Abstract
Deviations from Chargaff's 2nd parity rule, according to which A∼T and G∼C in single stranded DNA, have been associated with replication as well as with transcription in prokaryotes. Based on observations regarding mainly the transcription-replication co-linearity in a large number of prokaryotic species, we formulate the hypothesis that the replication procedure may follow different modes between genomes throughout which the skews clearly follow different patterns. We draw the conclusion that multiple functional sites of origin of replication may exist in the genomes of most archaea and in some exceptional cases of eubacteria, while in the majority of eubacteria, replication occurs through a single fixed origin.
INTRODUCTION
Under no strand bias conditions, i.e. when mutation rates are identical for complementary nucleotide substitutions (e.g. A to G and T to C), one can expect equimolarities of complementary nucleotides in single-stranded DNA (1). Chargaff was the first to report such an extended equidistribution in raw genomic sequences in 1951 (2) and this form of symmetry has since been widely referred to as ‘Chargaff 2nd Parity Rule’ (PR2).
Local violations of this parity have been observed in all known organisms and in bacteria they have been correlated with the positioning of the origin of replication. Differences in the synthesis of the leading and lagging strands have been proposed to be the reason behind the observed deviations, which also switch their direction at the point of the origin of replication (3). Relative nucleotide skews between complementary nucleotides [expressed as (A − T)/(A + T) and (G − C)/(G + C)] have been used in order to determine the position of the origin of replication in bacterial species (4–6), chloroplast genomes (7), viral and mitochondrial genomes (8). In particular, Frank and Lobry (9) have provided a very useful tool, which may predict probable origins of replication of bacterial genomes, using the nucleotide skews (see also http://pbil.univ-lyon1.fr/software/Oriloc).
Nonetheless, replication is not the only molecular process that seems to be affecting strand parity. Szybalski et al. (10) first noted the loading of mRNAs with purine residues. Bell and Forsdyke (11) demonstrated that the direction of transcription correlates with the well-reported complementary nucleotide skews and Lao and Forsdyke (12) went further to propose the necessity of avoiding the formation of extended secondary mRNA structures as an evolutionary pressure lying behind them. Touchon et al. (13) have also argued for transcription being the main source of PR2 violations but attribute the process to specific mutational biases. Recently, transcription coupled asymmetry of nucleotide transition rates was also reported in mammalian genomes (14,15).
Regarding the main cause of these asymmetries, various authors (16,17) have provided some solid evidence in favor of the mutationist view by demonstrating that the skews are mainly expressed in the third codon positions of genes as well as in non-coding regions where selective pressure is minimal. The deviations are found to be more intense in third codon positions of genes than in intergenic sequences (17). Single-stranded DNA is being exposed during the transcription as well as the replication process, and in that way asymmetric mutational pressures between leading and lagging strand can be shaping the nucleotide composition through both processes (18–21).
Another type of fundamental asymmetry, very common in bacterial genomes, is that of gene orientation. In most species, there seems to be a tendency for having the majority of genes transcribed in the replication direction (22–24). McInerney (25) observed that the codon usage variation in Borrelia burgdorferi is a result of this trend of strand asymmetry. Head-on collisions of the two polymerases can be by-passed as has been shown for Escherichia coli (26,27) but the evolutionary advantage remains in matters of transcription speed, especially in highly expressed genes (28). Rocha and Danchin (29) have brought evidence that it is essentiality and not expressiveness that drives genes to be encoded on the leading strand. In any case, when transcriptional activity is indispensable there is an evolutionary advantage that leads genes to be transcribed co-directionally with replication. The co-occurrence of the nucleotide asymmetries and gene-strand bias has been noticed previously (30) but not discussed in detail.
The origin of replication of certain genomes has proven difficult to be defined with or without the use of skews. Data referring to the localization of the origin of replication are lacking for the majority of the archaea while for some of them two or three functional sites have been reported (31,32) based on experimental data.
This work will attempt to focus on specific aspects of the correlation of skews with the processes of transcription and replication that have not yet been pointed out. Furthermore, it will attempt to argue on the possible existence of multiple origins of replication in specific bacterial genomes as a potential explanation regarding the observed skews as well as the failure to locate a single origin in certain species.
RESULTS
Parity violation in some distinct cases
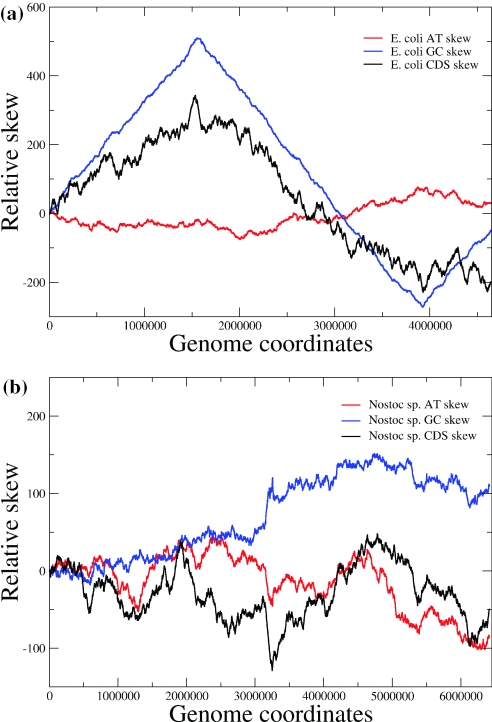
The well-studied genome of E.coli is one of the best examples where nucleotide skews are highly correlated with the location of the origin of replication (6). One can locate the origin of replication simply from observing a cumulative GC skew like the one presented in Figure 1a, where the CDS skew for the E.coli genome is also depicted. A CDS skew may be presented pictorially when one addresses the direction of transcription of genes in a manner resembling a walk along the DNA thread. The curve moves upwards for each gene encoded on the examined strand (denoted ‘+’ by convention) or downwards if the gene is encoded on the complementary ‘−’ strand. The space increment covered each time equal the gene's length. Thus, the shape of the CDS skew is indicative of specific trends in the gene orientation throughout the genome.
Figure 1.
Nucleotide and CDS skews for two different cases. (a) A typical eubacterium, E.coli, with clear-cut GC and CDS skew (b) Nostoc sp., a non-typical eubacterium, exhibiting fluctuating skews.
The significant correlation of the CDS skew with the formerly discussed, nucleotide ones (here GC skew only) is obvious in the case of E.coli. Through examination of all available genome skews as presented in the Oriloc website (http://pbil.univ-lyon1.fr/software/Oriloc), one may conclude that there are no cases of existence of the one type of skew without the other, while absence of a CDS skew is always followed by a subsequent absence of the nucleotide ones. It appears that the two types of skews are connected through a necessary condition. In the following, we will try to provide evidence advocating that this condition is also a sufficient one.
Based on the above observations, one can make some interesting conjectures on the genomes that do not have apparent nucleotide skews. The case of Nostoc sp. is characteristic. As shown in Figure 1b there are no clear-cut nucleotide skews in the genome of this organism. What is also interesting is the lack of a CDS skew, as the direction of transcription does not follow a special trend. Furthermore, examination of a large number of bacterial genomes (data from Oriloc) shows that the behaviour similar to Nostoc sp. is observed for a small number of eubacteria and for the majority of the archaea.
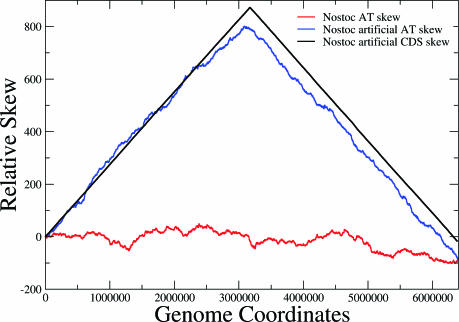
It is obvious from Figure 1b that the GC skews (and to a lesser extent the AT skews) follow the rudimentary CDS skew when this exists. We find reasonable to believe that the absence of nucleotide skews in Nostoc sp. and in most archaea is a direct consequence of the absence of a clear CDS skew. The above conjecture may be corroborated through the following findings. We have divided the complete genome of Nostoc sp. in parts, each including a gene and the adjacent non-coding spacer and have re-positioned them according to their transcription direction in order for a skewed distribution to be formed. This was carried out in the following way: an artificial genome has been constructed, containing the complete set of Nostoc sp. genes with all those encoded on the ‘+’ strand at the first half and all those of the ‘−’ strand at the second half. The relative positions of the non-coding spacers remained unchanged. An artificial, perfect CDS skew was thus created, using the complete set of genes of the Nostoc sp. genome. We then went on to check this artificial genome's nucleotide skews and compare them with the ones produced by the real genome. The results depicted in Figure 2 show that a very intense AT nucleotide skew is obtained from the ‘re-arranged’ genome, in contrast to the one produced by the real genome (the GC skew shape was very similar, data not shown). This may be held as evidence of the CDS skew being a sufficient condition for the observation of the nucleotide skews. Moreover, it constitutes a strong argument in favour of transcription-related mutational pressure being the main source of compositional asymmetry. If replication-related mutation bias were to be creating the nucleotide skews directly, it would be very unlikely that such an extensive manipulation of the genomic coordinates would lead to the formation of clear-cut nucleotide skews.
Figure 2.
AT skews obtained from the real genome of Nostoc sp. and an artificial one created when genes are rearranged in an order that produces a perfect CDS skew (all forward genes arranged sequentially, followed by all reverse complement genes). A clear-cut AT skew emerges from the artificial genome with the perfect CDS skew, which is also shown in black. The corresponding GC skews follow similar pattern and are not presented here for sake of simplicity.
But where lies the difference between bacteria that produce strong CDS skews, such as E.coli and others that do not, such as Nostoc sp. and the majority of the archaea? We believe that the existence of more than one functional origins of replication may come as a plausible explanation. In the case of the short eubacterial chromosomes, in contrast to eukaryotic ones, when more than one functional origins of replication exist, they will not have to be simultaneously active, rather one would be selected in a more or less stochastic way. Under this prospect and due to the alternations of the origins of the replication, no CDS skew would have emerged because of the lack of evolutionary advantages of co-directionality of transcription and replication. Thus, neither nucleotide skews would be observed. An inspection of Figure 1b, which corresponds to the skews produced by Nostoc sp., an exceptional eubacterial case, as well as the ones derived from most archaeal genomes (data not shown) provides the picture that one would expect under the above scenario.
The hypothesis of existence of multiple origins of replication in bacteria
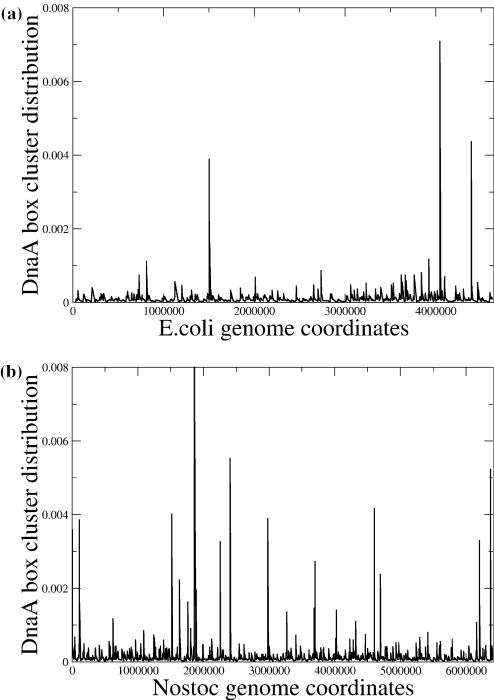
An additional point that can be raised for the possible existence of multiple origins of replication for Nostoc sp. may come from the localization of potential eubacterial DNA polymerase binding sites. Bacterial replication origins, although varying in size, contain (in the majority of cases) several DnaA boxes and clustering of these boxes may indicate the position of a functional origin (5). We have searched both the genomes of E.coli and Nostoc sp. for the E.coli's DnaA box consensus, which is 5′-TTATCCACA-3′, and represents a highly conserved sequence in the majority of bacterial species with small deviations depending on the G + C content of each genome. We performed our search allowing for all possible oligonucleotides that differed by no more than 1 nt from the consensus box. In Figure 3a and b, we have scanned the distribution of DnaA box clusters. The value r = 1/d as discussed in (5) was used in this case as a measure of the DnaA box cluster density, where with d is denoted the sum of the distances of each box to its two adjacent ones. Thus, high r-values indicate dense clustering of DnaA boxes in close vicinity, which would be primary candidates for sites, where replication could initiate.
Figure 3.
DnaA box clusters distribution for (a) E.coli and (b) Nostoc sp. Peaks correspond to multiple DnaA boxes occurring in close vicinity, indicating probable sites where replication may be initiated.
As one can see, the density of the clusters in the case of Nostoc sp. is considerably greater than the one of E.coli. Furthermore, the fact that the values of r are, in general, higher for Nostoc sp. comes as an additional indication that there exist multiple sites with high probability to serve as origins of replication in this genome.
The highest of the three distinct ‘spikes’ of the E.coli DnaA box-density distribution is the actual functional origin of replication. In the case of Nostoc sp., there are 14 ‘spikes’ with values higher than r = 0.002. The highest of these is located on a region where both AT and CDS skews reach a local maximum making this specific position a prime candidate for an origin of replication. However, neither this is an overall maximum, such as the one existing for E.coli (compare with Figure 1a), nor it is the only one with high clustering of DnaA boxes. Such sites exist throughout the genome of Nostoc sp. alongside other local maxima and minima of the nucleotide and CDS skews. Particularly, the second, third and fourth higher ‘spikes’ of the r-distribution also coincide with local AT and CDS skew maxima (compare Figures 1b and 3b). These observations may support the hypothesis of existence of multiple functional origins of replication in this specific genome.
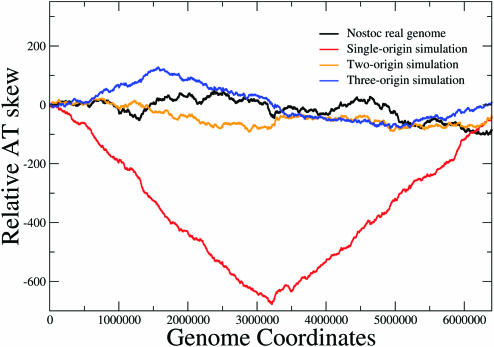
The effect, which multiple origins of replication would have on bacterial chromosomes, may be investigated through computational simulations. The simulations that follow incorporate the hypothesis that a gene rearrangement resulting in the gene being transcribed collinearly with the replication will be more likely than the opposite. This simple hypothesis was applied on the complete genome of Nostoc sp. The simulations were carried out as follows:
A supposed fixed origin of replication was set at the middle of the genome and leading and lagging strands were defined.
The genome was subjected to 500 ‘replication cycles’.
During each such cycle, every single gene was subjected to a strand switch, in a way that resembled strand transposition, with a probability of 1%.
About 90% of these rearrangements (0.9% of the total) were ‘selectively advantageous’, meaning that they led genes to be encoded on the leading strand as defined by the supposed origin of replication. The remaining 10% corresponded to rearrangements that led genes to switch from the leading to the lagging strand.
In all cases, positions and directions of non-coding segments remained un-altered. In this way, we were able to verify that the observed effects were exclusively due to the direction of genes. The above simple procedure was in position to reproduce the effect of the existence of a simple fixed origin with the only hypothesis being the selective advantage of genes encoded on the leading strand (see Figure 4, single-origin simulation). No specific mutation pressures were implemented. The choice of the probability parameters was arbitrary as was the number of replication cycles simulated. Several combinations of values for these parameters yield comparable results that do not change the main conclusion.
Figure 4.
AT skews obtained from the real genome of Nostoc sp. (black) and artificial ones produced through simulations assuming existence of a single (at 3.2 bp, red), two (at 0 and 3.2 Mb, orange) and three (at 0, 1.6 and 3.2 Mb, blue) distinct replication origins. A clear-cut pattern emerges for the simulated genome with a single replication origin, while for the two- and three-origin models the patterns resemble much the one of the real genome. All three simulations have reached the asymptotic limit (no significant changes are observed for higher number of iterations).
The produced simulated genome shows nucleotide skew patterns that resemble very much the ones exhibited by eubacteria, where one fixed origin is well established, like E.coli. It appears that a single origin of replication can lead to the emergence of clear-cut nucleotide skews, given that the leading strand is gradually enriched in genes. In order to verify that, two or more active origins of replication produce skew patterns similar to the ones observed for the natural genome of Nostoc sp. and most of the archaea, we carried out two additional simulations, but this time we used two and three alternating origins of replication, respectively. In this way, the leading and lagging strand partition is redefined every time the replication initiation site is altered. In both cases, the assumed origins of replication were activated (one at each replication cycle) randomly but equiprobably. Equiprobability was adopted for reasons of simplicity and it is checked that, if relaxed, the results change smoothly: the more one of the multiple sites becomes more probable, the more the resulting genome will resemble the one of the single origin simulation (data not shown). In Figure 4, one can observe the AT skews of all three artificial genomes alongside the one obtained from the organism's natural genome. The simple origin model leads to a pattern comparable with eubacterial genomes, which have fixed origins. On the other hand, the 2- and 3-origin models yield patterns that resemble the archaeal and the few eubacterial genomes, where there are no clear-cut nucleotide skews and throughout which the fluctuations appear to be random.
The above results may suggest a possible explanation for the emergence of CDS and nucleotide skews. This will be that given the selective advantage for genes to be encoded on the leading strand, a fixed origin of replication is a sufficient condition for the formation of the CDS skew. Subsequently, a skewed distribution of genes along the DNA strands is bound to cause the emergence of nucleotide skews. The compositional asymmetry is more likely to be a consequence of the transcription procedure as suggested by recent works (13,14), or of other gene sequence biases that are strand-specific.
Scale dependence of nucleotide skews. Quantifying the parity violation
Bell and Forsdyke (11) were the first to address the length dependence of the PR2 bias. They focused on length scales around the medium gene length and reached the conclusion that PR2 bias in bacteria is maximum, compared with a random sequence of identical composition, when measured at a window length of ∼1000 nt. The authors correlate this value with the processes of both transcription and recombination. However, the range of the examined length scales was not sufficient to lead them to any conclusions regarding correlation between PR2 bias and replication. We have taken up this task in complete bacterial genomes in the following way:
Each complete genome was read in overlapping windows of length l. The overlap step s was chosen to be 0.1 l at all times.
- For a given l the root-square deviation from PR2 was calculated for every segment i according to the following formula:
which is a form that incorporates both biases regardless of their sign. In a following step, the PR2(l) was calculated as the average over all PR2(l, i).
Finally, the value of l was increased by a given factor and the whole procedure was iterated.
We chose the l range to be between 103 and 105, so as not to exceed the size of small bacterial genomes and in order to be over the average gene length. Double logarithmic plots of l against PR2(l) produced linear fits in all cases. Through this procedure, we were able to ascribe to each species, one single value, which equaled to the slope of its log–log plot.
A correlation between the slope and the existence or absence of clear-cut PR2 skews is expected. Constant slopes are indicative of long regions with constant PR2 bias, a kind of homogeneity, which results in small absolute slope values. On the contrary, genomes throughout which the sign of the skews is fluctuating are expected to produce high absolute slope values near the random behaviour. In general, the absence of structure leads to steep slopes while the existence of long regions with constant skew sign is sufficient for the production of slopes values near zero.
We applied the above algorithm to obtain slopes for a total of 161 bacterial genomes, 18 of which where archaea. Slopes obtained varied from −0.47 to 0.002. A random sequence with no PR2 bias yielded, as expected, a slope of −0.5, while total bias would yield a constant zero slope, as is bound to produce every totally homogeneous sequence, meaning one with no fluctuations regarding nucleotide skews.
The majority of the examined archaeabacteria (17/18) produced slopes with absolute values higher than 0.3, while the majority of the eubacteria have slopes (absolute values) below this threshold. This particular trend seems to be correlated with the main difference in the skew pattern between eubacteria and archaea. Eubacteria have outright skews, which are probably reflections of the existence of a single origin of replication. On the other hand, the majority of the archaea lack a single, well-defined origin of replication and in some species there are experimental data corroborating the existence of more than one such sites (31,32). In Figure 5, the slopes obtained from some typical cases of eubacterial and archaebacterial genomes are depicted.
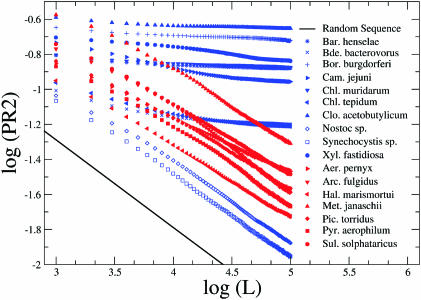
Figure 5.
Log-log plots of average root-squared PR2 deviations for two example sets of 10 eubacteria (blue) and 8 archaeabacteria (red). The majority of eubacteria produce slopes around zero while archaea appear to be close to the random behaviour as shown by the straight line in both panels. Two exceptional eubacterial cases (Nostoc sp. and Synechocystis, empty blue signs) share the behaviour of the archaea.
The question of the existence of a single origin of replication is once again emerging. The observations produced herein for a set of archaeal genomes in contrast to eubacterial ones may strengthen the aforementioned proposal of the existence of multiple origins of replication and the stochastic choice among them at each duplication event as a plausible explanation for the absence of clear-cut skews in archaebacteria and in some cases of eubacteria as well.
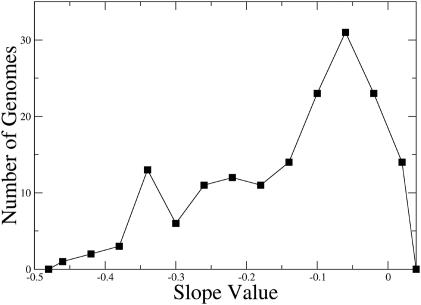
The behaviour of the examined bacterial genomes is better illustrated in Figure 6, where a distribution of the slope values is depicted for all genomes examined. One can observe two distinct and unequal peaks. The first and smaller one is located around high absolute values, which are representative of archaeal genomes bearing no clear-cut nucleotide skews. The second, located farther on the right around lower absolute values, is indicative of the eubacterial genome behaviour and is broader and higher due to the over-representation of eubacterial genomes in the examined collection. It is noticeable that the local minimum between these two peaks is located very close to −0.3. This particular value, as discussed above, seems to be a limit, drawing the borderline between two different modes of replication, one taking place through alternating multiple origins and the other through a single, fixed one. As simulations have indicated, intermediate slope values, corresponding to the ‘lump’ between the main peaks (−0.3 to −0.15), may be derived from genomes with more than one active origin, among which one is largely preferential.
Figure 6.
Distribution of log-log average root-square PR2 deviation slope values for 161 bacterial species. The two peaks exhibit the two distinct modes. The one located near −0.4 is representative of the archaea while the larger one closer to zero corresponds to the eubacteria. In general, species with slope values below the boundary value of −0.3 produce unclear skew patterns indicating an alternative mode of replication.
DISCUSSION
On whether replication or regional biases in gene sequences is the main reason lying behind the observed nucleotide skews, we have argued that neither may be disregarded. Although, among the possible regional mutational biases, we have only mentioned transcription-related ones as the main source of the parity violation, any gene sequence bias that is strand-related would be sufficient to support our proposed scenario. We believe that transcription might be the most probable cause, based on the following argument. Transcription-related nucleotide skews have been observed in higher eukaryotes (13). Nonetheless, in eukaryotes, there are clear boundaries between transcribed and non-transcribed regions, which cannot be defined in eubacterial genomes where overlapping genes are frequent. So we cannot carry out the analysis of regional skews around transcription start and end sites in eubacterial genomes. Even so, among the possible types of gene sequence bias, transcription seems to be the most probable, since it constitutes a common molecular process occurring very frequently throughout the cell cycle. Regarding the role of replication, it appears that a fixed replication origin may lead to the emergence of a CDS skew, due to the evolutionary advantage of the co-linearity of transcription and replication. In addition, establishing a CDS skew seems to be a sufficient condition for the emergence of nucleotide skews and this is the case in most eubacterial genomes as data from Oriloc may support. This was also shown herein, where the artificial production of a CDS skew in the genome of Nostoc sp. led to the emergence of clear-cut nucleotide skews.
Strand-biased mutational pressures inside genes are likely to produce local deviations from PR2. Furthermore, in order for this violation to be observed throughout the genome and not be confined locally in single transcriptional units, it is necessary that a strand bias is also existent. It is here that selection acts favouring those gene rearrangements that position genes on the leading strand. A fixed origin of replication is needed for selection to act this way. This hypothesis is able to incorporate the data referring to the emergence of CDS and nucleotide skews, as well as their correlation with the replication and transcription processes, in one simple framework.
Under this prospect, in the cases where both kinds of skews are absent, this may be ascribed to the presence of multiple origins of replication. These sites are likely to represent local maxima in cumulative nucleotide skew diagrams, as well as local maxima of CDS skew ‘walks’. Additionally, they are expected to be localized closely to clusters of DnaA boxes in eubacteria, which are conserved markers of DNA polymerase binding (see Figures 1b and 3b). Our results for Nostoc sp. seem to fit well with the above scenario, thus allowing us to support the proposal that multiple functional origins of replication may exist in this genome and are probable to be existing in other genomes, such as the ones of archaea, where clear-cut skews are not observed.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the Institute of Biology, NCSR ‘Demokritos’.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sueoka N. Intra-strand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995;40:318–325. doi: 10.1007/BF00163236. [DOI] [PubMed] [Google Scholar]
- 2.Chargaff E. Some recent studies on the composition and structure of nucleic acids. J. Cell Physiol. 1951;38:41–59. doi: 10.1002/jcp.1030380406. [DOI] [PubMed] [Google Scholar]
- 3.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 4.Lobry J.R. Origin of replication of Mycoplasma genitalium. Science. 1996;272:745–746. doi: 10.1126/science.272.5262.745. [DOI] [PubMed] [Google Scholar]
- 5.Mackiewicz P., Zakrzewska-Czerwinska J., Zawilak A., Dudek M.R., Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004;32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salzberg S.L., Salzberg A.J., Kerlavage A.R., Tomb J.F. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- 7.Morton B.R. Strand asymmetry and codon usage bias in the chloroplast genome of Euglena gracilis. Proc. Natl Acad. Sci. USA. 1999;96:5123–5128. doi: 10.1073/pnas.96.9.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grigoriev A. Strand-specific compositional asymmetries in double-stranded DNA viruses. Virus Res. 1999;60:1–19. doi: 10.1016/s0168-1702(98)00139-7. [DOI] [PubMed] [Google Scholar]
- 9.Frank A.C., Lobry J.R. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics. 2000;16:560–561. doi: 10.1093/bioinformatics/16.6.560. [DOI] [PubMed] [Google Scholar]
- 10.Szybalski W., Kubinski H., Sheldrick P. Pyrimidine clusters on the transcribing strand of DNA and their possible role in the initiation of RNA synthesis. Cold Spring Harb. Symp. Quant. Biol. 1966;31:123–127. doi: 10.1101/sqb.1966.031.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Bell S.J., Forsdyke D.R. Accounting units in DNA. J. Theor. Biol. 1999;197:51–61. doi: 10.1006/jtbi.1998.0857. [DOI] [PubMed] [Google Scholar]
- 12.Lao P.J., Forsdyke D.R. Thermophilic bacteria strictly obey Szybalski's transcription direction rule and politely purine-load RNAs with both adenine and guanine. Genome Res. 2000;10:228–236. doi: 10.1101/gr.10.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Touchon M., Arneodo A., d'Aubenton-Carafa Y., Thermes C. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res. 2004;32:4969–4978. doi: 10.1093/nar/gkh823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green P., Ewing B., Miller W., Thomas P.J., Green E.D. NISC Comparative Sequencing. Transcription-associated mutational asymmetry in mammalian evolution. Program Nature Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 15.Louie E., Ott J., Majewski J. Nucleotide frequency variation across human genes. Genome Res. 2003;13:2594–2601. doi: 10.1101/gr.1317703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tillier E.R., Collins R.A. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol. 2000;50:249–257. doi: 10.1007/s002399910029. [DOI] [PubMed] [Google Scholar]
- 17.Lobry J.R., Sueoka N. Asymmetric directional mutation pressures in bacteria. Genome Biol. 2002;3:research 0058.1–0058.14. doi: 10.1186/gb-2002-3-10-research0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beletskii A., Bhagwat A.S. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem. 1998;5:549–551. [PubMed] [Google Scholar]
- 19.Francino M.P., Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol. 2001;6:1147–1150. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- 20.Fijalkowska I.J., Jonczyk P., Tkaczyk M.M., Bialoskorska M., Schaaper R.M. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl Acad. Sci. USA. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frederico L.A., Kunkel T.A., Shaw B.R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 22.McLean M.J., Wolfe K.H., Devine K.M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 23.Brewer B.J. When polymerases collide: Replication and the transcriptional organization of the Escherichia coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 24.Zeigler D.R., Dean D.H. Orientation of genes in the Bacillus subtilis chromosome. Genetics. 1990;125:703–708. doi: 10.1093/genetics/125.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInerney J.O. Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc. Natl Acad. Sci. USA. 1998;95:10698–10703. doi: 10.1073/pnas.95.18.10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 27.Liu B., Alberts B.M. Head-on collision between DNA replication apparatus and RNA transcription complex. Science. 1995;267:1131–1137. doi: 10.1126/science.7855590. [DOI] [PubMed] [Google Scholar]
- 28.Guy L., Roten C.A. Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene. 2004;340:45–52. doi: 10.1016/j.gene.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 29.Rocha E.P.C., Danchin A. Essentiality, not expressiveness drives gene-strand bias in bacteria. Nature Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 30.Lopez P., Philippe H. Composition strand asymmetries in prokaryotic genomes: mutational bias and biased gene orientation. C. R. Acad. Sci. III. 2001;324:201–208. doi: 10.1016/s0764-4469(00)01298-1. [DOI] [PubMed] [Google Scholar]
- 31.Lundgren M., Andersson A., Chen L., Nilsson P., Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl Acad. Sci. USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson N.P., Dionne I., Lundgren M., Marsh V.L., Bernander R., Bell S.D. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]