regardless of whether their assay is a good surrogate for global DNA methylation or not. In our study, we have included this important validation with HPLC measurements of 5-methylcytosine content. Our MethyLight-based approach is the first to provide quantitative estimates of the degree of correlation between these repetitive element measurements and 5-methylcytosine content.
An important other innovation of our MethyLight-based methylation-specific assays is that the accuracy of these assays is not affected by deamination at CpG dinucleotides, whether by evolution or by bisulfite conversion, since our methylation-specific reactions only recognize fully methylated, non-deaminated CpG dinucleotides. The Yang method relies on an indirect subtraction to distinguish between evolutionary and bisulfite-mediated deamination.
Both COBRA and pyrosequencing require extensive post-PCR manipulation, thereby substantially increasing the labor intensiveness of the assay, and introducing the potential for contamination of future reactions by PCR products. The MethyLight assay is finished as soon as the PCR has been completed, requires no post-PCR processing and can easily be applied to hundreds or thousands of samples. These repetitive element reactions represent advances in the MethyLight assay not only for determining the methylation of individual repetitive elements, but also in serving as markers for global 5-methylcytosine content.
Pyrosequencing, COBRA and MethyLight provide different perspectives on DNA methylation determinations: pyrosequencing and COBRA are used to quantitatively measure methylation levels of individual CpGs within a given locus, while MethyLight assays measure the percentage of molecules in which all of the CpGs of a specific locus (usually between 4 and 10 CpGs) are either methylated or unmethylated. COBRA and pyrosequencing (18) platforms showed that Alu or LINE-1 methylation provides a marker for 5-Aza-CdR-mediated demethylation in human cancer cell lines. Unlike the MethyLight data shown here, it is indefinite whether COBRA and pyrosequencing assays could serve as surrogate markers for determining global methylation.
The measurements of repetitive element hypomethylation targeted by unmethylated MethyLight reactions were not significantly correlated with 5-methylcytosine content, although there was a trend of increasing levels of unmethylated repeat sequences with decreasing 5-methylcytosine content. The poor correlation may be due to relatively low numbers of completely unmethylated repetitive elements in vivo. In addition, these reactions generated low output fluorescence signals, and this may be due to difficulties in design and performance of MethyLight reactions specific for unmethylated sequences. These sequences have a higher A:T content than methylated sequences due to the fact that all unmethylated cytosines are converted to thymines after bisulfite conversion. This may reduce primer-template specificity and PCR efficiency.
The MethyLight-based Sat2 methylation data were significantly associated with HPLC-based global DNA methylation levels and also with Southern blot-based Sat2 hypomethylation scores. Therefore, Sat2 methylation analysis by MethyLight is a strong indicator of global DNA methylation measurements. This is in agreement with previous findings that the Southern blot-based Sat2 hypomethylation index is also statistically significantly associated with global DNA hypomethylation levels in numerous human cancer tissues (27,29,37,53)
We did not find a statistically significant correlation between Satα methylation by MethyLight and genomic 5-methylcytosine content. Previously, a significant association between Southern blot-determined Satα hypomethylation and global DNA methylation was shown in ovarian cancers, Wilms tumors and breast cancers (27,29,53). The discrepancy between these sets of data may be due to inherent assay differences. The MethyLight reaction design is based on a short DNA sequence and may not be specific toward the Chromosome 1 Satα sequence when compared with the hybridization probe used for the Southern blot assay. Moreover, the relatively high C(t) value for the Satα MethyLight reaction indicates that a relatively small number of Satα repeats contribute to the MethyLight measurement.
We also show here that Satα and Sat2 repeats are unmethylated in sperm. These data are consistent with previous findings of satellite hypomethylation in sperm based on Southern blot assays (26,33). However, the findings of Hassan et al. (49) demonstrated that there was sporadic methylation of Sat2 sequences in sperm, using bisulfite genomic sequencing. Partial Sat2 methylation may not be identified by this MethyLight assay, since the PCR specifically targets sequences containing multiple fully methylated CpGs.
The findings presented here also provide further insight into DNA hypomethylation in human cancers. Hypomethylation of diverse classes of repetitive elements, such as interspersed and tandemly repeated sequences, occurs together with regional CpG island hypermethylation in cancer cells (33,37). DNA hypomethylation is not restricted to satellite sequences and other tandem DNA repeats (6,54), but also includes the interspersed Alu and LINE elements. Repetitive element methylation, however, may be influenced by the local chromatin structure, especially for Alu and LINE-1 sequences, which are interspersed throughout the genome and can be embedded within genes. A study from Kondo and Issa (55) showed evidence of histone H3 lysine 9 methylation, a marker for inactive heterochromatin, at numerous Alu repeat sequences.
While Alu methylation was most closely associated with global methylation levels in our study, not all Alus are hypomethylated in human cancers. An Alu sequence located upstream of the extensively methylated CDKN2A promoter was found to be hypermethylated in cancer cell lines (22), and an Alu sequence located in intron 6 of TP53 showed extensive methylation (22,23). In support of this, a recent study showed that NBL2, a tandem repeat found in acrocentric chromosomes, can become either hypermethylated or hypomethylated in human cancers (54).
We also describe here the development of a novel strategy to design an Alu-based control reaction for measuring the levels of bisulfite-converted input DNA for MethyLight assays by which bisulfite and evolutionary deaminated sequences could be distinguished. The Alu-based control reaction shows fluorescence above background levels at ∼15 cycles earlier than the single-copy COL2A1 control reaction, suggesting an increased detection of four orders of magnitude (∼10 000 copies). This represents ∼1% of total Alu content. Using human colon cancer cells, Yang et al. (18) have deduced that a large number of CpGs in Alu sequences are evolutionarily deaminated, supporting our results in which a large number of deaminated Alu repeats were detected using this control reaction.
Although the Alu-based control reaction does not increase the sensitivity of the MethyLight methylation reactions and hence the sensitivity of methylation detection, it is useful for determining relative DNA amounts in specimens where the quantity and/or quality of DNA may be limited, such as formalin-fixed, paraffin-embedded tissues or free tumor DNA in plasma/serum. Therefore, a methylation detection sensitivity threshold for such samples can be determined. More importantly, the Alu control reaction is expected to be a more stable and reliable measure of input bisulfite-converted DNA levels than reaction toward a single-copy sequence when analyzing tumor samples with local amplifications or deletions, since single-copy genes may not always be present at diploid copy number levels in human cancers, where chromosomal alterations are frequent events. The high copy number of interspersed Alu repeats in the human genome makes it unlikely that cancer-related sequence abnormalities would substantially influence their PCR yield. The use of the Alu-based control reaction should therefore result in a more stable determination of PMR/PUR values. Moreover, Alu and other high copy repetitive element sequences may also be useful in measuring changes in gene dosage, such as gene amplifications.
In conclusion, the design and application of MethyLight assays to measure repetitive element methylation represent novel technical advancements. Their use as surrogate markers for global DNA methylation makes them attractive in analyzing the effects of DNA methylation on human disease in population-based studies.
Acknowledgments
The authors thank Dr Bert Vogelstein for providing the HCT116, HCT116 DNMT1−/−, HCT116 DNMT3B−/−, and HCT116 DNMT1−/−, DNMT3B−/− double knockout cells. This work was supported by NIH grants CA 81506 (to M.E.) and ES 011672 and CA 96958 (to P.W.L.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Laird P.W. The power and the promise of DNA methylation markers. Nature Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 3.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 4.Takai D., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 7.Gama-Sosa M.A., Slagel V.A., Trewyn R.W., Oxenhandler R., Kuo K.C., Gehrke C.W., Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg A.P., Gehrke C.W., Kuo K.C., Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 10.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 11.Jordan I.K., Rogozin I.B., Glazko G.V., Koonin E.V. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 12.Weiner A.M. SINEs and LINEs: the art of biting the hand that feeds you. Curr. Opin. Cell. Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 13.Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H.J. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Prak E.T., Kazazian H.H.J. Mobile elements and the human genome. Nature Rev. Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- 15.Kimura F., Seifert H.H., Florl A.R., Santourlidis S., Steinhoff C., Swiatkowski S., Mahotka C., Gerharz C.D., Schulz W.A. Decrease of DNA methyltransferase 1 expression relative to cell proliferation in transitional cell carcinoma. Int. J. Cancer. 2003;104:568–578. doi: 10.1002/ijc.10988. [DOI] [PubMed] [Google Scholar]
- 16.Florl A.R., Steinhoff C., Muller M., Seifert H.H., Hader C., Engers R., Ackermann R., Schulz W.A. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br. J. Cancer. 2004;91:985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalitchagorn K., Shuangshoti S., Hourpai N., Kongruttanachok N., Tangkijvanich P., Thong-Ngam D., Voravud N., Sriuranpong V., Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 18.Yang A.S., Estecio M.R.H., Doshi K., Kondo Y., Tajara E., Issa J.-P.J. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gama-Sosa M.A., Wang R.Y., Kuo K.C., Gehrke C.W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11:3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid C.W. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 1991;19:5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kochanek S., Renz D., Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993;12:1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisenberger D.J., Velicescu M., Cheng J.C., Gonzales F.A., Liang G., Jones P.A. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- 23.Magewu A.N., Jones P.A. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol. Cell. Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., Lin C.C. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 25.Jeanpierre M. Human satellites 2 and 3. Ann. Genet. 1994;37:163–171. [PubMed] [Google Scholar]
- 26.Narayan A., Ji W., Zhang X.Y., Marrogi A., Graff J.R., Baylin S.B., Ehrlich M. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int. J. Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 27.Qu G., Dubeau L., Narayan A., Yu M.C., Ehrlich M. Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutat. Res. 1999;423:91–101. doi: 10.1016/s0027-5107(98)00229-2. [DOI] [PubMed] [Google Scholar]
- 28.Qu G.Z., Grundy P.E., Narayan A., Ehrlich M. Frequent hypomethylation in Wilms tumors of pericentromeric DNA in chromosomes 1 and 16. Cancer Genet. Cytogenet. 1999;109:34–39. doi: 10.1016/s0165-4608(98)00143-5. [DOI] [PubMed] [Google Scholar]
- 29.Ehrlich M., Hopkins N.E., Jiang G., Dome J.S., Yu M.C., Woods C.B., Tomlinson G.E., Chintagumpala M., Champagne M., Dillerg L., et al. Satellite DNA hypomethylation in karyotyped Wilms tumors. Cancer Genet. Cytogenet. 2003;141:97–105. doi: 10.1016/s0165-4608(02)00668-4. [DOI] [PubMed] [Google Scholar]
- 30.Xu G.L., Bestor T.H., Bourc'his D., Hsieh C.L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J.J., Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 31.Hansen R.S., Wijmenga C., Luo P., Stanek A.M., Canfield T.K., Weemaes C.M., Gartler S.M. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrlich M., Gama-Sosa M.A., Huang L.H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widschwendter M., Jiang G., Woods C., Muller H.M., Fiegl H., Goebel G., Marth C., Muller-Holzner E., Zeimet A.G., Laird P.W., et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 34.Xiong Z., Laird P.W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:e32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widschwendter M., Siegmund K.D., Muller H.M., Fiegl H., Marth C., Muller-Holzner E., Jones P.A., Laird P.W. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 37.Ehrlich M., Jiang G., Fiala E., Dome J.S., Yu M.C., Long T.I., Youn B., Sohn O.S., Widschwendter M., Tomlinson G.E., et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;26:6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 38.Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Danenberg P.V., Laird P.W. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 39.Eads C.A., Lord R.V., Wickramasinghe K., Long T.I., Kurumboor S.K., Bernstein L., Peters J.H., DeMeester S.R., DeMeester T.R., Skinner K.A., et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 40.Siegmund K.D., Laird P.W. Analysis of complex methylation data. Methods. 2002;27:170–178. doi: 10.1016/s1046-2023(02)00071-3. [DOI] [PubMed] [Google Scholar]
- 41.Stroun M., Lyautey J., Lederrey C., Mulcahy H.E., Anker P. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: evidence for a preferential release from viable cells? Ann. N.Y. Acad. Sci. 2001;945:258–264. doi: 10.1111/j.1749-6632.2001.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 42.Zeschnigk M., Bohringer S., Price E.A., Onadim Z., Masshofer L., Lohmann D.R. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umetani N., de Maat M.F.G., Mori T., Takeuchi H., Hoon D.S.B. Synthesis of universal control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochem. Biophys. Res. Commun. 2005;329:219–223. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
- 44.Garmendia C., Bernad A., Esteban J.A., Blanco L., Salas M. The bacteriophage phi 29 DNA polymerase, a proofreading enzyme. J. Biol. Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 45.Blanco L., Bernad A., Lazaro J.M., Martin G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 46.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeanpierre M., Turleau C., Aurias A., Prieur M., Ledeist F., Fischer A., Viegas-Pequignot E. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 48.Tuck-Muller C.M., Narayan A., Tsien F., Smeets D.F., Sawyer J., Fiala E.S., Sohn O.S., Ehrlich M. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet. Cell Genet. 2000;89:121–128. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 49.Hassan K.M., Norwood T., Gimelli G., Gartler S.M., Hansen R.S. Satellite 2 methylation patterns in normal and ICF syndrome cells and association of hypomethylation with advanced replication. Hum. Genet. 2001;109:452–462. doi: 10.1007/s004390100590. [DOI] [PubMed] [Google Scholar]
- 50.Rhee I., Jair K.W., Yen R.W., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 51.Rhee I., Bachman K.E., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 52.Kim G.D., Ni J., Kelesoglu N., Roberts R.J., Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson K., Yu M.C., Arakawa K., Fiala E., Youn B., Fiegl H., Muller-Holzner E., Widschwendter M., Ehrlich M. DNA hypomethylation is prevalent even in low-grade breast tumors. Cancer Biol. Ther. 2004;3:1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 54.Nishiyama R., Qi L., Tsumagri K., Weissbecker K., Dubeau L., Champagne M., Sikka S., Nagai H., Ehrlich M. A DNA repeat, NBL2, is hypermethylated in some cancers but hypomethylated in others. Cancer Biol. Ther. 2005;4:440–448. doi: 10.4161/cbt.4.4.1622. [DOI] [PubMed] [Google Scholar]
- 55.Kondo Y., Issa J.P. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 2003;278:27658–27662. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]