Abstract
The expression of the catalytic subunit (hTERT) represents the limiting factor for telomerase activity. Previously, we detected a transcriptional repressor effect of the proximal exonic region (first two exons) of the hTERT gene. To better understand the mechanism involved and to identify a potential repressor, we further characterized this region. The addition of the hTERT proximal exonic region downstream of the hTERT minimal promoter strongly reduced promoter transcriptional activity in all cells tested (tumor, normal and immortalized). This exonic region also significantly inhibited the transcriptional activity of the CMV and CDKN2A promoters, regardless of the cell type. Therefore, the repressor effect of hTERT exonic region is neither cell nor promoter-dependent. However, the distance between the promoter and the exonic region can modulate this repressor effect, suggesting that nucleosome positioning plays a role in transcriptional repression. We showed by electrophoretic mobility shift assay that CCCTC-binding factor (CTCF) binds to the proximal exonic region of hTERT. Chromatin immunoprecipitaion assays confirmed the binding of CTCF to this region. CTCF is bound to hTERT in cells in which hTERT is not expressed, but not in telomerase-positive ones. Moreover, the transcriptional downregulation of CTCF by RNA interference derepressed hTERT gene expression in normal telomerase-negative cells. Our results suggest that CTCF participates in key cellular mechanisms underlying immortality by regulating hTERT gene expression.
INTRODUCTION
Telomeres are specific sequences composed of tandem repeats of the TTAGGG sequence at the end of human chromosomes (1). Possible functions of this non-coding DNA include prevention of chromosome degradation, end-to-end fusions, rearrangements and chromosome loss (2). In normal cells, each division is associated with telomere shortening (3). Telomerase is a ribonucleoprotein complex in which the catalytic subunit, hTERT, uses a specific RNA, hTERC, as a template for the addition of telomeric repeat sequences to the ends of chromosomes (4–6). Telomerase activity is detectable during embryogenesis in a variety of fetal tissues (7), while in adult humans, telomerase activity is not detectable in most somatic cells (8). In contrast, highly proliferative cells, such as germ cells and stem cells, as well as 85–95% of cancers express telomerase (9).
The presence of the two major subunits, hTERT and hTERC, is sufficient to reconstitute telomerase activity in vitro (10), but additional telomerase-associated proteins have been identified in vivo (11,12). It has been shown that expression of hTERT is sufficient to restore telomerase activity in telomerase-negative cells (13–15). Expression of the hTERT gene is highly regulated and correlates with telomerase activity (16,17). The genomic organization of the hTERT gene and features of its promoter have been described by several groups and a region encompassing the 283 bp upstream of the translation site, designated as the core promoter, is essential for transcriptional activity (18–20). Specific binding sites for activators and repressors of transcription have been identified in the hTERT promoter sequence (21–35). Additional regulatory elements have been identified distant from the 5′ flanking region of the hTERT promoter (36). Moreover, studies of RNA processing revealed complex splicing patterns in different cell types (16) which suggest regulation of hTERT translation by alternative splicing (37–39).
In telomerase-positive cell lines, the transient transfection of the hTERT promoter yields a high level of transcriptional activity, similar to that induced by the SV40 early promoter (40,41). This is in stark contrast with the low endogenous hTERT mRNA levels detected in telomerase-positive cell lines which are as low as 0.2–6 copies/cell (36,42). In transient transfection assays the proximal exonic region (first two exons) of the hTERT gene was shown to contain repressor elements (41). The regulation of hTERT expression, therefore, appears to be rather complex.
Based upon the published reports we hypothesize that the proximal exonic region of the hTERT gene plays an important role in the regulation of hTERT transcription. Conceptually, this might be due to the distance between the core promoter and the transcriptional start site and might involve binding of repressors, as well as histone acetylation. We therefore set out to explore the effect of the distance between the core promoter and the transcription start site on hTERT transcription in telomerase-positive and -negative cells. In addition, we investigated the interaction of CCCTC-binding factor (CTCF), a ubiquitously expressed 11 zinc finger protein, with the first exon of the hTERT gene.
MATERIALS AND METHODS
Cell culture
The human tumor cell lines (HeLa, cervical adenocarcinoma; SW480, colorectal adenocarcinoma; NCCIT, teratocarcinoma; OVCAR-3, adenocarcinoma of the ovary; and U2-OS, osteosarcoma) and normal human fibroblasts (BJ) were obtained from ATCC. All cells were grown in the medium recommended by ATCC. The GM847, HLF and HLF/hTERT cells were kindly provided by Dr Joachim Lingner (ISREC, Epalinges, Switzerland). They were cultured in DMEM supplemented with 10% heat inactivated fetal bovine serum (FBS). The cell lines, HeLa, SW480, NCCIT and OVCAR-3, are telomerase-positive, whereas the U2-OS cell line is telomerase-negative. The normal human fibroblasts HLF and BJ are telomerase-negative. GM847 is an SV40 immortalized, but telomerase-negative cell line derived from normal fibroblasts. HLF/hTERT cells were obtained through stable transfection of HLF cells with hTERT cDNA construct, express hTERT and exhibit telomerase activity.
Plasmid construction
pTERT–297 contains the hTERT minimal promoter (41). The pTERT–297/ex1 vector contains the hTERT minimal promoter and 80 bp of the first exon. To construct this vector, an hTERT fragment was generated by PCR and cloned into the pGL3 basic vector (Promega, Madison, WI) opened previously with SacI and HindIII. The primers used for PCR amplification of the hTERT fragment also contained the SacI and the HindIII sites (in bold) and were FW-5′-GGCTGCGAGCTCCAGGCCGGGCTCCCAGTGGAT-3′ (beginning of the exon1) and REV-5′-GGCAAGCTTCGAACGTGGCCAGCGGCAGCACCTC-3′ (end of the exon1).
A 1804 bp fragment, containing the hygromycine resistance gene and the FRT site, was deleted by PvuII digestion from the pcDNA5/FRT vector (Invitrogen, Basel, Switzerland). The resulting 3266 bp vector was named pcDNA5. The 1983 bp firefly luciferase sequence was obtained through digestion of the pGL3 basic vector (Promega, Madison, WI) with NheI and BamHI and was subcloned into pcDNA5. This new construct contained the CMV promoter upstream of the firefly luciferase gene and was named pCMVluc. We further deleted the CMV promoter from the pCMVluc vector with BglII and NheI, and inserted two other promoters, hTERT or the CDKN2A promoter amplified by PCR. These constructs were named pTERTluc and pCDKN2Aluc, respectively.
After digestion of the pCMVluc, pTERTluc and pCDKN2Aluc vectors with NheI and HindIII, the PCR fragment containing the hTERT proximal exonic region (first exon and 885 bp of the second exon, without the first intron) and flanking with the NheI and the HindIII restriction sites, was subcloned into these vectors to create the pCMVtertluc, the pTERTtertluc and the pCDKN2Atertluc vectors, respectively.
Three different sequences (44, 100 and 200 bp), which did not contain motifs with transcriptional interest, were got from the pcDNA5 vector. These plasmid fragments were generated by PCR using primers containing the NheI restriction site: (in bold) 5′-GCTGGCGCTAGCTCATAGCTCACGCTGTA-3′ and 5′-GCCTGCTAGCCCTACACCGAACTGAGATAC-3′ generated the 44 bp insert; 5′-TAACGTGCTAGCGCGCCTTATCCGGTAACT-3′ and 5′-GTCGCTAGCCTGCTAATCCTGTTACCAGTGGC-3′ generated the 100 bp insert; 5′-TAACGTGCTAGCGCTTTCTCATAGCTCACGCT-3′ and 5′-GCTGCTAGCCTGCTAATCCTGTTACCAGTGGC-3′ generated the 200 bp insert. Then, these sequences were inserted into the pCMVtertluc digested with NheI, to create the pCMV44tertluc, pCMV100tertluc and pCMV200tertluc constructs, respectively. The same inserts were subcloned into the pTERTtertluc and the pCDKN2Atertluc vectors to create the plasmids pTERT44tertluc, pTERT100tertluc, pTERT200tertluc, pCDKN2A44tertluc, pCDKN2A100tertluc and pCDKN2A200tertluc.
All these constructs have been confirmed by sequencing on an ABI Prism 3100 sequencer (Applied Biosystems, Foster City, CA).
Transient transfection and luciferase assay
Cells were seeded at a concentration of 300 000/3.8 cm2 for HeLa, NCCIT, OVCAR-3, U2-OS and GM847, and 100 000 cells/3.8 cm2 for HLF, HLF/hTERT and BJ, and cultured overnight. Transient transfection of luciferase reporter plasmids were carried out using lipofectin plus reagent for HeLa, or LipofectAMINE for all other cell lines according to the manufacturer's protocols (Invitrogen, Basel, Switzerland). All experiments were performed at least in triplicate. The Renilla luciferase reporter vector (Promega, Madison, WI) was co-transfected as an internal control for transfection efficiency. Briefly, HeLa cells were exposed to a transfection mixture containing 0.6 µg of luciferase reporter plasmids and 0.6 µg of internal control vector mix with 0.75 µl of lipofectine for 3 h at 37°C. Then, 1 ml of growth media supplemented with 15% HI-FBS was added to the cells. The cells were harvested 48 h after transfection. The other cell lines were exposed to a transfection mixture containing 0.8 µg of luciferase reporter plasmids and 0.8 µg of internal control vector with 4 µl of lipofectAMINE for 5 h at 37°C. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). To compare the results, the mean values of relative luciferase activity obtained in the three experiments were used. The transcription levels detected with the different constructs were compared with the transcription level of the pGL3-control vector containing the firefly luciferase gene under the control of the SV40 early promoter and to the transcription level of the pGL3 basic vector, a promoterless and enhancerless luciferase vector.
Small interfering RNA (siRNA) transfection assay
Cells were seeded in 12-well plates at a concentration of 100 000/well for HeLa and 30 000 cells/well for BJ. The cells were then cultured overnight. Co-transfections of siRNA (50 nM) and plasmids (1 µg/well) were carried out in duplicate using the JetSI-ENDO Cationic Transfection reagent (4 µl/well) (Polyplus-transfection, Illkirch, France) according to the manufacturer's instructions. siRNA for CTCF 5′-(UUGGUUCGGCAUCGUCGUU)dTT-3′ was synthesized by Qiagen-Xeragon (Germantown, MD). siRNAs for silencing-control and for luciferase pGL3 were provided in the jetsilencing control kit Luciferase pGL3 (Polyplus-transfection, Illkirch, France). BJ cells, transfected only with siRNAs, were harvested 48 h post transfection. The hTERT gene expression was analyzed by RT–PCR and TRAP assay (43). HeLa cells were co-transfected with siRNAs and either pTERT/−297 or pTERT−297/ex1, and luciferase assays were performed 48 h later.
RNA extraction and hTERT RT–PCR analysis
RNAs were extracted from cells transfected with siRNA, using the TRIzol LS Reagent (Invitrogen, Basel, Switzerland). Platinum quantitative RT–PCR Thermoscript one-step system (Invitrogen) was used to amplify the hTERT mRNA using the primers LT5 5′-CGGAAGAGTGTCTGGAGCAA-3′ (exon3) and LT6 5′-GGATGAAGCGGAGTCTGGA-3′ (exon4), with co-amplification of β-actin with the primers FW 5′-AGGCCAACCGCGAGAAGATGA-3′ and REV 5′-GCCGTGGTGGTGAAGCTGTAG-3′. The hTERT splicing variants (α, β, α/β and FL) were determined by RT–PCR using the primers FW-SP 5′-GCCTGAGCTGTACTTTGTCAA-3′ and REV-SP 5′-CGCAAACAGCTTGTTCTCCATGTC-3′. The FL variant was obtained with the primers FW-FL 5′-CGCCTGAGCTGTACTTTGTCA-3′ and REV-FL 5′-CGGCTGGAGGTCTGTCAAG-3′. RT–PCR products were analyzed on 2% agarose gel. The FL and the β-spliced variant fragments were extracted from the gel, purified with the QIAquick gel extraction kit (Qiagen) and sequenced on an ABI Prism 3100 sequencer (Applied Biosystems, Foster City, CA).
Electrophoretic mobility shift assay (EMSA) and dimethyl sulfate (DMS)-methylation interference analysis
Different fragments of the hTERT promoter and proximal exonic region were synthesized by PCR with the following primers: F1-FW-5′-AGCCCCTCCCCTTCCTTTC-3′ and F1-RV-5′-CGGGTCCCCGCGCTGCACCA-3′; F2-FW-5′-GTCCGGCTGGGGTTGAG-3′ and F2-RV-5′-GCACGCTGGTGGTGAAGG-3′; F3-FW-5′-GCCCGAGTGCTGCAGAGG-3′ and F3-RV-5′-GGGAGCCACCAGCACAAAGA-3′; F4-FW-5′-CGTGGGGGCTGCTGCTG-3′ and F4-RV-5′-ACGCCGGCCTCCCTGAC-3′. EMSA was performed as described earlier (44). Briefly, PCR fragments were end-labeled using 32P-γ-ATP and T4 polynucleotide kinase (New England Biolabs). Protein–DNA complexes were allowed to form by incubation for 30 min at ambient temperature in phosphate-buffered saline (PBS) with 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% Nonidet P-40, 10% glycerol and poly(dI–dC). Full-length CTCF and the Zn-fingers DNA-binding domain of CTCF were translated in vitro using the TnT kit (Promega, WI). Protein–DNA complexes were resolved from unbound DNA probe in 5% native polyacrylamide gel in 0.5× TBE.
DMS-methylation interference analysis was performed with full-length CTCF and the F1 and F3 PCR fragments amplified by sense and antisense primers, F1-FW and F1-RW, and F3-FW and F3-RV, respectively. Fragments were subcloned to pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) and sequence verified. Then, fragments containing the region F1 or F3 were amplified by PCR using 32P 5′ end-labeled M13 forward or reverse primers in order to obtain labeling on either the sense or the antisense strand. Methylation of guanine residues was done by DMS, using conditions that modified only a single nucleotide per molecule fragment. After binding to high concentrations of CTCF, EMSA was performed. Free and bound probes were cut and extracted from the gel, cleaved at modified nucleotides with piperidine and then analyzed on a sequencing gel as described previously (45).
The 122 bp F1wt and 103 bp F3wt fragments, containing the CTCF-binding sites located in the first and second exon of hTERT, respectively, were synthesized by PCR. Mutant fragments F1mut and F3mut were prepared by mutagenesis using synthetic oligonucleotides of 122 and 103 bp, respectively, with subsequent PCR amplification with the same primers as for wild-type fragments. Mutations were confirmed by sequencing of the PCR fragments. Eleven and five bases were changed in the F1 and F3 fragments, respectively.
Chromatin immunoprecipitaion (ChIP) assay
To crosslink proteins to DNA, 12 ml of a 1% formaldehyde solution in culture medium was added to the cells cultured to 50% confluency, and the cells were incubated 15 min at room temperature. Then, 1.3 ml of a 1.25 M glycine solution was added to the medium to stop the reaction. After 5 min, the cells were centrifuged and resuspended in 10 ml of ice-cold PBS containing protease inhibitors. One million cells were pelleted, resuspended in 200 µl of SDS lysis buffer (1% SDS, 10 nM EDTA and 50 mM Tris–HCl, pH 8.1) and incubated on ice for 10 min. The lysate was sonicated to shear DNA. The chromatin solution was diluted in 1800 µl of the ChIP dilution buffer (0.01% SDS; 1.1% Triton X-100; 1.2 mM EDTA; 16.7 mM Tris–HCl, pH 8.1; 167 mM NaCl; and protease inhibitors) for further immunoprecipitation or stored at 4°C to be directly uncrosslinked and purified (DNA input fraction). Magnetic beads, 80 µl (Dynabeads M-280 Sheep anti-Rabbit IgG, Dynal biotech, Oslo, Norway) were washed three times with 1 ml of blocking solution (1× PBS; 5 mg/ml BSA; 3% of a 1 mg/ml sonicated herring sperm DNA solution; and protease inhibitors). Half of the beads were suspended in 250 µl of the blocking solution either with 2 µg of the rabbit polyclonal anti-CTCF antibody (Upstate biotechnology, Lake Placid, NY) or without antibody, and rocked for 4 h at 4°C. The beads were then washed three times with 1 ml of the blocking solution, and then, added to 1 ml of the diluted chromatin solution and incubated overnight at 4°C. They were then washed twice with 500 µl of each washing solutions: low salt solution (0.1% SDS; 10% Triton X-100; 2 mM EDTA; 20 mM Tris–HCl, pH 8.1; and 150 mM NaCl), high salt solution (0.1% SDS; 10% Triton X-100; 2 mM EDTA; 20 mM Tris–HCl, pH 8.1; and 500 mM NaCl), LiCl solution (0.25 M LiCl; 1% NP40; 1% deoxycholate; 1 mM EDTA; and 10 mM Tris–HCl, pH 8.1) and TE (10 mM Tris–HCl; and 1 mM EDTA, pH 8.0). The eluate was then resuspended in 50 µl of the elution buffer (1% SDS; and 0.1 M NaHCO3) and incubated 10 min at 65°C. To reverse protein–DNA crosslinks, eluates were incubated at 65°C for 4 h in 120 µl of a solution containing 1% of SDS and 0.3 M NaCl. Then 180 µl of ATL buffer (Qiagen, Basel, Switzerland) and 20 µl of proteinase K were added to samples, and incubation was performed overnight at 55°C. Immunoprecipitated DNA was recovered by extraction with the DNeasy tissue kit (Qiagen, Basel, Switzerland). Purified DNA was analyzed by PCR with specific primers for the amplification of either the first exon of the hTERT gene to generate a 299 bp fragment (ChIP-ex-FW 5′-CCCTGCTGCGCAGCCACTAC-3′ and ChIP-ex-RV 5′-TCGGGCCACCAGCTCCTTCA-3′) or the H19 gene as a control (ChIP-H19 FW 5′-CTCCTTCGGTCTCACCGCCTGGAT-3′ and ChIP-H19-RV 5′-CCTTAGACGGAGTCGGAGCTG-3′). PCR products were analyzed on 2% agarose gel.
Western blot analysis
Approximately 40 µg of total cell extracts prepared from transfected cells in 5× Passive lysis buffer (Promega) were boiled 5 min in loading buffer (125 mmol/l Tris–HCl, pH 6.8, 40 g/l SDS, 200 g/l glycerol and 0.05 g/l bromphenol blue) for 5 min and then loaded onto a 8% SDS–PAGE. After electrophoresis, proteins were transferred onto PVDF membrane. Immunodetection was performed using a rabbit polyclonal anti-CTCF antibody (Upstate biotechnology, Lake Placid, NY). Protein bands were visualized using ECL-Plus kit (Amersham Pharmacia Biotech, Switzerland) according to the manufacturer's instructions. Mouse antibody against β-actin (AC-15, Sigma-Aldrich, Switzerland) in western blot was used as a control for protein concentration.
RESULTS
The inhibitory effect of the proximal exonic region of the hTERT gene is cell type-independent
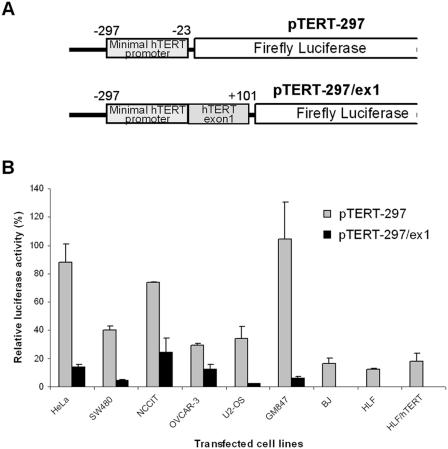
In earlier studies on HeLa cells we found the proximal exonic region to repress hTERT expression (41). We further studied the effect of this regulatory region in telomerase-positive, -negative and immortalized cells. The intronic region was omitted, to avoid transcript variation resulting from alternative splicing (41). As shown in Figure 1, the hTERT minimal promoter exhibited a high level of transcriptional activity in 4 telomerase-positive tumor cell lines, in U2-OS and in GM847 cells (30–120%). In contrast, the promoter activity was significantly lower in the normal telomerase-negative and in the HLF/hTERT cells (12–18%). A markedly decreased activity (7-fold in the HeLa and SW480, 3-fold in OVCAR-3 and NCCIT, 16-fold in U2-OS and GM847 cells) was observed when the first exon of the hTERT gene was included in the reporter construct (Figure 1). In telomerase-negative and HLF/hTERT immortalized cells, transcriptional activity was no longer detected. These results demonstrate that the inhibitory effect of the hTERT proximal exonic region is not cell type-dependent.
Figure 1.
Transcriptional activity of the hTERT promoter in different cell lines. (A) Schematic representation of the luciferase reporter plasmids pTERT-297 and pTERT-297/ex1. (B) These two reporters were transfected into four telomerase-positive cancer cell lines (HeLa, SW480, OVCAR-3 and NCCIT), one telomerase-negative sarcoma line (U2-OS), and two normal telomerase-negative (BJ and HLF), and two immortalized (GM847 immortalized in vitro with SV40 and HLF/hTERT that exogenously expressed hTERT) fibroblast cell lines. For all experiments, 100% of the luciferase activity is represented by the pGL3-control vector activity containing the SV40 enhancer/promoter.
The proximal exonic region of the hTERT gene has a repressor effect on different promoters
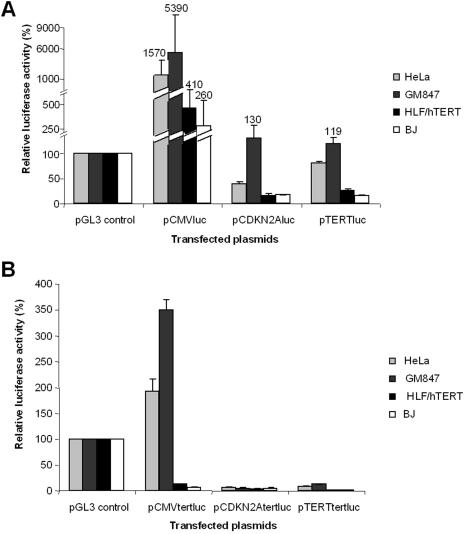
To determine if the repressor effect of the proximal exonic region is promoter specific, constructs containing this region but directed either by the CMV or CDKN2A promoters were transiently transfected into the following cell lines: HeLa, GM847, HLF/hTERT and BJ cells. In HeLa and GM847 cells, the CMV promoter had a 15- and 53-fold higher activity than the reference SV40 promoter, whereas the transcriptional activity of hTERT and CDKN2A promoters was significantly lower (Figure 2A). Relative to the SV40 promoter, the activity of the CDKN2A promoter was higher in GM847 cells than in HeLa cells, with levels increased 1.3- and 0.4-fold respectively. In HLF/hTERT and BJ cells, the activity of the CMV promoter was 4.1- and 2.6-fold higher than the SV40 promoter (Figure 2A). These results show that each promoter is more active in HeLa and GM847 than in the two other cell lines. We also studied the effect of the proximal exonic region of hTERT (first exon and 885 bp of the second exon) on the transcriptional activity of these promoters (Figure 2B). In HeLa and GM847 cells, the exonic region reduced CMV promoter activity by 8.1- and 15.4-fold, respectively. In the same cells, CDKN2A promoter activity was reduced 5- and 23-fold and hTERT promoter activity was reduced 10- and 9.4-fold. In HLF/hTERT and BJ, the activity of the CDKN2A and hTERT promoters was barely detectable, while the CMV promoter was 33- and 22-fold decreased (Figure 2B). Therefore, the strong repressor effect of the proximal exonic region of the hTERT gene is both promoter- and cell type-independent.
Figure 2.
Transcriptional activity of different promoters in HeLa, GM847, HLF/hTERT and BJ cell lines. (A) Activities of luciferase reporter plasmids containing the CMV, CDKN2A and hTERT promoter in these cell lines. (B) Activities of luciferase reporter plasmids containing the hTERT exon 1 and 885 bp of the exon 2, without the first intron, driven by the CMV, CDKN2A and hTERT promoter in these cell lines. For all experiments, 100% of the luciferase activity is represented by the pGL3-control vector activity containing the SV40 enhancer/promoter.
The level of inhibition by the exonic hTERT region is dependent on the distance between the promoter and the exonic region
In order to further analyze the repressive effect of the proximal exonic region of the hTERT gene, short plasmid sequences (44, 100 and 200 bp) were inserted between the promoter and the exonic region. Relative to the constructions without insert, a 2.8-fold decrease in promoter activity was observed after insertion of the 44 bp fragment, a 1.6-fold reduction with the introduction of the 100 bp fragment, while no decrease or even a slight increase in expression was detected when the 200 bp fragment was inserted (Table 1). The same pattern was observed for HeLa and GM847 cells, regardless of which promoter was used. In HLF/hTERT and BJ cells, similar observations were made for the CMV promoter. For the CDKN2A and hTERT promoters, transcriptional activity was undetectable. We conclude that increasing the distance between the promoter and the exonic region, by either 44 or 100 bp, leads to significantly stronger inhibition of transcriptional activity by the hTERT proximal exonic region. A return to a basal level of transcriptional activity was observed when the 200 bp sequence was inserted, which could suggest a link with the nucleosome positioning.
Table 1.
Relative luciferase activity after insertion of a plasmid sequence between a promoter and the hTERT proximal exonic region
| Transfected plasmid | Cell lines | |||||||
|---|---|---|---|---|---|---|---|---|
| HeLa | GM847 | HLF/hTERT | BJ | |||||
| A | B | A | B | A | B | A | B | |
| PTERT tertluc | 8 | 100 | 12.6 | 100 | NS | — | NS | — |
| PTERT 44 tertluc | 2.5 | 31 | 9.5 | 75 | NS | — | NS | — |
| PTERT 100 tertluc | 4.7 | 59 | 6.6 | 52 | NS | — | NS | — |
| PTERT 200 luc | 10.9 | 136 | 7.6 | 60 | NS | — | NS | — |
| pCDKN2A tertluc | 6.7 | 100 | 5.6 | 100 | NS | — | NS | — |
| pCDKN2A 44 tertluc | 2.13 | 30 | 2.2 | 39 | NS | — | NS | — |
| pCDKN2A 100 tertluc | 4.2 | 60 | 3.1 | 55 | NS | — | NS | - |
| pCDKN2A 200 tertluc | 6.9 | 100 | 2.3 | 40 | NS | — | NS | — |
| pCMV tertluc | 193 | 100 | 350 | 100 | 12.5 | 100 | 7.1 | 100 |
| pCMV 44 tertluc | 75 | 40 | 136 | 35 | 3.1 | 25 | 3.55 | 50 |
| pCMV 100 tertluc | 150 | 80 | 277 | 71 | 5.4 | 43 | 6.67 | 94 |
| pCMV 200 tertluc | 250 | 134 | 600 | 150 | 13 | 104 | 7.17 | 101 |
A, % of luciferase activity relative to the SV40 early promoter (100%). Representative experiment of at least three independent transfections.
B, % of luciferase activity relative to the plasmid without insertion (100%).
NS, non-significant value, at the background level.
CTCF interacts in vitro and in vivo with the hTERT proximal exonic region
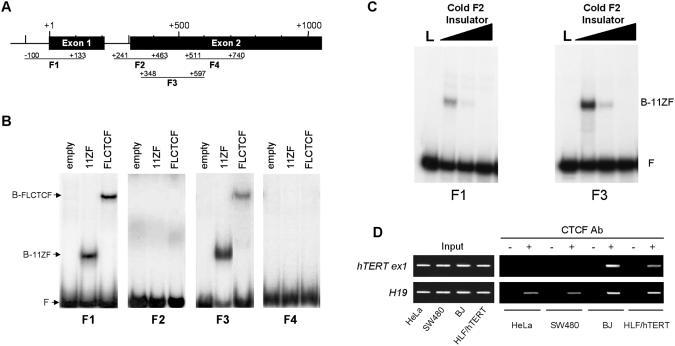
CTCF, a ubiquitously expressed 11 zinc finger nuclear phosphoprotein, binds to GC-rich sequences and inhibits transcription when located downstream of a transcriptional start site (46). Since the exonic region of the hTERT gene is GC-rich and its repressor effect is not cell type-specific, we considered CTCF a plausible candidate for involvement in the inhibition of hTERT expression. (47). In vitro binding of fragments of the proximal exonic region to full-length CTCF protein was tested by EMSA (Figure 3B). The −100 to +133 bp (fragment F1, located in the exon 1) and +348 to +597 bp (fragment F3, located in the exon 2) sequences of the hTERT gene showed retarded migration (Figures 3A and 3B). Mobility shifts were not observed with two other regions tested. These results demonstrate that CTCF binds in vitro to the first two exons of the hTERT gene. CTCF-binding specificity was further confirmed by competition assay with F2 chicken globin insulator (48). Both F1 and F3 fragments were successfully competed by a strong CTCF-binding site and 100-fold excess of competitor completely abrogate CTCF binding to hTERT (Figure 3C).
Figure 3.
In vitro and in vivo interactions of CTCF with hTERT gene. (A) Schematic localization in the exons 1 and 2 of the hTERT gen of the PCR fragments used in EMSA (the ATG is considered as +1). (B) EMSA was carried out with either control lysate (empty), lysate containing the 11 Zn-fingers of CTCF DNA-binding domain (11ZF), or lysate containing the in vitro translated full-length CTCF (FLCTCF). The positions of the bound CTCF–DNA complexes, containing either the 11ZF domain or the FLCTCF, are indicated on the left by B-11ZF and B-FLCTCF, respectively. Free DNA probe (F) is also indicated. (C) Competition assay of CTCF 11ZF using radioactive probes of F1 and F3 hTERT fragments and excess of cold F2 chicken globin insulator. L is in vitro translated luciferase and was used as a negative control. Lanes covered with black triangle on top are 0-, 10- and 100-fold molar excess of competitor, respectively. Bound and free bands are indicated as in (B). (D) ChIP assay using anti-CTCF antibody on two telomerase-positive cell lines (HeLa and SW480), one immortalized cell line (HLF/hTERT) and one telomerase-negative cell line (BJ). PCR amplification of the test fragments (hTERT exon1 and H19) using as template DNA input fraction and DNA recovered from immunoprecipitated fractions bound (plus) versus unbound (minus) by the anti-CTCF antibody.
To determine if CTCF binds in vivo to this region, we performed ChIP assays. Two telomerase-positive cell lines (HeLa and SW480), one immortalized cell line (HLF/hTERT) and one telomerase-negative cell line (BJ) were tested. The experiments were carried out with and without CTCF-specific antibody for the immunoprecipitation. Then, PCRs were performed at different number of cycles to amplify CTCF-hTERT exon 1 site and H19 ICR4 site, which was used as a positive control (44). PCRs were performed at conditions in which the samples from controls without antibody always showed negligible levels of background amplification. Amplification of H19 ICR4 fragment can be seen in all DNA fractions obtained with CTCF antibodies but not in negative controls suggesting the ChIP assay was very specific. In contrast to the H19 ICR4, the hTERT first exon fragment was found only in immunoprecipitated DNA from BJ and HLF/hTERT cells (Figure 3D). These results indicate that CTCF binds in vivo to the first exon of hTERT in BJ normal cells, as well as in HLF/hTERT that do not express endogenous hTERT, but not in telomerase-positive ones.
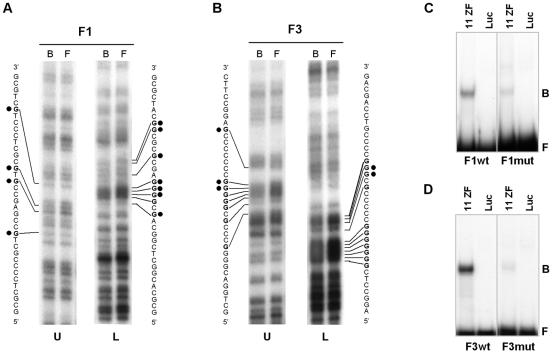
A DMS-methylation interference assay was used to determine potential contact guanine nucleotides recognized by CTCF in the first two exons of the hTERT gene (Figure 4A and B). CTCF-contacting guanines were located within the DNA sequence from positions +4 to +39 and +422 to +440 downstream from the ATG translational start site. These CTCF recognition sequences are GC-rich (83% for exon 1 and 100% for exon 2) and both contain CpG sites. To confirm that contact guanines were correctly identified, we performed site-directed mutagenesis within the F1 and F3 regions. Contact guanines, marked by black dots on Figure 4A and B, were substituted by adenines. EMSA comparison of wild-type (F1wt and F3wt) and mutant (F1mut and F3mut) fragments showed at least 90% reduction of the CTCF binding upon mutation of part of the CTCF-contact guanines in F1 and F3 (Figure 4C and D, respectively). The F2 fragment also contained the binding sequence +422 to +440, but did not exhibit retarded migration by EMSA (Figure 3B). This might be due to location of the CTCF-binding site at the extremity of the F2 fragment.
Figure 4.
Identification of contact guanines specifically recognized by CTCF in the proximal exonic region of the hTERT gene. (A) DMS-methylation interference analysis was carried out with the hTERT F1 fragment. Top (U) and bottom (L) strands are shown respectively. Bound (B) and free (F) probes are indicated on top of each lane. Contact guanines are indicated in boldface. Guanines substituted for adenines in site-directed mutagenesis are indicated by black dots (B) DMS-methylation interference analysis was carried out with the hTERT F3 fragment. All labels are as in (A). Guanines substituted for adenines in site-directed mutagenesis are indicated by black dots. Eleven and five bases were changed in the F1 and F3 fragments, respectively. (C) EMSA of the wt (F1wt) and mutant (F1mut) fragments F1, located in the first exon of the hTERT gene. (D) EMSA of the wt (F3wt) and mutant (F3mut) fragments F3, located in the second exon of the hTERT gene. Luciferase control (Luc) and DNA-binding domain of CTCF (11ZF) were used in the assay. Free (F) and bound (B) probes are indicated.
Binding of CTCF represses hTERT transcription
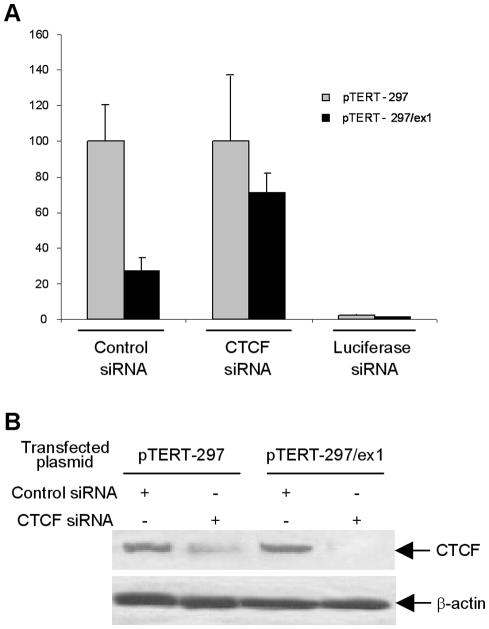
To determine if CTCF binding to the hTERT exon 1 is responsible for the hTERT transcriptional inhibition, we examined the effect of CTCF depletion with siRNA against CTCF. siRNA activity was examined by co-transfection of HeLa cells with luciferase expressing constructs pTERT–297 or pTERT–297/ex1. Western blot analysis of cell lysates showed a dramatic reduction of CTCF expression upon co-transfection of the siRNA, but not cells transfected with a control siRNA (Figure 5B). Consistent with our proposed role for CTCF as a negative regulator of hTERT expression, co-transfection of the pTERT–297/ex1 construct (promoter and first exon of hTERT) with CTCF siRNA resulted in a 3-fold increase in exogenous hTERT promoter activity, when compared with cells co-transfected with a control siRNA (Figure 5A).
Figure 5.
Depletion of CTCF by siRNA results in the relief of inhibition of hTERT promoter by the hTERT exonic region. (A) Luciferase reporter plasmids containing the hTERT minimal promoter, with or without the proximal exonic region, are co-transfected into HeLa cells with either unrelated siRNA (control siRNA), or CTCF siRNA or luciferase siRNA. In each experiment, the activity is calculated considering the activity of hTERT promoter as 100%. pTERT-297, minimal hTERT promoter; pTERT-297/ex1, minimal promoter and first exon of the hTERT gene. (B) Western blot showing the decrease of CTCF protein in cells treated with the specific siRNA for CTCF. β-Actin was used as internal control to normalize the total protein amount.
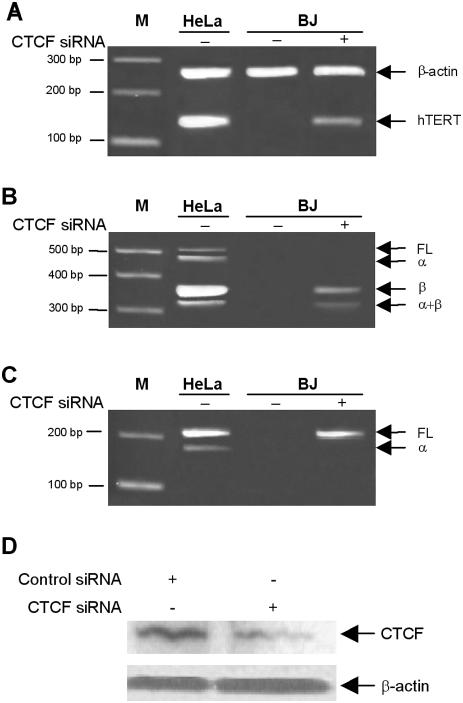
To provide additional evidence for the role of CTCF as a repressor of hTERT gene expression, we transfected CTCF siRNA in BJ, a telomerase-negative human non-immortalized fibroblast. A strong decrease of CTCF level was observed by western blotting after 48 h transfection (Figure 6D). The hTERT messenger was detected in the extract of BJ cells treated with siRNA for CTCF (Figure 6), and this with three different sets of primers, which amplify full-length (FL) hTERT mRNA and the α deletion, or the four different forms in the same time (FL, α-, β- and α/β-deletions). Each amplification was sequenced, confirming the presence of FL and β-deletion forms of the hTERT mRNA. Nevertheless, no positive TRAP assay was obtained in these conditions (data not shown).
Figure 6.
Depletion of CTCF by siRNA results in transcriptional activation of hTERT in telomerase-negative cells. (A) RT–PCR of hTERT from extracted RNA of HeLa (telomerase-postive) and BJ (telomerase-negative) cells treated with either control siRNA (minus) or CTCF siRNA (plus). RT–PCR was performed using primer pair LT5 and LT6 to amplify a 145 bp segment common to all hTERT transcripts. (B) Expression of spliced variants of hTERT mRNA. The positions of the full-length (457 bp), α-spliced (421 bp), β-spliced (275 bp) and α+β-spliced (239 bp) variants are indicated on the right. (C) Amplification of FL and α-spliced variant only. (D) Western blot showing the decrease of CTCF protein in cells treated with the specific siRNA for CTCF. β-Actin was used as internal control to normalize the total protein amount.
DISCUSSION
In a previous study, we detected a repressor effect of the proximal exonic region (first two exons) of the hTERT gene on its transcription (41). To better understand the mechanism involved and to identify a potential repressor, we further characterized the effects of this region of the hTERT gene.
Reporter gene constructs, composed of the hTERT minimal core promoter with or without the exonic region, were transiently transfected in normal telomerase-negative cells (BJ and HLF), telomerase-negative sarcoma cells (U2-OS), telomerase-positive cancer cells (HeLa, SW480; NCCIT and OVCAR-3) and immortalized cell lines (telomerase-negative GM847 and telomerase-positive HLF/hTERT). The transcriptional activity of the hTERT core promoter was significantly higher in telomerase-postive cancer cells than in normal cells, very high in telomerase-negative GM847, but weak in telomerase-positive HLF/hTERT. Transcriptional activity of the hTERT minimal promoter was significantly up-regulated in GM847 cells, obtained through transfection with the SV40 large T-antigen, and immortalized through the alternative lengthening telomere (49), in contrast to HLF/hTERT cell line, which was obtained by infection of primary fibroblast HLF cells with a MSCV-hTERT retrovirus. HLF/hTERT cells express high levels of exogenous hTERT, but the extension of life span does not change the phenotypic properties of the cells because telomerase, unlike oncogenes, does not cause growth deregulation (50). This could explain why, in transient transfection experiments, we did not observe transcriptional activation of the hTERT minimal core promoter in HLF/hTERT cells, as occurred in tumor and transformed cells. Thus, the presence of hTERT protein or telomerase complex does not itself activate hTERT gene transcription.
The addition of the proximal exonic region of hTERT downstream of the hTERT minimal promoter strongly reduced promoter transcriptional activity in all cell lines tested. Furthermore, this exonic region significantly inhibited the transcriptional activity of the CMV and CDKN2A promoters, regardless of the cell type. Therefore, the repressor effect of the hTERT exonic region is not cell- or promoter-dependent. The decrease in the transcriptional promoter activity in presence of the hTERT exonic region was higher in HLF/hTERT or BJ cells than in HeLa or GM847 cells.
Sequence insertion was used to show how structural and regulatory phenomena could involve nucleosome positioning (51,52). Insertion of DNA sequences of 44 or 100 bp between a promoter and the hTERT exonic region resulted in reduced transcriptional expression, whereas a relatively constant level of transcription was observed with spacer DNAs of 200 bp. Thus, proper arrangement of nucleosome positioning within the hTERT exonic region might inoculate the activity, and perhaps accessibility, of the presumed repressor(s). Another explanation for this observation could be that the repressor(s) forms a bulky complex, which physically interferes with the RNA polymerase II complex from the hTERT promoter. As a function of the spatial configuration of these two large complexes, this disturbance might be more or less pronounced.
CTCF was considered to be a good candidate for hTERT transcriptional inhibition: CTCF is expressed in almost all cell types, binds GC-rich sequences, represses transcriptional activity when it is localized downstream of a transcriptional start site and its activity depends on the chromatin structure (44–46,53). We hypothesized that CTCF might be the repressor involved. EMSA showed that CTCF binds to two distinct sequences in exons 1 and 2 of hTERT. The methylation interference experiment permitted the identification of CTCF-binding sequences within the hTERT first exon at +4 to +39 positions and within the hTERT second exon at +422 to +440 positions (relative to the ATG translational start codon). Mutations within these core sequences eliminated binding of CTCF, confirming the identity of the two CTCF-binding sites. Moreover, as one can see in Figure 4A and B, single contact guanine modification had a relatively low impact on CTCF binding, suggesting some evolutionary pressure to keep the CTCF-binding sites present regardless of stochastic point mutations. In the case of the site F1, within the first exon of hTERT, we had to substitute 11 contact guanines in order to eliminate CTCF binding to the site, which is so far unprecedented. Interestingly, and consistent with the description of some other CTCF-binding sites (54), most of the CTCF-contact nucleotides were located on a single DNA strand.
Many contact guanines are part of CpG sites suggesting that CTCF binding might be regulated by DNA methylation. ChIP assays confirmed that CTCF binds to the first exon of hTERT in BJ and HLF/hTERT cells in which the endogenous hTERT gene is not expressed, but not in HeLa and SW480 cells which express hTERT. Moreover, in HeLa cells, the reduction of CTCF expression by RNA interference stimulated the transcriptional activity of the reporter that contained the first exon of hTERT. In BJ cells, siRNA against CTCF induced the transcription of hTERT mRNA. These results show that CTCF is involved in the hTERT transcriptional repression. Human hTERT is the first gene implicated in immortality that might be controlled by CTCF.
In addition to CTCF, several negative regulators of hTERT transcription have been identified, including p53, WT-1, Mad1, myeloid-specific zinc finger protein 2 (MZF-2), and others (55). Overexpression of wild-type p53 in MCF-7 cells can downregulate hTERT mRNA expression, but inhibition of p53 activity does not reactivate hTERT expression (56). WT-1 can inhibit hTERT transcription (30) and Mad1 can regulate the ability of c-Myc to activate hTERT (57). Although all these factors can negatively regulate hTERT, none of them appear to be responsible for the downregulation of hTERT in the majority of telomerase-negative cells. To our knowledge, CTCF is the only known factor that can negatively regulate hTERT transcription independently of the cell type. Indeed, CTCF is one such factor that is highly conserved, ubiquitously expressed, and possesses versatile regulatory functions (57). CTCF is a DNA-binding protein that uses various combinations of its 11 zinc fingers to recognize a variety of unrelated DNA sequences (45). It plays a major role in transcriptional activation of the APPB promoter (58), silencing of c-myc (45), insulation of β-globin gene (48,59) and imprinting control of the H19 region (44). Concerning the hTERT gene, CTCF binds to a region located within the first two exons and just downstream the promoter. Silencing of hTERT could be the consequence of either inhibition of the transcription–initiation complex or blockage of transcription elongation.
Our results suggest a model in which CTCF participates in key cellular mechanisms underlying immortality by selectively regulating hTERT gene expression. We postulate that, in telomerase-negative normal cells, CTCF binds to the hTERT proximal exonic region and thus blocks transcription. The fact that the hTERT first exon sequences are hypermethylated in telomerase-positive cells (60) could explain the failure of CTCF to bind to these sequences.
Acknowledgments
We wish to thank Dr Phil Shaw for critical reading of the manuscript. This work was funded by a grant from the Swiss National Science Foundation (grant number: 3100AO-101732). Funding to pay the Open Access publication charges for this article was provided by the University of Lausanne.
Conflict of interest statement. None declared.
REFERENCES
- 1.Greider C.W. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 2.Greider C.W. Chromosome first aid. Cell. 1991;67:645–647. doi: 10.1016/0092-8674(91)90058-7. [DOI] [PubMed] [Google Scholar]
- 3.Allsopp R.C., Vaziri H., Patterson C., Goldstein S., Younglai E.V., Futcher A.B., Greider C.W., Harley C.B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng J., Funk W.D., Wang S.S., Weinrich S.L., Avilion A.A., Chiu C.P., Adams R.R., Chang E., Allsopp R.C., Yu J. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 5.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 6.Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 7.Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W., Shay J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Yasumoto S., Kunimura C., Kikuchi K., Tahara H., Ohji H., Yamamoto H., Ide T., Utakoji T. Telomerase activity in normal human epithelial cells. Oncogene. 1996;13:433–439. [PubMed] [Google Scholar]
- 9.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 10.Beattie T.L., Zhou W., Robinson M.O., Harrington L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 11.Collins K., Kobayashi R., Greider C.W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama J., Saito M., Nakamura H., Matsuura A., Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 13.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 14.Counter C.M., Meyerson M., Eaton E.N., Ellisen L.W., Caddle S.D., Haber D.A., Weinberg R.A. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 15.Weinrich S.L., Pruzan R., Ma L., Ouellette M., Tesmer V.M., Holt S.E., Bodnar A.G., Lichtsteiner S., Kim N.W., Trager J.B., et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 16.Kilian A., Bowtell D.D., Abud H.E., Hime G.R., Venter D.J., Keese P.K., Duncan E.L., Reddel R.R., Jefferson R.A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 17.Meyerson M., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 18.Horikawa I., Cable P.L., Afshari C., Barrett J.C. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59:826–830. [PubMed] [Google Scholar]
- 19.Takakura M., Kyo S., Kanaya T., Hirano H., Takeda J., Yutsudo M., Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 20.Wick M., Zubov D., Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto K., Kyo S., Takakura M., Kanaya T., Kitagawa Y., Itoh H., Takahashi M., Inoue M. Identification and characterization of negative regulatory elements of the human telomerase catalytic subunit (hTERT) gene promoter: possible role of MZF-2 in transcriptional repression of hTERT. Nucleic Acids Res. 2000;28:2557–2562. doi: 10.1093/nar/28.13.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goueli B.S., Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22:8042–8047. doi: 10.1038/sj.onc.1206847. [DOI] [PubMed] [Google Scholar]
- 23.Goueli B.S., Janknecht R. Upregulation of the catalytic telomerase subunit by the transcription factor ER81 and oncogenic HER2/Neu, Ras, or Raf. Mol. Cell. Biol. 2004;24:25–35. doi: 10.1128/MCB.24.1.25-35.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunes C., Lichtsteiner S., Vasserot A.P., Englert C. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 2000;60:2116–2121. [PubMed] [Google Scholar]
- 25.Kyo S., Takakura M., Taira T., Kanaya T., Itoh H., Yutsudo M., Ariga H., Inoue M. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Cao Y., Berndt M.C., Funder J.W., Liu J.P. Molecular interactions between telomerase and the tumor suppressor protein p53 in vitro. Oncogene. 1999;18:6785–6794. doi: 10.1038/sj.onc.1203061. [DOI] [PubMed] [Google Scholar]
- 27.Lv J., Liu H., Wang Q., Tang Z., Hou L., Zhang B. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem. Biophys. Res. Commun. 2003;311:506–513. doi: 10.1016/j.bbrc.2003.09.235. [DOI] [PubMed] [Google Scholar]
- 28.Misiti S., Nanni S., Fontemaggi G., Cong Y.S., Wen J., Hirte H.W., Piaggio G., Sacchi A., Pontecorvi A., Bacchetti S., et al. Induction of hTERT expression and telomerase activity by estrogens in human ovary epithelium cells. Mol. Cell. Biol. 2000;20:3764–3771. doi: 10.1128/mcb.20.11.3764-3771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishi H., Nakada T., Kyo S., Inoue M., Shay J.W., Isaka K. Hypoxia-inducible factor 1 mediates upregulation of telomerase (hTERT) Mol. Cell. Biol. 2004;24:6076–6083. doi: 10.1128/MCB.24.13.6076-6083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh S., Song Y., Yim J., Kim T.K. The Wilms' tumor 1 tumor suppressor gene represses transcription of the human telomerase reverse transcriptase gene. J. Biol. Chem. 1999;274:37473–37478. doi: 10.1074/jbc.274.52.37473. [DOI] [PubMed] [Google Scholar]
- 31.Wu K.J., Grandori C., Amacker M., Simon-Vermot N., Polack A., Lingner J., Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nature Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 32.Xu D., Wang Q., Gruber A., Bjorkholm M., Chen Z., Zaid A., Selivanova G., Peterson C., Wiman K.G., Pisa P. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–5133. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 33.Xu D., Popov N., Hou M., Wang Q., Bjorkholm M., Gruber A., Menkel A.R., Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl Acad. Sci. USA. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yago M., Ohki R., Hatakeyama S., Fujita T., Ishikawa F. Variant forms of upstream stimulatory factors (USFs) control the promoter activity of hTERT, the human gene encoding the catalytic subunit of telomerase. FEBS Lett. 2002;520:40–46. doi: 10.1016/s0014-5793(02)02757-6. [DOI] [PubMed] [Google Scholar]
- 35.Yatabe N., Kyo S., Maida Y., Nishi H., Nakamura M., Kanaya T., Tanaka M., Isaka K., Ogawa S., Inoue M. HIF-1-mediated activation of telomerase in cervical cancer cells. Oncogene. 2004;23:3708–3715. doi: 10.1038/sj.onc.1207460. [DOI] [PubMed] [Google Scholar]
- 36.Ducrest A.L., Amacker M., Mathieu Y.D., Cuthbert A.P., Trott D.A., Newbold R.F., Nabholz M., Lingner J. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61:7594–7602. [PubMed] [Google Scholar]
- 37.Liu W.J., Zhang Y.W., Zhang Z.X., Ding J. Alternative splicing of human telomerase reverse transcriptase may not be involved in telomerase regulation during all-trans-retinoic acid-induced HL-60 cell differentiation. J. Pharmacol. Sci. 2004;96:106–114. doi: 10.1254/jphs.fp0030600. [DOI] [PubMed] [Google Scholar]
- 38.Ulaner G.A., Hu J.F., Vu T.H., Giudice L.C., Hoffman A.R. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 39.Yi X., White D.M., Aisner D.L., Baur J.A., Wright W.E., Shay J.W. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cong Y.S., Wen J., Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 41.Renaud S., Bosman F.T., Benhattar J. Implication of the exon region in the regulation of the human telomerase reverse transcriptase gene promoter. Biochem. Biophys. Res. Commun. 2003;300:47–54. doi: 10.1016/s0006-291x(02)02775-4. [DOI] [PubMed] [Google Scholar]
- 42.Yi X., Shay J.W., Wright W.E. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan P., Bosman F.T., Benhattar J. Tissue quality is an important determinant of telomerase activity as measured by TRAP assay. Biotechniques. 1998;25:660–662. doi: 10.2144/98254dt05. [DOI] [PubMed] [Google Scholar]
- 44.Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 45.Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lobanenkov V.V., Nicolas R.H., Adler V.V., Paterson H., Klenova E.M., Polotskaja A.V., Goodwin G.H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 48.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 49.Hwang S.P., Kucherlapati R.S. Events preceding stable integration of SV40 genomes in a human cell line. Somatic. Cell Genet. 1983;9:457–468. doi: 10.1007/BF01543046. [DOI] [PubMed] [Google Scholar]
- 50.Harley C.B. Telomerase is not an oncogene. Oncogene. 2002;21:494–502. doi: 10.1038/sj.onc.1205076. [DOI] [PubMed] [Google Scholar]
- 51.Nishikawa J., Amano M., Fukue Y., Tanaka S., Kishi H., Hirota Y., Yoda K., Ohyama T. Left-handedly curved DNA regulates accessibility to cis-DNA elements in chromatin. Nucleic Acids Res. 2003;31:6651–6662. doi: 10.1093/nar/gkg854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pina B., Barettino D., Truss M., Beato M. Structural features of a regulatory nucleosome. J. Mol. Biol. 1990;216:975–990. doi: 10.1016/S0022-2836(99)80015-1. [DOI] [PubMed] [Google Scholar]
- 53.Lutz M., Burke L.J., Barreto G., Goeman F., Greb H., Arnold R., Schultheiss H., Brehm A., Kouzarides T., Lobanenkov V., et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nature Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 55.Horikawa I., Barrett J.C. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24:1167–1176. doi: 10.1093/carcin/bgg085. [DOI] [PubMed] [Google Scholar]
- 56.Lin S.Y., Elledge S.J. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 57.Ohlsson R., Renkawitz R., Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 58.Vostrov A.A., Quitschke W.W. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 59.Farrell C.M., West A.G., Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guilleret I., Benhattar J. Unusual distribution of DNA methylation within the hTERT CpG island in tissues and cell lines. Biochem. Biophys. Res. Commun. 2004;325:1037–1043. doi: 10.1016/j.bbrc.2004.10.137. [DOI] [PubMed] [Google Scholar]