Abstract
In Trypanosoma brucei, DNA recombination is crucial in antigenic variation, a strategy for evading the mammalian host immune system found in a wide variety of pathogens. T.brucei has the capacity to encode >1000 antigenically distinct variant surface glycoproteins (VSGs). By ensuring that only one VSG is expressed on the cell surface at one time, and by periodically switching the VSG gene that is expressed, T.brucei can evade immune killing for prolonged periods. Much of VSG switching appears to rely on a widely conserved DNA repair pathway called homologous recombination, driven by RAD51. Here, we demonstrate that T.brucei encodes a further five RAD51-related proteins, more than has been identified in other single-celled eukaryotes to date. We have investigated the roles of two of the RAD51-related proteins in T.brucei, and show that they contribute to DNA repair, homologous recombination and RAD51 function in the cell. Surprisingly, however, only one of the two proteins contributes to VSG switching, suggesting that the family of diverged RAD51 proteins present in T.brucei have assumed specialized functions in homologous recombination, analogous to related proteins in metazoan eukaryotes.
INTRODUCTION
DNA recombination is central to the survival and functioning of all organisms. Recombination is one of a number of DNA repair pathways that reverse genotoxic damage (1), it is intimately linked to DNA replication (2,3), it can provide a means for telomere maintenance (4) and is crucial in genetic exchange (5). Some organisms harness recombination reactions for specific genetic functions; prominent examples include vertebrate immunoglobulin and T-cell receptor gene re-arrangements (6), and mating type gene switching in fungi (7). In pathogenic organisms, including bacteria, fungi and protozoans, recombination can be central to strategies for dealing with host immunity (8,9). African trypanosomes, including Trypanosoma brucei, are protistan parasites of mammals belonging to a deeply branching eukaryotic order termed the kinetoplastida (10), and are responsible for devastating disease in sub-Saharan Africa. T.brucei is an exclusively extracellular parasite of the mammalian bloodstream and tissue fluids, and is subject to immune attack throughout an infection. To avoid being eliminated by the immune response, T.brucei uses antigenic variation of its major surface antigen, the variant surface glycoprotein (VSG). Related strategies of antigenic variation have evolved in a diverse range of pathogens (11).
In T.brucei each cell expresses one antigenic form of VSG at one time and switches this to another VSG during antigenic variation [for recent reviews see (12–14)]. Switching of the expressed VSG gene is not elicited by the host immune response and can occur by two mechanisms, which are considered to be distinct. To ensure expression of a single VSG coat, VSG genes are transcribed only when present in a telomeric transcription unit termed the expression site (ES) (15,16). Because T.brucei contains ∼20 mammalian ESs, a VSG switch can be effected by silencing the actively transcribed ES and activating one of the silent ESs (17,18). The mechanisms and factors which control this transcriptional, or in situ, switching reaction are not understood. However, it appears to be a co-ordinated process (19), and the active ES is found in a sub-nuclear domain, termed the ES body (20,21), from which the inactive ESs appear to be excluded.
Outside the ESs, the T.brucei genome contains >1000 VSG genes (22), which are transcriptionally silent in situ, but can be activated by recombination into the ESs. A number of such recombination reactions have been described. The most commonly observed are gene conversions, where a copy is generated from a silent VSG and displaces the VSG resident in the ES (23,24). Less commonly observed are crossover, or reciprocal, recombination reactions where chromosome ends, containing the active ES and a silent VSG, are exchanged (25,26). Finally, segmental gene conversion reactions have also been described where novel VSG genes are constructed from the open reading frames (ORFs) of multiple silent VSGs or VSG pseudogenes (27–29).
In contrast to transcriptional switching, an understanding of the factors that contribute to VSG switching by recombination is now emerging. Mutation of the T.brucei gene encoding RAD51, the eukaryotic enzyme that catalyses DNA strand exchange during homologous recombination (30), reduces the frequency of VSG switching (31). In contrast, mutation of KU70 or KU80, which together act as a heterodimer in other eukaryotes to co-ordinate a distinct form of DNA double-strand break (DSB) repair termed non-homologous end-joining (32), does not affect the frequency of VSG switching (33). These data imply that at least some of the reactions that contribute to T.brucei antigenic variation utilize homologous recombination. However, detailed analysis of the VSG switching mechanisms used in RAD51 mutants suggests this may not represent the whole picture. First, VSG switching by gene conversion (31), and other forms of recombination (34), can be detected in T.brucei RAD51 mutants, although at a reduced level. This suggests that RAD51-independent recombination pathways operate in T.brucei and can act in VSG switching. Second, the reduced VSG switching frequency in RAD51 mutants appears, surprisingly, to involve a reduction in both gene conversion and transcriptional switching reactions, despite a lack of evidence that the latter mechanism involves recombination. More recent work has also suggested that the link between VSG switching and homologous recombination may not be straightforward. T.brucei MRE11 mutants are impaired comparably with RAD51 mutants in homologous recombination and display elevated levels of chromosomal re-arrangements (35,36), phenotypes consistent with T.brucei MRE11 either catalysing or regulating recombination in similar ways to other eukaryotes (37). Despite this, we found no evidence that MRE11 acts in VSG switching (35). We have also characterized the genes that act in mismatch repair (MMR) in T.brucei, and show that this DNA repair process, as in other organisms, influences homologous recombination (38,39). However, like MRE11, we found no evidence that MMR influences VSG switching.
To assess more directly the role of RAD51 and homologous recombination in VSG switching, this work has examined the functions of two genes putatively more intimately linked to RAD51 function than either MRE11 or MMR. Although Rad51 and its eubacterial homologue, RecA can alone catalyse homology-directed DNA strand exchange in vitro, other factors are needed in vivo (40,41). Prominent amongst these are multiple Rad51-like proteins encoded by most eukaryotes. A meiosis-specific protein named Dmc1 has been described in many, but not all, eukaryotes and is highly related to Rad51 in both sequence and function (42,43). Variable numbers of more distantly related Rad51-like proteins, often called Rad51 paralogues (44,45), are found in most eukaryotes. Saccharomyces cerevisiae contains two such Rad51-like proteins, Rad55 and Rad57 (46). These are found also in Schizosaccharomyces pombe (which may have a further Rad51 paralogue, Rlp1) (47), whereas another fungus, Ustilago maydis, encodes a single, highly diverged Rad51 paralogue, Rec2 (48). Vertebrates, plants and insects encode an expanded repertoire of Rad51-like factors relative to fungi. Five Rad51 paralogues have been described in vertebrates (44) and in Arabidopsis thaliana (49), and are named Xrcc2, Xrcc3, Rad51B (Rad51L1), Rad51C (Rad51L2) and Rad51D (Rad51L3). Drosophila, in contrast, encodes a subtly distinct repertoire of Rad51-like proteins: Rad51 (named SpnA) and four RAD51 paralogues (SpnB, SpnD, Rad51C and Rad51D) have been described, but Dmc1 is absent (50,51).
The roles of the Rad51 paralogues have been best characterized to date in yeast and vertebrates. Mutants of yeast Rad55 and Rad57 display cold-dependent DNA damage sensitivity, impaired recombination (52,53) and a defect in Rad51 sub-nuclear localization during DSB repair that is most marked during meiosis (54–56). Mutations of most vertebrate Rad51 paralogues are embryonically lethal (57–59), but mitotic repair, recombination and Rad51 localization defects have been described in mutants of vertebrate cell lines (60–66), which also display chromosome instability (58,67). in vitro and in vivo studies suggest that yeast Rad55 and Rad57 form a stable heterodimer (52,68) that mediates the formation of, or stabilizes, Rad51 nucleoprotein filaments on single-stranded DNA (69,70). In contrast, the vertebrate Rad51 paralogues form two complexes, one composed of Xrcc3 and Rad51C, and the other composed of Rad51B, Rad51C, Rad51D and Xrcc2 (71–74). In vitro assays suggest that at least some vertebrate Rad51-like proteins can act as mediators of Rad51 DNA strand exchange (75–77), analogous to yeast Rad55–Rad57. However, considerable work suggests that they may play roles beyond this (Discussion), adopting discrete roles at either early (78) or late (79,80) steps in homologous recombination, or at discrete regions of the genome (81). Whether or not vertebrate Rad51-like functions, and sub-cellular complexes, are conserved in Arabidopsis and Drosophila is not yet clear. Mutations in four of the Arabidopsis Rad51 paralogues are developmentally viable but result in increased sensitivity to DNA cross-linking agents (49,82,83). In Drosophila, SpnB mutants display modestly increased sensitivity to ionizing radiation (50). During meiosis, discrete functions of the proteins are apparent in both organisms. In Arabidopsis, mutations in Rad51C and Xrcc3, though not Rad51B or Xrcc2, impair meiosis (49,82,83). Indeed, Drosophila SpnD may be truly meiosis-specific, since mutants are impaired in meiosis but do not display detectably increased sensitivity to DNA damage (84,85).
Here, we show that T.brucei encodes a repertoire of RAD51-like proteins more similar to metazoans than to fungi. Mutations in two of the proteins resulted in reduced levels of recombination, altered the sub-nuclear localization of RAD51 following DNA damage and sensitized the parasite to induced DNA damage. Surprisingly, only one of the two proteins contributes to antigenic variation, suggesting distinct functions of the T.brucei RAD51 paralogues in homologous recombination.
MATERIALS AND METHODS
Trypanosoma brucei strains, growth and transformation
T.brucei bloodstream cells of strain 3174, a derivative of MITat1.2a (86), were used throughout and grown at 37°C in HMI-9 medium (87). Growth rates in vitro were determined by diluting the T.brucei cell lines to 5 × 104 cells ml−1 and counting concentrations thereafter with a haemocytometer (bright-line; Sigma); in vivo growth rates were assayed as described previously (31). Electroporation of DNA constructs was conducted as described previously (34,38). In generating RAD51-3 mutants, transformants were selected with 0.5 or 1.0 µg ml−1 puromycin and 2.5 µg ml−1 blasticidin, while RAD51-5 mutants were selected with 0.25 µg ml−1 puromycin and 2.5 µg ml−1 blasticidin. To assay recombination, 5 × 107 cells of each T.brucei cell line, grown to a maximum density of 2 × 106 cells ml−1, were transformed with 5 µg of tub-BLE-tub (pRM450) that had been digested with NotI and ApaI. Electroporated cells were recovered in non-selective media for three generations (24 h for wild-type and +/− mutants; 45 and 39 h for the rad51-3−/− and rad51-5−/− mutants, respectively) before applying selection with 2 µg ml−1 phleomycin. For wild-type and +/− cells, 5 × 106 electroporated cells were plated over 24 wells in 1.5 ml well−1 of antibiotic selective media. In contrast, 2 × 107 cells were plated over 48 wells for the rad51-3−/− and rad51-5−/− mutants. Three independent transformations were performed for each cell line.
Generation and analysis of RAD51-3 and RAD51-5 mutants and re-expressors
Constructs for gene disruption contained expression cassettes for puromycin N-acetyl transferase (puromycin) or blasticidin S deaminase (blasticidin) flanked by 5′ and 3′ sequences derived from the RAD51-3 or RAD51-5 ORFs and were used over two successive rounds of transformation to disrupt both alleles of the two genes. For RAD51-3, the 5′ flank was 336 bp of sequence beginning 9 bp downstream of the predicted start codon and was PCR-amplified from MITat1.2 genomic DNA using Herculase DNA polymerase (Stratagene) with the primers RAD57-LHF-For (5′-GATACACTAGAATTCCAATGCTCCTCACTTACCCC; EcoRI underlined) and RAD57-LHR-Rev (5′-GGTAAGCTTGTTAACAAGAATATCAAGACTCCGGC; HindIII, HpaI underlined); the 3′ flank was 368 bp, ending 3 bp upstream of the stop codon, and was amplified with RAD57-RHF-For (5′-ACAAAGCTTGATATCCAAATGGCACACGTATGGGG; HindIII, EcoRV underlined) and RAD57-RHF-Rev (5′-ACTGGCAGACTCGAGAGTGGGTCGGAAAACATCGC; XhoI underlined). For RAD51-5, the 5′ flank was 200 bp of sequence beginning immediately downstream of the start codon and was PCR-amplified with RECA-LHF-For (5′-GATACACTAGAATTCATGTCTGTGTGTCCTCCACC; EcoRI underlined) and RECA-LHR-Rev (5′-GGTAAGCTTGTTAACAACACCTCCAGGACTGTCCC; HindIII, HpaI underlined); the 3′ flank was 225 bp, ending 65 bp upstream of the stop codon, and was amplified with RECA-RHF-For (5′-ACAAAGCTTGATATCTCTGTTGGTGGTAGTGTGGC; HindIII, EcoRV underlined) and RECA-RHF-Rev (5′-ACTGGCAGACTCGAGATACG TGTTTTGCAG CCCCG; XhoI underlined). For each gene, the 5′ and 3′ flanks were cloned together into EcoRI–XhoI digested pBluescript (Stratagene). Gene disruption constructs were then made by cloning blasticidin (BSD) or puromycin (PUR) resistance cassettes into the HpaI sites between the two flanks, generating the constructs RAD51-3::BSD or RAD51-5::BSD, and RAD51-3::PUR or RAD51-5::PUR, respectively; the resistance cassettes consisted of the antibiotic resistance ORF flanked by processing sequences derived from actin or tubulin intergenic sequences. All the gene disruption constructs were digested with NotI and ApaI before transformation.
Re-expression of RAD51-3 and RAD51-5 in their respective −/− mutants was performed by PCR amplifying the genes' ORFs with Herculase polymerase and cloning them into EcoRV-digested construct pRM481. Here, the genes were flanked upstream by, successively, a 400 bp actin intergenic sequence and a phleomycin resistance gene (BLE). Surrounding these sequences were targeting flanks that allowed RAD51-3 and RAD51-5 to be integrated, following transformation of the pRM481-derived constructs, into the tubulin array, replacing an α-tubulin ORF. In this location, expression of the genes relies on read-through transcription and mRNA processing by the pRM481-derived upstream actin sequence and downstream β-tubulin sequence. Integration of RAD51-3 and RAD51-5 into the tubulin locus was confirmed by Southern analysis of transformant genomic DNA digested with BstXI or SpeI, respectively.
Drug resistant transformants were analysed by probing Southern blots of restriction digested genomic DNA that had been separated on 0.8% agarose gels using α-32P radiolabelled DNA fragments. For RT–PCR, total RNA was prepared from T.brucei cell lines using an RNeasy kit (Qiagen) and cDNA was synthesized with random hexamers and Superscript reverse transcriptase (Life Technologies). For RAD51-3, the primers RAD57-KO5′ (5′-TGTAACAACACTTTGCCG) and RAD57-KO3′ (5′-CCGTGACTTTCAATAACGCC) were used, giving a product of 669 bp if intact RNA is present; for RAD51-5, primers RECA- LHF-For and RECA-U2 (5′-GGGATTGAGAGAGGACGGG) were used, giving a 435 bp product on intact RNA. RNA Polymerase (Pol) I control primers have been described before (26).
Assaying DNA damage sensitivity and VSG switching frequency
Sensitivities of the T.brucei cell lines to methyl methanesulphonate (MMS) were assayed essentially as described before (33,35). Cells were grown to a maximum density of 1 × 106 cells ml−1, and plated in 96-well plates at a concentration of 1 cell well−1 in increasing concentrations of MMS, plus a no drug control. For each cell line, at each drug concentration, four repetitions were performed. The numbers of wells with viable parasite populations were scored initially after 7 days, and monitored for further growth up to 20 days.
Determination of VSG switching frequency was performed as described before (31,86), but with some modifications. To allow VSG switched variants to arise, all cell lines were diluted from media containing hygromycin (5 µg ml−1) and G418 (2.5 µg ml−1) into non-selective media at a starting concentration of 4 × 104 cells ml−1. The cells were then grown for nine generations, taking into account the increased generation times of the −/− mutants, before injection into immunized mice. For the wild-type and +/− mutants 4 × 107 cells were injected, whereas 8 × 107 −/− mutant cells were injected. Switched variants were recovered in all cases 24 h later, and VSG switching frequency calculations take account of the smaller numbers of divisions the −/− mutants undergo in this time.
RAD51 immunolocalization
To examine the localization of RAD51 following DNA damage, 5 ml cultures of the cell lines were inoculated at 106 cells ml−1 in media containing phleomycin. After 18 h growth, 1.5 ml was centrifuged at 1500 g for 10 min at room temperature, resuspended in 1 ml of phosphate-buffered saline (PBS, pH 7.4), centrifuged again and resuspended in 40 µl of PBS. Samples (20 µl) were allowed to settle on glass slides, air-dried and soaked in methanol for 10 min at room temperature. The slides were then washed in PBS containing 0.2% Tween-20 for 5 min and placed in a humid chamber. Polyclonal antiserum (1 ml), raised in lop-eared rabbits (Diagnostics Scotland, UK) against bacterially expressed, histidine-tagged T.brucei RAD51 and diluted 1:500 in PBS containing 1% Tween-20 and 3% BSA (PBS-T-BSA), was added to each slide for 90 min at room temperature. The slides were rinsed with PBS-T-BSA, soaked twice in fresh PBS-T-BSA for 5 min, then returned to the humid chamber, where 0.5 ml of the fluorescein isothiocyanate (FITC) conjugated goat-derived anti-rabbit IgG (Sigma), diluted 1:100 in PBS-T-BSA, was added for 30 min at room temperature. The slides were then rinsed with PBS-T-BSA, again soaked twice in PBS-T for 5 min, before being mounted in Vectashield with 4,6-diamidino-2-phenylindole (DAPI; Vector laboratories). Microscopic analysis was performed using an Axioskop 2 microscope (Zeiss) and images obtained with Openlab software (Improvision). Two researchers scored independently 100–120 cells for the presence and number of visible sub-nuclear foci.
Accession numbers of kinetoplastid RAD51-related proteins analysed in this study
T.brucei RAD51: XP_828893/AAD51713. T.cruzi RAD51: XP_814843/AAZ94621. L.major RAD51: CAJ05032/AAC16334. T.brucei DMC1: XP_827266/EAN76936. T.cruzi DMC1: XP_821533/XP_818873. L.major DMC1: AAZ14711/AAC16335. T.brucei RAD51-3: XP_828338. T.cruzi RAD51-3: XP_820915. L.major RAD51-3: CAJ06683. T.brucei RAD51-4: XP_828775. T.cruzi RAD51-4: XP_807204/XP_809595. L.major RAD51-4: CAJ02433. T.brucei RAD51-5: EAN78580. T.cruzi RAD51-5: XP_811996/XP_807606. T.brucei RAD51-6: AAX81032. T.cruzi RAD51-6: XP_806422. L.major RAD51-6: AAZ09495.
RESULTS
Trypanosoma brucei encodes six RAD51-related proteins
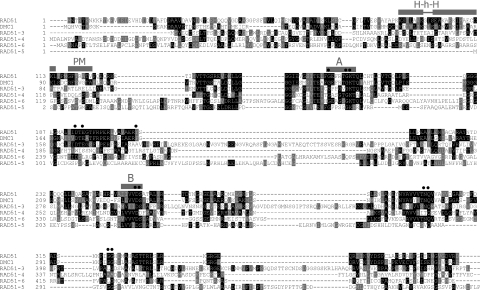
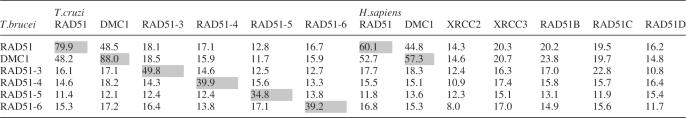
The largely complete genome sequence of T.brucei and two related trypanosomatid parasites (T.cruzi and L.major) has been determined recently. From these sequences (88), six single copy RAD51-related genes could be identified in both T.brucei (Figure 1) and T.cruzi (Table 1), and five in L.major (data not shown). One gene, described previously (31), encodes RAD51 itself; the predicted amino acid sequence of this is highly conserved with other eukaryotes (60–80% sequence identity), and homologues have been shown to be transcriptionally induced by DNA damage in L.major (89) and Plasmodium falciparum (90). Another gene is highly related to T.brucei RAD51 (49% sequence identity; Figure 1) but groups closely with Dmc1 from other eukaryotes in phylogenetic analysis (data not shown). All the other genes are more distantly related to RAD51 (10–17% sequence identity with T.brucei RAD51). Sequence alignments show the relationship between these proteins, RAD51 and DMC1. The central core of all the proteins, which is conserved between eukaryotic Rad51 and eubacterial RecA (91), contains Walker A and B box nucleotide binding motifs. In contrast, further residues that are known to be important for the structure and enzymatic activity of Rad51 and RecA (92) are not well conserved beyond T.brucei RAD51 and DMC1. Outside the RecA-related core, a helix–hairpin–helix motif involved in DNA binding (93,94) and a short motif for polymerization of RAD51 in the nucleoprotein filament (94,95) have been described: both could be identified in T.brucei RAD51 and probably also in DMC1, but were not apparent in the four other polypeptides. Attempting to assign functional identities to the diverged T.brucei predicted proteins based on this analysis, or on pairwise sequence comparisons with RAD51-like proteins from other eukaryotes (Table 1), or by phylogenetic analysis (data not shown), is difficult and so we have given the genes systematic names. Nevertheless, comparison of the predicted T.brucei proteins with those in T.cruzi (Table 1) identifies clear orthologues, showing that the functions of these factors are conserved in at least these two kinetoplastids.
Figure 1.
A comparison of RAD51-related proteins in T.brucei. T.brucei RAD51 amino acid sequence (top) is compared with the T.brucei RAD51-like predicted polypeptides; the alignment was carried out using ClustalW (111), manually adjusted and coloured using Boxshade 3.21 (residues identical in 30% or more sequences are boxed black, conserved residues are boxed grey). Walker A (A) and B (B) boxes conserved in all the proteins are indicated, as are a putative polymerization motif (PM), a helix–hairpin–helix motif (H–h–H) and catalytic and structurally conserved residues (closed circles) in RAD51 and DMC1.
Table 1.
Percent amino acid sequence identities comparing RAD51-like proteins from T.brucei and T.cruzi, and from T.brucei and Homo sapiens
Likely orthologous proteins are indicated by grey shading.
T.brucei RAD51-3 and RAD51-5 are involved in DNA repair and recombination
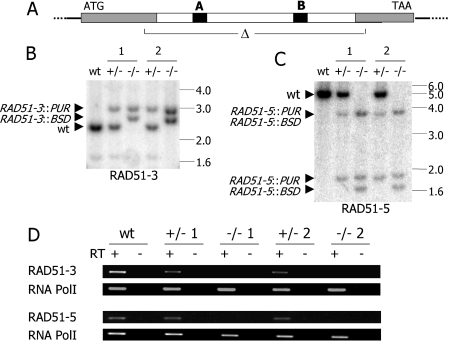
To examine the roles of the divergent RAD51-like genes in T.brucei, we made mutants of RAD51-3, which is marginally the most related gene to T.brucei RAD51 (17% amino acid sequence identity), and RAD51-5, the most distantly related (10% identity). In each case, the central core of the ORF (including the Walker A and B boxes) was replaced with antibiotic resistance cassettes. Following transformation of such constructs, homologous recombination using the 5′ and 3′ ends of the ORF allowed recombination into the T.brucei genome, disrupting the endogenous alleles (Figure 2A) in a similar manner to RAD51 mutants we generated previously (31). Using two rounds of transformation we isolated heterozygous (+/−) and homozygous (−/−) mutants for both RAD51-3 and RAD51-5 in bloodstream form cells of T.brucei line 3174.2 (86). Southern analysis demonstrated that the allele disruptions had occurred as predicted (Figure 2B and C), and the absence of intact RNA transcripts in the −/− mutants was confirmed by RT–PCR (Figure 2D).
Figure 2.
Generation of RAD51-3 and RAD51-5 mutants in bloodstream form T.brucei. (A) For both genes, the central core of the ORF, containing the Walker (A) and (B) boxes (black) was deleted (open triangle) by and replaced with cassettes encoding either blasticidin (RAD51-3::BSD/RAD51-5::BSD) or puromycin resistance (RAD51-3::PUR/RAD51-5::PUR). (B and C) Southern blots of wild-type (WT), two heterozygous mutants (+/− 1, 2) and two homozygous mutants (−/− 1, 2) probed with the ORFs of RAD51-3 or RAD51-5. RAD51-3 mutants' genomic DNA was digested with BstXI, while SpeI was used for RAD51-5 mutants. Positions of size markers (in kb) are indicated, and hybridizing fragments from the WT genes, prior to disruption, or the disrupted alleles are marked by arrow heads. (D) RT–PCR on cDNA from WT, +/− or −/− cells using primers specific for RAD51-3 or RAD51-5; RNA Pol I-specific primers were used to demonstrate intact cDNA was present.
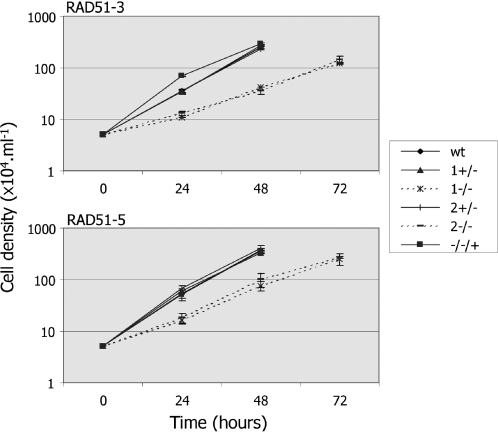
Although the rad51-3−/− and rad51-5−/− mutants are viable, each displayed an increased population doubling time in vitro relative to wild-type cells or their +/− antecedents (Figure 3). Wild-type population numbers doubled on average in 8.2 (±0.4) h, compared with 8.55 (±0.15) h and 7.8 (±0.2) h for the RAD51-3+/− and RAD51-5+/− cells, respectively. In contrast, rad51-3−/− mutants doubled in 15.35 (±0.35) h, and rad51-5−/− mutants in 12.7 (±0.1) h. This level of growth impairment is at least comparable with, and may even be more severe than, that of T.brucei rad51−/− mutants (31). It is possible in kinetoplastids to define the cell cycle stage of individual cells by DNA staining following fixation (96). By doing this, we found that the increased population doubling time of the rad51-3−/−, rad51-5−/− or rad51−/− cells was not due to accumulation of cells at a single cell cycle stage (data not shown), and perhaps represents higher levels of cell death in the mutants. Despite their in vitro growth impairment, the rad51-3−/− and rad51-5−/− mutants were still able to establish infections in rodent hosts, as do rad51−/− mutants (31), with increased population doubling times relative to wild-type or +/− cells still apparent (data not shown).
Figure 3.
Growth of RAD51-3 and RAD51-5 mutants in vitro. Growth of wild-type (WT), two heterozygous mutants (+/− 1, 2) and two homozygous mutants (−/− 1, 2) of RAD51-3 and RAD51-5 were compared by measuring cell densities at the given times. Growth of rad51-3−/− and rad51-5−/− cell lines in which an intact copy of the mutated gene was re-expressed from the tubulin locus (−/−/+) is shown also.
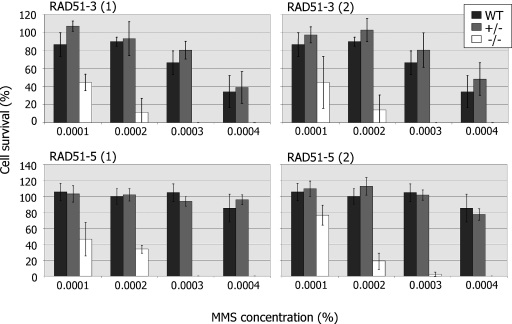
To determine whether or not RAD51-3 and RAD51-5 contribute to DNA repair in T.brucei, we next assayed the ability of the mutants to grow from single cells at various concentrations of MMS (Figure 4). The results for either gene were very similar: disruption of a single allele caused no increased sensitivity to MMS relative to wild-type cells, whereas disruption of both alleles caused a markedly increased sensitivity to MMS. The level of this phenotype is comparable with that described previously for rad51−/− mutants (35), and indicates that both proteins contribute to the repair of MMS-induced DNA damage. Indeed, both the −/− mutants display increased sensitivity to phleomycin also (data not shown), indicating a general repair deficiency.
Figure 4.
RAD51-3 or RAD51-5 mutation leads to DNA damage sensitivity. The survival of wild-type (WT) T.brucei in the presence of increasing concentrations of MMS was compared with heterozygous (+/−) and homozygous (−/−) mutants of both RAD51-3 and RAD51-5 by plating each cell line at one cell well−1 in 96-well plates. The average number of wells containing growing parasite populations after 7–20 days is plotted as a percentage compared with the average number of wells surviving in the absence of MMS. SD is indicated and the data are presented for two (1 and 2) independent heterozygous and homozygous mutants for each gene.
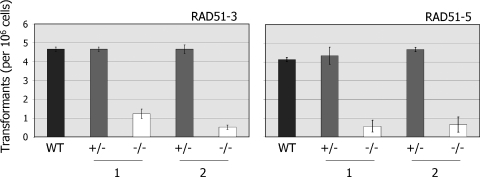
To date, Rad51 paralogues have been shown to influence the efficiency and profile of mitotic homologous recombination only in yeast and vertebrates. To test this in T.brucei, we compared the efficiency with which the rad51-3−/− and rad51-5−/− mutants generate antibiotic resistant transformants following electroporation relative to wild-type and +/− cells. To do this, we transformed all the cell lines with the construct tub-BLE-tub, which encodes the bleomycin resistance protein and normally should confer phleomycin resistance after integrating in the tubulin locus via homologous recombination on terminal sequences derived from α- and β-tubulin intergenic equences (35). In a number of previous studies we have shown that the number of transformants generated by electroporation of this and other constructs is a reliable measure of recombination efficiency in T.brucei, where non-homologous recombination has never been observed (33,34,38,97). The results of this analysis are shown in Figure 5, and demonstrate that the rad51-3−/− and rad51-5−/− mutations resulted in impaired, though not abolished, recombination. The rad51-3−/− mutant cell lines showed between a 3.8- and 9-fold reduction in transformation efficiency relative to their +/− antecedents or wild-type cells, whilst the two rad51-5−/− cell lines displayed 6.7- and 7.7-fold reductions. The extent of this impairment appears not to be as severe as that observed in rad51−/− mutants, where a 7- to 16-fold reduction in recombination efficiency was observed for a range of constructs (34), but is more severe than we observed for mre11−/− mutants (35). Since transformants were still detected in the −/− mutants, we examined the patterns of plasmid integration by Southern analysis. In T.brucei rad51−/− mutants most integrations occur by homologous recombination, but small numbers of microhomology-dependent reactions can also be found (34). In all cases in the rad51-3−/− and rad51-5−/− mutants, however, plasmid integrations had occurred by homologous recombination (data not shown), and no microhomology-dependent reactions were observed.
Figure 5.
RAD51-3 or RAD51-5 mutation impairs recombination. The number of transformants recovered (per 106 cells put on antibiotic selection) when the construct tub-BLE-tub was electroporated into wild-type (WT) T.brucei, or into two independent heterozygous (+/− 1, 2) or homozygous (−/− 1, 2) mutants of RAD51-3 or RAD51-5, is shown. Values are averages from three experiments, and SDs are indicated.
Mutation of RAD51-3 or RAD51-5 attenuates the ability of T.brucei to form DNA damage-induced RAD51 sub-nuclear foci
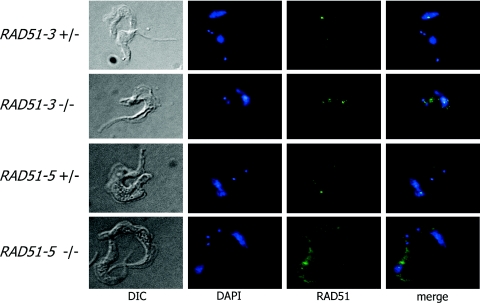
If T.brucei RAD51-3 and RAD51-5 are functionally equivalent to the Rad51 paralogues in other organisms, at least a component of their cellular functions should be to contribute to RAD51-mediated recombination. To test this, we examined RAD51 localization in the rad51-3−/− and rad51-5−/− mutants relative to wild-type and +/− T.brucei before and after induced DNA damage. In yeast and vertebrates, many forms of DNA damage have been shown to cause a re-localization of Rad51 into sub-nuclear foci. The formation of such foci is believed to be a component of eukaryotic cells' response to DNA damage, and they are thought to represent a site of repair in which many factors co-localize [for reviews see (45,98)]. Moreover, the formation of RAD51 foci is dependent on a range of factors, including the Rad51 paralogues (62,65,66).
To assess the localization of T.brucei RAD51 following DNA damage, immunolocalization was conducted using antiserum recognizing T.brucei RAD51 on methanol-fixed bloodstream form cells that had been exposed to phleomycin, which causes DSBs (99). A full discussion of this analysis will be published elsewhere (K. Norrby, C. Proudfoot and R. McCulloch, manuscript in preparation), but Table 2 shows the percentage of cells displaying foci and the number of foci detectable in individual cells, while Figure 6 shows examples of such foci. In RAD51-3+/−, RAD51-5+/− and wild-type cells, discrete sub-nuclear RAD51 loci could readily be detected following phleomycin treatment. At this concentration of drug (1.5 µg ml−1), a majority of wild-type and +/− cells displayed foci (only 22–35% showed no foci), whereas in the absence of phleomycin only 1% of cells showed any sub-nuclear localization of RAD51 (data not shown). Most cells contained between 1 and 3 RAD51 foci, although small numbers of cells containing larger numbers were detectable. No difference in the proportion of cells containing foci was discernible between the wild-type and RAD51-3+/− or RAD51-5+/− cells. In contrast, the formation of RAD51 foci was attenuated in both the rad51-3−/− and rad51-5−/− mutants. For the −/− mutants, RAD51 localization was examined at an equivalent phleomycin concentration to the wild-type and +/− cells and also at reduced concentrations. This was done because the mutants are more sensitive to phleomycin-induced damage, and we have found that at high concentrations (>5 µg ml−1), RAD51 foci are undetectable in wild-type cells, possibly because the cells are severely compromised (data not shown). Phleomycin (0.3 and 0.5 µg ml−1) was used for the rad51-3−/− and rad51-5−/− mutants, respectively, because these levels caused comparable levels of cell death or growth retardation relative to wild-type and +/− cells grown in 1.5 µg ml−1 phleomycin (data not shown). Irrespective of the drug concentration used, in both −/− mutants a majority of cells (52–82%) did not have RAD51 foci and, where foci were detected, this was rarely more than one or two (Table 2). In fact, this may be an overestimate, since for both −/− mutants, but particularly rad51-5−/−, higher levels of cytoplasmic staining were apparent following phleomycin treatment, and some staining may have been counted as foci.
Table 2.
Percentages of RAD51-3 or RAD51-5 mutant cells displaying RAD51 foci following 18 h growth in phleomycin (BLE; concentrations are in µg ml−1), and the number of foci that are visible
| [BLE] | No. of foci | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | L | ||
| RAD51-3 | |||||||||
| Wild type | 1.5 | 35.0 | 28.2 | 21.4 | 10.3 | 3.4 | 3.4 | 0.9 | 0.9 |
| 1+/− | 1.5 | 27.4 | 30.8 | 18.8 | 15.4 | 6.0 | 1.7 | 0.0 | 0.0 |
| 1−/− | 0.3 | 81.7 | 7.3 | 5.5 | 5.5 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1−/− | 1.5 | 63.8 | 18.1 | 12.1 | 3.4 | 2.6 | 0.0 | 0.0 | 0.0 |
| 2+/− | 1.5 | 22.2 | 26.5 | 18.8 | 14.5 | 4.3 | 1.7 | 0.9 | 1.7 |
| 2−/− | 0.3 | 59.0 | 19.7 | 12.0 | 4.3 | 0.9 | 0.9 | 0.0 | 0.0 |
| 2−/− | 1.5 | 52.1 | 24.8 | 8.5 | 0.9 | 1.7 | 0.9 | 0.0 | 0.9 |
| RAD51-5 | |||||||||
| Wild type | 1.5 | 22.3 | 16.1 | 24.1 | 17.9 | 11.6 | 4.5 | 1.8 | 1.8 |
| 1+/− | 1.5 | 19.8 | 24.3 | 33.3 | 14.4 | 5.4 | 1.8 | 0.9 | 0.0 |
| 1−/− | 0.5 | 67.3 | 18.3 | 16.3 | 5.8 | 1.0 | 0.0 | 0.0 | 0.0 |
| 1−/− | 1.5 | 59.5 | 18.0 | 9.0 | 2.7 | 0.9 | 0.9 | 0.0 | 0.9 |
| 2+/− | 1.5 | 27.9 | 32.4 | 19.8 | 13.5 | 2.7 | 4.5 | 3.6 | 3.6 |
| 2−/− | 0.5 | 64.0 | 22.5 | 7.2 | 3.6 | 0.9 | 0.0 | 0.9 | 0.9 |
| 2−/− | 1.5 | 64.9 | 23.4 | 3.6 | 1.8 | 2.7 | 0.0 | 0.0 | 0.0 |
L represents >6 foci, +/− denotes heterozygous mutants, and −/− denotes homozygous mutants).
Figure 6.
Examples of RAD51 foci in T.brucei. Examples of RAD51-3 or RAD51-5 heterozygous (+/−) or homozygous (−/−) mutants with discernible sub-nuclear RAD51 foci following 18 h growth in 1.5 µg ml−1 phleomycin are shown. Cells were visualized by differential interference contrast (DIC; left), the DNA was stained with DAPI (centre left), and RAD51 (centre right) was visualized using a polyclonal anti-RAD51 antiserum and FITC conjugated goat-derived anti-rabbit IgG secondary. Merged DAPI and RAD51 images are also shown (right).
RAD51-3 contributes to T.brucei antigenic variation, but RAD51-5 does not
To examine the roles of RAD51-3 and RAD51-5 in T.brucei VSG switching, the mutants were made in the strain 3174. We have described this strain before (31,33,35,38,86). Briefly, it expresses genes encoding resistance to G418 and hygromycin from the active ES (encoding VSG221) and allows the frequency of VSG switching to be calculated using an in vivo selection regime, which also generates clonal switched variants in which the type of switching event used can be assessed via the antibiotic resistance genes. Figure 7 shows the VSG switching frequencies determined for mutants of the two RAD51-like genes relative to wild-type cells, while Figure 8 shows the profiles of VSG switching events that the cell lines employ. In doing this analysis, we re-introduced intact copies of the RAD51-3 or RAD51-5 genes into one of the two independently generated rad51-3−/− or rad51-5−/− mutants, respectively. In each case, re-expression reverted the strong growth impairment that we observed, resulting in population doubling times in the two re-expression cell lines that were indistinguishable from wild-type or +/− cells (Figure 3).
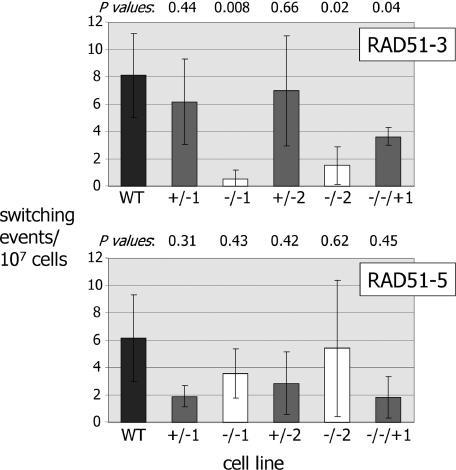
Figure 7.
Effect of RAD51-3 or RAD51-5 mutation on the frequency of VSG switching. VSG switching frequencies for wild-type T.brucei cells (3174.2; WT) were compared with two independently generated heterozygous (+/− 1, 2) and homozygous mutants (−/− 1, 2) of either RAD51-3 or RAD51-5, in addition to rad51-3−/− and rad51-5−/− cell lines in which an intact copy of the mutated gene was re-expressed from the tubulin locus (−/−/+). Means of at least three independent experiments, plus SDs, are shown. P-values (determined by Student paired t-tests) of +/− cells' VSG switching frequencies relative to wt cells are indicated above the graph, as are the −/− cells' frequencies relative to the +/− cells from which they are derived, and the −/−/+ cells relative to the −/− mutants.
Figure 8.
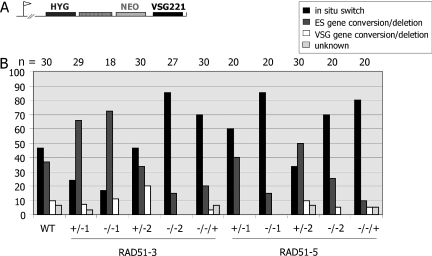
Effect of RAD51-3 or RAD51-5 mutation on the VSG switching profile. (A) The form of VSG switching used in each cell line (see text) was determined by assaying for the expression of hygromycin and G418 resistance from the antibiotic marker genes (HYG and NEO, respectively) inserted around VSG-associated 70 bp repeat sequences (hatched box) in the active VSG221 ES (arrow denotes promoter) in strain 3174, as well as using PCR to assess whether HYG, NEO or VSG221 had been removed by gene conversion. (B) The percentage of clones that had undergone VSG switching by in situ switching, ES gene conversion/deletion or VSG gene conversion/deletion (see text) for each cell line are shown; the number of clones analysed (n) is indicated above the graph.
As we have observed before, there was considerable variation in the VSG switching frequencies determined by this assay, reflecting the randomness of the process and the low rate (∼2–11 × 10−7 switchers cell−1 generation−1) at which VSG switch variants arise in the Lister 427 strain from which 3174 wild-type cells are derived (31,33,35,38,86). However, it was apparent that disruption of RAD51-3 led to a reduction in VSG switching frequency in each of the two independently generated rad51-3−/− mutants. The average switch frequencies for the mutants were 0.5 × 10−7 and 1.5 × 10−7 switches cell−1 generation−1, which represents statistically significant reductions (Figure 7) relative to either the +/− cells from which they are derived (average switch frequencies, 6.1 and 7.0 × 10−7, respectively), the wild-type cells (8.1 × 10−7) or the RAD51-3 re-expressor (3.6 × 10−7). In fact, the calculated switch frequencies for the rad51-3−/− mutants may be over-estimates, as we consistently failed to recover any switched variants in some of these experiments (3 out of 8 repetitions for rad51-3−/−1, and 3 out of 6 for rad51-3−/−2; negative experiments were excluded from the frequency calculations, however). This was not seen for any other cell lines, and suggests that the switch frequency for these mutants may be at the threshold of detection under the assay conditions we have employed.
In contrast to RAD51-3, we found no evidence that RAD51-5 contributes to VSG switching. The calculated VSG switching frequencies for the two rad51-5−/− mutants (averages of 3.6 × 10−7 and 5.4 × 10−7 switches.cell−1.generation−1) were not statistically significantly different from the wild-type cells (P-values 0.50 and 0.89, respectively), or from the +/− cells from which they were derived (Figure 7; averages of 1.9 × 10−7 and 2.8 × 10−7, respectively) or the RAD51-5 re-expressor (1.8 × 10−7).
To determine whether or not disruption of either of the RAD51-like genes alters the profile of VSG switching mechanisms in T.brucei, a number of variant clones from each of the cell lines were assayed for the type of VSG switching event that they had undergone. In this assay, we can distinguish three types of switching event (Figure 8A). Firstly, transcriptional, or in situ, switches are identified by retention of VSG221 and the antibiotic resistance genes. Secondly, events in which VSG221 and the antibiotic resistance genes are deleted represent either gene conversions that encompass all these markers (86), or deletions of these genes accompanied by a switch in transcription to another ES (100) (we term these ES gene conversion/deletions). Thirdly, events in which the hygromycin resistance gene is retained and continues to be expressed, but the G418 resistance and VSG221 genes are deleted, most probably represent gene conversions of the VSG that extend to the 70 bp repeats, although may represent deletions of the ES on the 70 bp repeats that elicit a transcriptional switch to another ES (termed VSG gene conversion/deletion). Very occasionally (3% of switch variants), we observed events that are not readily explained by these switching reactions, and we termed these ‘unknown’. Despite considerable fluctuation in the data (Figure 8B), in general terms in the wild-type, RAD51-3+/−, RAD51-5+/−, RAD51-3 re-expressor and RAD51-5 re-expressor cell lines in situ switches slightly predominated (24–80% of switching events) over ES gene conversion/deletions (10–66%), whilst VSG gene conversion/deletions were relatively rare (0–20%). In the homozygous mutants of each gene, all of the different VSG switching events could be detected. Variations in the proportion of the events in these mutants (for instance, between rad51-3−/− mutants 1 and 2; Figure 8B) most probably reflect fluctuations in sampling, rather than phenotypic differences. This indicates that both transcription- and recombination-based switching still occur. Indeed, the recombination event seen most rarely (VSG gene conversion/deletion) was still detected readily, indicating that recombination accounts for a significant proportion of VSG switching. For the rad51-5−/− mutants, this is not surprising, as no change in VSG switch frequency was observed. For the rad51-3−/− mutants, which had impaired VSG switching, this is reminiscent of what we observed previously for rad51−/− mutants, where no change in the ratio of transcription- and recombination-based switching was found (31), and suggests that the lowered VSG switching frequency in these mutants is not due to a selective reduction in recombination-based switching, as might be expected for proteins which act in homologous recombination.
DISCUSSION
Work from vertebrates, plants, insects and fungi indicates that eukaryotic organisms encode a complex repertoire of Rad51-like proteins. Here, we add to that picture by showing that T.brucei, a highly divergent eukaryote (10), encodes six RAD51-related proteins. We show that two of the distantly related T.brucei RAD51-like proteins, like RAD51 itself (31,34), act in mitotic DNA repair and recombination. Furthermore, both RAD51-like proteins contribute to the formation of RAD51 sub-nuclear foci following DNA damage. To date, the formation of such foci has been observed in fungi (101), vertebrates (45) and nematodes (102), and the finding that it occurs also in T.brucei suggests that it is a widely conserved eukaryotic response to DNA damage (98).
Although Rad51 and Dmc1, a meiosis-specific homologue of Rad51 (43), are highly conserved between eukaryotes, further Rad51-like proteins are highly diverged. This is apparent in the low levels of sequence identity amongst the proteins, and also differences in the numbers of such proteins encoded by eukaryote genomes. Most metazoans appear to encode relatively large families of diverged Rad51-like proteins: vertebrates (44,45) and Arabidopsis (49) contain five, whilst Drosophila (50) contains four. In contrast, fungi appear to encode a smaller repertoire (103) with one, two and three diverged Rad51-like proteins described in U.maydis, S.cerevisiae and S.pombe, respectively. The presence of four distantly related RAD51-like genes in T.brucei and T.cruzi is more reminiscent of metazoan organisms, and illustrates that the number of such genes an organism possesses cannot simply be related to genome size or multicellularity. Indeed, this is strikingly demonstrated by the fact that Caenorhabditis elegans encodes only Rad51, and no Rad51-like proteins (104). By understanding the similarities and differences in the roles of these proteins in different organisms, and their interactions with each other and with other recombination factors, including Rad51, we will understand the complex evolutionary history of the recombination machinery in eukaryotes. For example, it remains unclear whether or not the diverged Rad51-like proteins (and protein complexes) are truly paralogues (44), as they are often referred to. Moreover, it is unknown if they provide essentially the same functions in different organisms, or if their roles (and presence or absence) are tailored to the unique biology of each organism.
Distinct functions for RAD51-3 and RAD51-5 in T.brucei antigenic variation
A critical function of DNA recombination in T.brucei is in the switching of VSGs during antigenic variation. We have shown previously that this process is influenced by RAD51 (31), and now demonstrate that a RAD51-like protein, RAD51-3, has a similar role. This provides further evidence that at least some VSG switching is driven not by a recombination machinery dedicated specifically towards antigenic variation, but by general homologous recombination proteins that act in ‘housekeeping’ repair. An orthologue of RAD51-3 (and, indeed, of each T.brucei RAD51-like protein) could be identified in T.cruzi, demonstrating that this factor has not evolved for VSG switching.
We show also here that a second RAD51-like protein, RAD51-5, does not detectably contribute to T.brucei antigenic variation. It is possible that this phenotype is explained by genetic redundancy, where one of the other RAD51-like proteins can assume RAD51-5's function when it is absent. Indeed, it is notable that RAD51-5 appears not to be present in L.major, a related kinetoplastid parasite. Alternatively, it may be that RAD51-3 has a more central function in homologous recombination than RAD51-5, and any effect on VSG switching caused by mutating RAD51-5 is too subtle to be detected by our assay. However, rad51-3−/− and rad51-5−/− mutants both displayed very similar levels of impairment in DNA repair, recombination of transformed DNA constructs and RAD51 foci formation, indicating that either of the above explanations extend only to homologous recombination during VSG switching. We favour a third explanation, that the distinction between rad51-3 and rad51-5 mutants in VSG switching indicates that the proteins have distinct roles in homologous recombination. None of our analysis to date allows us to say if RAD51-3 and RAD51-5 operate in the same pathway in the cell, or if they interact. However, unlike in S.cerevisiae, where Rad55 and Rad57 form a stable heterodimer that acts as a single co-factor in Rad51-catalysed recombination (46), considerable evidence from metazoans points to functional diversification of the Rad51 paralogues. In mammals, Xrcc3 has been shown to localize to DSBs prior to, and independently from, Rad51, suggesting that it may prepare breaks for RAD51 binding (78). Furthermore, mammalian Rad51D, but not Rad51C or Xrcc2, interacts with telomeres and mutation leads to telomere dysfunction, suggesting that it can act in telomere maintenance (81). Rad51C and Xrcc3 have been shown to be involved in Holliday junction processing in human cell extracts (80), suggesting a late role in homologous recombination, which is consistent with altered profiles of homologous recombination reactions observed in Xrcc3 mutant hamster cells (79). Finally, specific roles of a sub-set of the diverged Rad51-like proteins may be seen also during meiotic recombination in plants and yeast (49,84,85). If we are correct for T.brucei, then this functional diversification is common to many eukaryotes, not just to some metazoans.
In T.brucei we still know very little about the detailed biochemistry of the recombination reactions that drive VSG switching, and therefore a number of scenarios might be envisaged to explain the distinction in functions between RAD51-3 and RAD51-5. For example, we do not know the nature of the event(s) that initiates VSG switching. It has been suggested that this is most probably a DSB (105,106), and an endonuclease to generate this lesion has been postulated (106). However, perhaps RAD51-5 acts at an early step in recombination by binding DSBs, and VSG switching is initiated by a distinct DNA lesion and thus bypasses the need for RAD51-5. Such a scenario may also explain the lack of a role for T.brucei MRE11 (35): the initiating lesion may not require MRE11 to contribute to processing the DNA ends (107), or may not activate the DNA damage signalling functions that have been ascribed to this protein in other organisms (108). An alternative scenario is that RAD51-5 is a component of the T.brucei counterpart of the human Rad51C–Xrcc3 complex, which contributes to Holliday junction resolution (80). It has been pointed out before that it would be advantageous for VSG switching to proceed by a synthesis-dependent strand annealing mechanism, as this would avoid the formation of Holliday junction intermediates that could lead to lethal translocations of silent VSG arrays into the ES (11,105,106). Finally, we might also envisage that RAD51-3 has a predominantly (sub) telomeric function, akin to human Rad51D (81), and perhaps directs T.brucei DSB repair in this environment, while RAD51-5 acts in interstitial repair. The sub-telomeres of T.brucei, in common with many pathogens (9), are where antigenic variation is conducted: VSGs are transcribed at the ends of the ESs, recombination must occur here for VSG switching and, it is now clear, the large majority of the silent VSG archive is sub-telomeric (22).
Does transcriptional VSG switching involve recombination?
A striking finding from this work is that the VSG switching phenotype we described previously in T.brucei rad51−/− mutants (31) is re-capitulated in rad51-3−/− mutants: the impairment in VSG switching is of a similar magnitude, and there is no compelling evidence that this impairment arises owing to a selective reduction in recombination-based VSG switching over transcription-based switching. Given that rad51-5−/− and mre11−/− mutants (35) do not cause a reduction in VSG switching frequency, but have similar levels of growth impairment to rad51−/− and rad51-3−/− cells both in vitro and in vivo, the VSG switching phenotype cannot be explained as a growth artefact. As we have argued before for RAD51 (31), this genetic implication of RAD51-3 in transcriptional switching does not necessarily infer a direct catalytic role for recombination, but offers independent evidence that recombination factors may contribute to the process. Much of the work to date on VSG transcriptional switching has focused on the mechanisms that limit full transcription to 1 of the 20 or so ESs (20), rather than the switching process itself. Transcriptional switching appears to be a co-ordinated reaction, since Borst and co-workers (19) have identified putative switching intermediates in which two ES are co-transcribed unstably and switch to expressing one or other ES at very high rates. It is intriguing that this intermediate can involve two ESs but not three (109), and that the two ESs are found rather close together in the nucleus. Furthermore, treatments that cause nuclear DNA damage or replication arrest result in increased transcription from the silent ESs, as well as other silent loci (109,110). These data are not incompatible with RAD51 and RAD51-3 contributing to the reaction, although it is unlikely to be this simple. For instance, transcriptional (and recombination) switching events could still be detected in the rad51 and rad51-3 mutants, suggesting (at the very least) that other factors must be able to assume their functions. Moreover, some of the DNA damage which causes altered ES expression (e.g. alkylation or interstrand cross-links) is most probably recognized and repaired by DNA repair pathways other than homologous recombination. Nevertheless, RAD51 and RAD51-3 are the only genes to date whose mutation has been shown to have any effect on VSG switching, and it is possible that the regulation and execution of antigenic variation has links with cellular repair pathways that we are only beginning to understand.
Acknowledgments
The authors thank Marshall Stark and Peter Burton for critical reading of the manuscript. R.McC. is a Royal Society University Research Fellow, and we thank the Wellcome Trust and Medical Research Council for financial support. Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.West S.C. Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 2.Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 3.Michel B., Grompone G., Flores M.J., Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maser R.S., DePinho R.A. Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair (Amst.) 2004;3:979–988. doi: 10.1016/j.dnarep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity. 2002;88:125–141. doi: 10.1038/sj.hdy.6800034. [DOI] [PubMed] [Google Scholar]
- 6.Roth D.B. Restraining the V(D)J recombinase. Nature Rev. Immunol. 2003;3:656–666. doi: 10.1038/nri1152. [DOI] [PubMed] [Google Scholar]
- 7.Haber J.E. Switching of Saccharomyces mating-type genes. In: Craig N.L., Berg D.E., editors. Mobile DNA II. Washington: ASM Press; 2002. pp. 927–953. [Google Scholar]
- 8.Deitsch K.W., Moxon E.R., Wellems T.E. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol. Mol. Biol. Rev. 1997;61:281–293. doi: 10.1128/mmbr.61.3.281-293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry J.D., Ginger M.L., Burton P., McCulloch R. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 2003;33:29–45. doi: 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 10.Dacks J.B., Doolittle W.F. Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell. 2001;107:419–425. doi: 10.1016/s0092-8674(01)00584-0. [DOI] [PubMed] [Google Scholar]
- 11.Barry J.D., McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 12.Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr. Mol. Med. 2004;4:563–576. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- 13.Pays E., Vanhamme L., Perez-Morga D. Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries. Curr. Opin. Microbiol. 2004;7:369–374. doi: 10.1016/j.mib.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 14.McCulloch R. Antigenic variation in African trypanosomes: monitoring progress. Trends Parasitol. 2004;20:117–121. doi: 10.1016/j.pt.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Becker M., Aitcheson N., Byles E., Wickstead B., Louis E., Rudenko G. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 2004;14:2319–2329. doi: 10.1101/gr.2955304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kooter J.M., van der Spek H.J., Wagter R., d'Oliveira C.E., van der H.F., Johnson P.J., Borst P. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T.brucei. Cell. 1987;51:261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- 17.Borst P., Ulbert S. Control of VSG gene expression sites. Mol. Biochem. Parasitol. 2001;114:17–27. doi: 10.1016/s0166-6851(01)00243-2. [DOI] [PubMed] [Google Scholar]
- 18.Vanhamme L., Lecordier L., Pays E. Control and function of the bloodstream variant surface glycoprotein expression sites in Trypanosoma brucei. Int. J. Parasitol. 2001;31:523–531. doi: 10.1016/s0020-7519(01)00143-6. [DOI] [PubMed] [Google Scholar]
- 19.Chaves I., Rudenko G., Dirks-Mulder A., Cross M., Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- 21.Navarro M., Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 22.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 23.Hoeijmakers J.H., Frasch A.C., Bernards A., Borst P., Cross G.A. Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature. 1980;284:78–80. doi: 10.1038/284078a0. [DOI] [PubMed] [Google Scholar]
- 24.Robinson N.P., Burman N., Melville S.E., Barry J.D. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell. Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pays E., Guyaux M., Aerts D., Van Meirvenne N., Steinert M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature. 1985;316:562–564. doi: 10.1038/316562a0. [DOI] [PubMed] [Google Scholar]
- 26.Rudenko G., McCulloch R., Dirks-Mulder A., Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- 27.Roth C.W., Longacre S., Raibaud A., Baltz T., Eisen H. The use of incomplete genes for the construction of a Trypanosoma equiperdum variant surface glycoprotein gene. EMBO J. 1986;5:1065–1070. doi: 10.1002/j.1460-2075.1986.tb04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longacre S., Eisen H. Expression of whole and hybrid genes in Trypanosoma equiperdum antigenic variation. EMBO J. 1986;5:1057–1063. doi: 10.1002/j.1460-2075.1986.tb04322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamper S.M., Barbet A.F. Surface epitope variation via mosaic gene formation is potential key to long-term survival of Trypanosoma brucei. Mol. Biochem. Parasitol. 1992;53:33–44. doi: 10.1016/0166-6851(92)90004-4. [DOI] [PubMed] [Google Scholar]
- 30.Conway A.B., Lynch T.W., Zhang Y., Fortin G.S., Fung C.W., Symington L.S., Rice P.A. Crystal structure of a Rad51 filament. Nature Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 31.McCulloch R., Barry J.D. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nature Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 33.Conway C., McCulloch R., Ginger M.L., Robinson N.P., Browitt A., Barry J.D. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem. 2002;277:21269–21277. doi: 10.1074/jbc.M200550200. [DOI] [PubMed] [Google Scholar]
- 34.Conway C., Proudfoot C., Burton P., Barry J.D., McCulloch R. Two pathways of homologous recombination in Trypanosoma brucei. Mol. Microbiol. 2002;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- 35.Robinson N.P., McCulloch R., Conway C., Browitt A., Barry J.D. Inactivation of Mre11 does not affect VSG gene duplication mediated by homologous recombination in Trypanosoma brucei. J. Biol. Chem. 2002;277:26185–26193. doi: 10.1074/jbc.M203205200. [DOI] [PubMed] [Google Scholar]
- 36.Tan K.S., Leal S.T., Cross G.A. Trypanosoma brucei MRE11 is non-essential but influences growth, homologous recombination and DNA double-strand break repair. Mol. Biochem. Parasitol. 2002;125:11–21. doi: 10.1016/s0166-6851(02)00165-2. [DOI] [PubMed] [Google Scholar]
- 37.Assenmacher N., Hopfner K.P. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 38.Bell J.S., McCulloch R. Mismatch repair regulates homologous recombination, but has little influence on antigenic variation, in Trypanosoma brucei. J. Biol. Chem. 2003;278:45182–45188. doi: 10.1074/jbc.M308123200. [DOI] [PubMed] [Google Scholar]
- 39.Bell J.S., Harvey T.I., Sims A.M., McCulloch R. Characterization of components of the mismatch repair machinery in Trypanosoma brucei. Mol. Microbiol. 2004;51:159–173. doi: 10.1046/j.1365-2958.2003.03804.x. [DOI] [PubMed] [Google Scholar]
- 40.Sung P., Krejci L., Van Komen S., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 41.Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop D.K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E.coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 43.Sauvageau S., Ploquin M., Masson J.Y. Exploring the multiple facets of the meiotic recombinase Dmc1. Bioessays. 2004;26:1151–1155. doi: 10.1002/bies.20150. [DOI] [PubMed] [Google Scholar]
- 44.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 45.Tarsounas M., Davies A.A., West S.C. RAD51 localization and activation following DNA damage. Philos. Trans. R Soc. Lond. B. Biol. Sci. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 47.Khasanov F.K., Salakhova A.F., Chepurnaja O.V., Korolev V.G., Bashkirov V.I. Identification and characterization of the rlp1+, the novel Rad51 paralog in the fission yeast Schizosaccharomyces pombe. DNA Repair (Amst.) 2004;3:1363–1374. doi: 10.1016/j.dnarep.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Bennett R.L., Holloman W.K. A RecA homologue in Ustilago maydis that is distinct and evolutionarily distant from Rad51 actively promotes DNA pairing reactions in the absence of auxiliary factors. Biochemistry. 2001;40:2942–2953. doi: 10.1021/bi002494i. [DOI] [PubMed] [Google Scholar]
- 49.Bleuyard J.Y., Gallego M.E., Savigny F., White C.I. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005;41:533–545. doi: 10.1111/j.1365-313X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- 50.Staeva-Vieira E., Yoo S., Lehmann R. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 2003;22:5863–5874. doi: 10.1093/emboj/cdg564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radford S.J., Sekelsky J.J. Taking Drosophila Rad51 for a SPiN. Nature Struct. Mol. Biol. 2004;11:9–10. doi: 10.1038/nsmb0104-9. [DOI] [PubMed] [Google Scholar]
- 52.Hays S.L., Firmenich A.A., Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl Acad. Sci. USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugawara N., Ivanov E.L., Fishman-Lobell J., Ray B.L., Wu X., Haber J.E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 54.Gasior S.L., Olivares H., Ear U., Hari D.M., Weichselbaum R., Bishop D.K. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl Acad. Sci. USA. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasior S.L., Wong A.K., Kora Y., Shinohara A., Bishop D.K. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyazaki T., Bressan D.A., Shinohara M., Haber J.E., Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu Z., Smith S., Wang L., Rice M.C., Kmiec E.B. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(−/−) background. Mol. Cell. Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smiraldo P.G., Gruver A.M., Osborn J.C., Pittman D.L. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 59.Deans B., Griffin C.S., Maconochie M., Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lio Y.C., Schild D., Brenneman M.A., Redpath J.L., Chen D.J. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J. Biol. Chem. 2004;279:42313–42320. doi: 10.1074/jbc.M405212200. [DOI] [PubMed] [Google Scholar]
- 61.Liu N., Lamerdin J.E., Tebbs R.S., Schild D., Tucker J.D., Shen M.R., Brookman K.W., Siciliano M.J., Walter C.A., Fan W., et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 62.Takata M., Sasaki M.S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L.H., Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson R.D., Liu N., Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 64.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bishop D.K., Ear U., Bhattacharyya A., Calderone C., Beckett M., Weichselbaum R.R., Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 66.van Veelen L.R., Essers J., van de Rakt M.W., Odijk H., Pastink A., Zdzienicka M.Z., Paulusma C.C., Kanaar R. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat. Res. 2005;574:34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 67.Cui X., Brenneman M., Meyne J., Oshimura M., Goodwin E.H., Chen D.J. The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat. Res. 1999;434:75–88. doi: 10.1016/s0921-8777(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 68.Johnson R.D., Symington L.S. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 70.Fortin G.S., Symington L.S. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51–DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu N., Schild D., Thelen M.P., Thompson L.H. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Masson J.Y., Tarsounas M.C., Stasiak A.Z., Stasiak A., Shah R., McIlwraith M.J., Benson F.E., West S.C. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller K.A., Sawicka D., Barsky D., Albala J.S. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004;32:169–178. doi: 10.1093/nar/gkg925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schild D., Lio Y.C., Collins D.W., Tsomondo T., Chen D.J. Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 75.Shim K.S., Schmutte C., Tombline G., Heinen C.D., Fishel R. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J. Biol. Chem. 2004;279:30385–30394. doi: 10.1074/jbc.M306066200. [DOI] [PubMed] [Google Scholar]
- 76.Lio Y.C., Mazin A.V., Kowalczykowski S.C., Chen D.J. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- 77.Sigurdsson S., Van Komen S., Bussen W., Schild D., Albala J.S., Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Forget A.L., Bennett B.T., Knight K.L. Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51. J. Cell. Biochem. 2004;93:429–436. doi: 10.1002/jcb.20232. [DOI] [PubMed] [Google Scholar]
- 79.Brenneman M.A., Wagener B.M., Miller C.A., Allen C., Nickoloff J.A. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 80.Liu Y., Masson J.Y., Shah R., O'Regan P., West S.C. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 81.Tarsounas M., Munoz P., Claas A., Smiraldo P.G., Pittman D.L., Blasco M.A., West S.C. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 82.Bleuyard J.Y., White C.I. The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 2004;23:439–449. doi: 10.1038/sj.emboj.7600055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Osakabe K., Abe K., Yamanouchi H., Takyuu T., Yoshioka T., Ito Y., Kato T., Tabata S., Kurei S., Yoshioka Y., et al. Arabidopsis Rad51B is important for double-strand DNA breaks repair in somatic cells. Plant Mol. Biol. 2005;57:819–833. doi: 10.1007/s11103-005-2187-1. [DOI] [PubMed] [Google Scholar]
- 84.Abdu U., Gonzalez-Reyes A., Ghabrial A., Schupbach T. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics. 2003;165:197–204. doi: 10.1093/genetics/165.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghabrial A., Ray R.P., Schupbach T. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 1998;12:2711–2723. doi: 10.1101/gad.12.17.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCulloch R., Rudenko G., Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in variant surface glycoprotein expression sites lacking 70-base-pair repeat sequences. Mol. Cell. Biol. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hirumi H., Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 88.El Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N., Ghedin E., Worthey E.A., Delcher A.L., Blandin G., et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 89.McKean P.G., Keen J.K., Smith D.F., Benson F.E. Identification and characterisation of a RAD51 gene from Leishmania major. Mol. Biochem. Parasitol. 2001;115:209–216. doi: 10.1016/s0166-6851(01)00288-2. [DOI] [PubMed] [Google Scholar]
- 90.Bhattacharyya M.K., Kumar N. Identification and molecular characterisation of DNA damaging agent induced expression of Plasmodium falciparum recombination protein PfRad51. Int. J. Parasitol. 2003;33:1385–1392. doi: 10.1016/s0020-7519(03)00212-1. [DOI] [PubMed] [Google Scholar]
- 91.Dunderdale H.J., West S.C. Recombination genes and proteins. Curr. Opin. Genet. Dev. 1994;4:221–228. doi: 10.1016/s0959-437x(05)80048-6. [DOI] [PubMed] [Google Scholar]
- 92.Story R.M., Bishop D.K., Kleckner N., Steitz T.A. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- 93.Shao X., Grishin N.V. Common fold in helix–hairpin–helix proteins. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin D.S., Pellegrini L., Daniels D.S., Yelent B., Craig L., Bates D., Yu D.S., Shivji M.K., Hitomi C., Arvai A.S., et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 96.McKean P.G. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr. Opin. Microbiol. 2003;6:600–607. doi: 10.1016/j.mib.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 97.McCulloch R., Vassella E., Burton P., Boshart M., Barry J.D. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 2004:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- 98.Lisby M., Rothstein R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 99.Favaudon V. On the mechanism of reductive activation in the mode of action of some anticancer drugs. Biochimie. 1982;64:457–475. doi: 10.1016/s0300-9084(82)80162-4. [DOI] [PubMed] [Google Scholar]
- 100.Cross M., Taylor M.C., Borst P. Frequent loss of the active site during variant surface glycoprotein expression site switching in vitro in Trypanosoma brucei. Mol. Cell. Biol. 1998;18:198–205. doi: 10.1128/mcb.18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kojic M., Zhou Q., Lisby M., Holloman W.K. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol. Cell. Biol. 2005;25:2547–2557. doi: 10.1128/MCB.25.7.2547-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martin J.S., Winkelmann N., Petalcorin M.I., McIlwraith M.J., Boulton S.J. RAD-51-dependent and- independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Richard G.F., Kerrest A., Lafontaine I., Dujon B. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol. Biol. Evol. 2005;22:1011–1023. doi: 10.1093/molbev/msi083. [DOI] [PubMed] [Google Scholar]
- 104.Takanami T., Mori A., Takahashi H., Horiuchi S., Higashitani A. Caenorhabditis elegans Ce-rdh-1/rad-51 functions after double-strand break formation of meiotic recombination. Chromosome Res. 2003;11:125–135. doi: 10.1023/a:1022863814686. [DOI] [PubMed] [Google Scholar]
- 105.Borst P., Rudenko G., Taylor M.C., Blundell P.A., van Leeuwen F., Bitter W., Cross M., McCulloch R. Antigenic variation in trypanosomes. Arch. Med. Res. 1996;27:379–388. [PubMed] [Google Scholar]
- 106.Barry J.D. The relative significance of mechanisms of antigenic variation in African trypanosomes. Parasitol. Today. 1997;13:212–218. doi: 10.1016/s0169-4758(97)01039-9. [DOI] [PubMed] [Google Scholar]
- 107.Llorente B., Symington L.S. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee J.H., Paull T.T. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 109.Ulbert S., Chaves I., Borst P. Expression site activation in Trypanosoma brucei with three marked variant surface glycoprotein gene expression sites. Mol. Biochem. Parasitol. 2002;120:225–235. doi: 10.1016/s0166-6851(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 110.Sheader K., te V.D., Rudenko G. Bloodstream form-specific up-regulation of silent vsg expression sites and procyclin in Trypanosoma brucei after inhibition of DNA synthesis or DNA damage. J. Biol. Chem. 2004;279:13363–13374. doi: 10.1074/jbc.M312307200. [DOI] [PubMed] [Google Scholar]
- 111.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]