Abstract
Background
We report the characterisation of the variable large protein (vlp) gene expressed by clinical isolate A1 of Borrelia recurrentis; the agent of the life-threatening disease louse-borne relapsing fever.
Methods
The major vlp protein of this isolate was characterised and a DNA probe created. Use of this together with standard molecular methods was used to determine the location of the vlp1B. recurrentis A1 gene in both this and other isolates.
Results
This isolate was found to carry silent and expressed copies of the vlp1B. recurrentis A1 gene on plasmids of 54 kbp and 24 kbp respectively, whereas a different isolate, A17, had only the silent vlp1B. recurrentis A17 on a 54 kbp plasmid. Silent and expressed vlp1 have identical mature protein coding regions but have different 5' regions, both containing different potential lipoprotein leader sequences. Only one form of vlp1 is transcribed in the A1 isolate of B. recurrentis, yet both 5' upstream sequences of this vlp1 gene possess features of bacterial promoters.
Conclusion
Taken together these results suggest that antigenic variation in B. recurrentis may result from recombination of variable large and small protein genes at the junction between lipoprotein leader sequence and mature protein coding region. However, this hypothetical model needs to be validated by further identification of expressed and silent variant protein genes in other B. recurrentis isolates.
Background
Expression of a variant surface antigen is utilised by a wide variety of pathogenic micro-organisms to evade the immune defences of the host, thereby prolonging the duration of infection and increasing the likelihood of transmission. In the early part of the last century microbiologists first observed this process in relapsing fever caused by Borreliae as the spirochaetes recovered from each relapse differed serologically. The classical molecular description of this phenomenon comes from studies of Borrelia hermsii, an aetiologic agent of tick-borne relapsing fever. Variable major proteins are the dominant surface antigen [1], which are involved in evasion of the host's immune system as Borreliae emerging at each relapse express a different antigenic form [2]. Detailed molecular analysis has revealed that the corresponding genes are arranged into silent and expressed copies on different plasmids [3,4]. Antigenic variation occurs by replacement of the entire open reading frame of the expressed gene by a previously silent one, or by activation of a previously silent downstream gene [3-8]. Recently the variant major proteins, formerly denoted Vmp, have been divided into two groups based on size, namely the variant large proteins (encoded by vlp genes of typical size 1 kbp) and variant small proteins (encoded by vsp genes of typical size 0.6 kbp).
The present study considers the related organism B. recurrentis, the causative agent of louse-borne relapsing fever (LBRF). LBRF is a systemic inflammatory disease, which, if untreated, is associated with mortality rates of up to 40% [9]. This is reduced to 2–4% by use of antibiotics. Although the disease is relatively localised at present, with an endemic focus in Ethiopia, the potential for epidemics remains. Indeed the most recent outbreak occurred in the Sudan [10]. The disease is characterised by high fevers due to production of inflammatory mediators by the host in response to spirochaetal toxins [11,12]. It has been shown that the major pro-inflammatory molecule of B. recurrentis clinical isolate A1 is highly homologous to Vlp12B. hermsii HS1, and is here referred to as Vlp1B. recurrentis A1[13].
Studies of the molecular biology of B. recurrentis have been hampered by the lack of an animal model. The first cultivation of a clinical isolate in vitro was achieved in 1993 [14] and to date eighteen clinical isolates have been characterised by their plasmid profile and by the expression of a putative Vlp or Vsp [15]. As the first step to study antigenic variation in B. recurrentis, we report the complete organisation and characterisation of vlp1B. recurrentis A1 gene coding for the variant surface antigen of this isolate.
Methods
Borrelia recurrentis isolates
Isolates A1 and A17 from Ethiopian patients with LBRF and were subcultured in vitro as previously described [14,15]. There was no change in protein profiles or plasmid DNA banding patterns during this investigation compared with those observed for each isolate following original isolation from patients described previously [14,15].
Nucleic acid purification
Plasmid and chromosome-rich DNA fractions were prepared as described [30]. Total RNA was purified using the TRI Reagent method [31] (Sigma) and resuspended in RNase-free water.
Probe labelling
The 715 bp PCR product amplified from part of the expressed vlp1B. recurrentis A1 gene [13] was labelled with a32P-dCTP by random-priming (Amersham-Pharmacia Biotech). Oligonucleotide probes (LLS1, CACCA CTATCATCATCATCAAT and LLS2, TCATCACCATCACCAGCACC) were labelled with ?32P-ATP by enzymatic phosphorylation with polynucleotide kinase (Roche Molecular Biochemicals; the sequences corresponding to the probes LLS1 and LLS2 are double-underlined in Fig. 2). All radioactive probes were purified (Chromaspin-10, Clontech) prior to hybridisation.
Figure 2.
Ungapped DNA sequence alignment of the two forms of vlp1B. recurrentis A1 The upper row represents the sequence derived from the slower migrating 54 kbp plasmid of figure 1C. The sequence corresponding to the UP element consensus found in the bottom row sequence is italicised. The putative '-35' and '-10' elements and ribosome binding sites are underlined. Translation start sites are shown in bold. Region complementary to LLS1 and LLS2 probes are double-underlined. Accession number for both sequences are given in the methods section.
Library construction and screening
Plasmid DNA from isolate A1 was digested to completion with HindIII. Fragments of relevant sizes were gel purified using QIAEXII kit (Qiagen) and ligated in pBluescript linearised with HindIII. A portion of the ligation reaction was used to transform library efficient E. coli DH5a strain (GIBCO-BRL). Colonies were lifted onto Hybond-N membranes (Amersham-Pharmacia Biotech) and hybridised at high stringency with the 715 bp vlp1 probe as described [32]. After a second round of screening, positive clones were sequenced on an ABI377 automated sequencer using BigDye Terminator (Applied Biosystem). Sequences were assembled and analysed using the Wisconsin Genetics Computer Group, Madison, WI software package.
Southern hybridisation
Eight hundred nanograms of DNA were digested with a range of restriction enzymes. Digestion products were fractionated on a 1% agarose TAE gel. After electrophoresis the gel was treated as described [32] prior to capillary transfer onto Hybond-N membranes (Amersham-Pharmacia Biotech) in 10 × SSC overnight. Following UV-crosslinking, blots were hybridised in Rapid-Hyb solution (Amersham-Pharmacia Biotech) and washed using high stringency conditions (0.5 × SSC/0.1% SDS and 0.1 × SSC/0.1% SDS at 65°C each for 15 minutes).
Field inversion gel electrophoresis
One microgram of plasmid DNA from isolates A1 or A17 were separated by field inversion gel electrophoresis on a 1% agarose, 0.5 × TBE gel using the predefined programme P1 to separate DNA fragments between 150 kbp and 5 kbp (Biorad).
Northern hybridisation
One and a half microgram of total RNA were separated on a formaldehyde agarose gel and transferred onto a Hybond-N membrane in 10 × SSC. Following UV-crosslinking, blots were hybridised in Rapid-Hyb solution (Amersham-Pharmacia Biotech) and washed using high stringency. When labelled oligonucleotides were used as probes, hybridisations were carried out at 1°C below the calculated melting temperature.
Database accession numbers
Expressed vlp1B. recurrentis A1: AJ237608; Silent vlp1B. recurrentis A1: AJ237609.
Results
vlp1B. recurrentis A1 is plasmid encoded
A probe encoding a 715 bp portion of the expressed vlp1B. recurrentis A1, generated by microsequencing [13] was used to characterise vlp1 in two distinct B. recurrentis isolates. Plasmids and chromosome-rich extracts from isolate A1 were digested with several restriction endonucleases. This was followed by Southern hybridisation with the 715 bp vlp1 probe. Restriction enzymes that do not cleave the region of DNA encompassing the hybridisation probe were used.
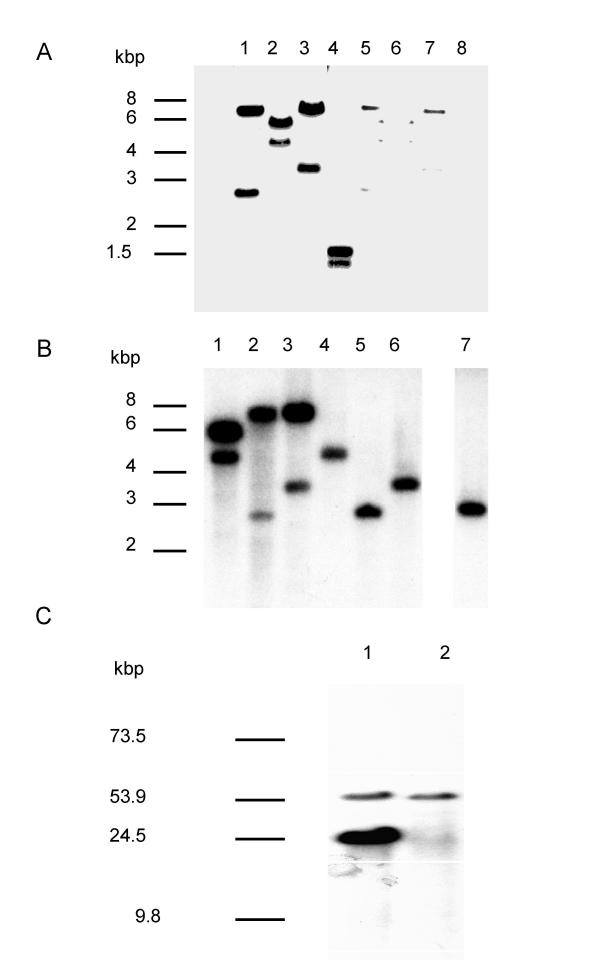
As shown in figure 1A, two bands were detected in the plasmid extract under high stringency hybridisation conditions. A third band of 43 kbp was observed at lower stringency conditions (data not shown). In contrast, the chromosomal-rich extract gave only weak hybridisation signals, probably caused by contaminating plasmid DNA. When the same experiment was performed on total and plasmid DNA from isolate A17 under high stringency, only one band could be detected (fig. 1B; lane 4 to 7).
Figure 1.
vlp1B. recurrentis A1 is plasmid encoded and duplicated in isolate A1. A: Plasmid (lane 1 to 4) and chromosome-rich (lane 5 to 8) DNA were digested with EcoRI (lane 1 and 5), HindIII (lane 2 and 6), XbaI (lane 3 and 7), and DraI (lane 4 and 8), transferred to a membrane and hybridised under high stringency with the 715 bp vlp1 probe. B: Plasmid DNA from isolate A1 (lane 1 to 3) and isolate A17 (lanes 4 to 6) and total DNA from isolate A17 (lane 7) were digested with HindIII (lane 1 and 4), EcoRI (lane 2, 5, and 7), XbaI (lane 3 and 6), and treated as described above. C: Intact plasmids from isolate A1 (lane 1) and isolate A17 (lane 2) were separated by field inversion gel electrophoresis (predefined program P1, Biorad) and treated as described above.
To confirm these findings, plasmids from both isolates were separated using field inversion gel electrophoresis (FIGE). Ethidium bromide staining revealed six major plasmid species in the range of 10 to over 100 kbp (not shown). Hybridisation under high stringency with the vlp1 probe detected a common band between isolate A1 and A17 corresponding to a plasmid of about 54 kbp (fig. 1C). This was also detected by the vlp1 probe in 10 further isolates possessing the 54 kbp plasmid. However, an extra band was detected in isolate A1 corresponding to a plasmid of about 24 kbp. Similarly, this 24 kbp band was also present in three other isolates falling into the same group of B. recurrentis[15]. Densitometry analysis showed that the signal intensity of the 24 kbp plasmid was about 3 to 4 times higher than the intensity given by the 54 kbp plasmid. These results show that vlp1B. recurrentis is plasmid-encoded and in isolate A1 the two vlp1 loci are localised on different plasmids. These results also raise the possibility that silent and expressed copies of vlp1 are present in B. recurrentis A1, whereas isolate A17 possesses only the silent copy.
Sequence of the vlp1B. recurrentis A1 loci
To investigate further the vlp1B. recurrentis loci we screened a partial library of isolate A1 plasmid DNA with the vlp1 probe. Positive clones of two different sizes were obtained, corresponding to thevlp1 loci carried on the 54 and 24 kbp plasmids depicted in figure 1C. The ungapped alignment of the two copies of vlp1B. recurrentis A1 is presented in figure 2; sequence 1 is derived from the slower migrating 54 kbp plasmid. Overall these two sequences are about 65% A/T rich in keeping with the low G/C content of borrelial DNA [16]. There is an extensive region (1461 bp) of strict identity that starts in the vlp1B. recurrentis open reading frame and extends beyond the stop codon. An additional feature of this region is the presence downstream of the open reading frame starting at nt1385 of a 300 bp segment that possesses 73% identity with the vlp1B. recurrentis A1 gene itself. Analysis of the translated sequences showed that the N-terminal component is not conserved, the identity between the two sequences starts one residue before the unique cysteine. Both N-terminal sequences code for a putative lipoprotein leader signal sequence [17,18], consistent with the finding that Vlp1B. recurrentis A1 has a lipid modification [13]. The divergence in the 5' end of the vlp1 coding region extends upstream where the two sequences are highly dissimilar. Interestingly, the upstream region of sequence 2 is richer in pyrimidine than that of sequence 1 (pyrimidine content of 59% and 34% respectively). However, both upstream regions possess a potential ribosome binding site and imperfect -10 and -35 regions, the consensus recognition site for s 70 RNA polymerase. In addition sequence 2 has a nearly perfect consensus UP element [19], the binding site for the C-terminal domain of the RNA polymerase, and an unusually long uninterrupted tract of 42 dT. Of note, another potential translational start is found in sequence 2 at nucleotide 191, with the -10 region starting at position 141 (not shown).
Taken together these results show that the two vlp1B. recurrentis A1 genes code for the same mature protein but have a different lipoprotein leader sequence. In addition, the presence of putative promoter features in upstream regions of both genes leaves the question of silent and expressed copies unresolved.
vlp1B. recurrentis is organised into silent and expressed copies
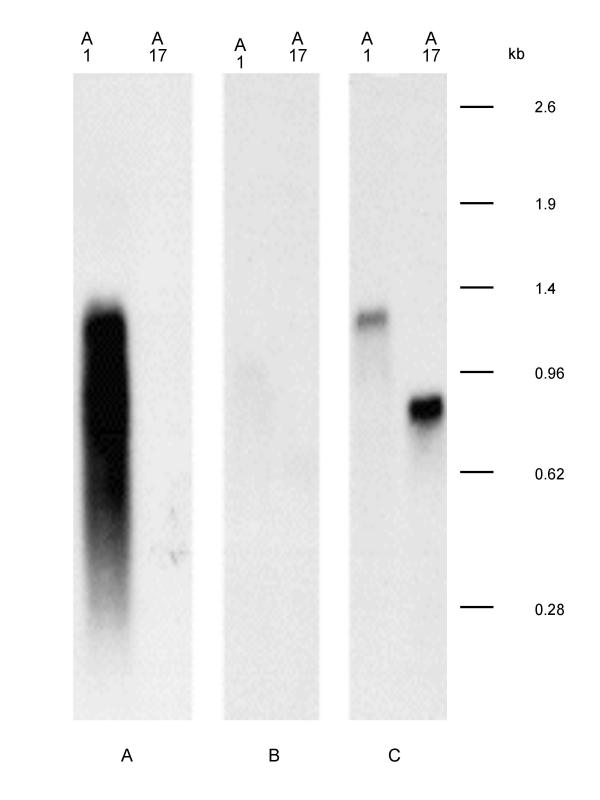
The total RNA were extracted from isolates A1 and A17 respectively, and Northern hybridisation was performed using oligonucleotide probes specific for the two lipoprotein leader signal sequences (LLS1 & LLS2), and the partial vlp1 probe. When the vlp1 probe was used, a strong hybridisation signal corresponding to a mRNA of around 1.2 kb was obtained only from isolate A1 (Fig. 3). This illustrates that vlp1B. recurrentis A1 is expressed only in isolate A1, but not in A17. No signal was detected when the oligonucleotide probe LLS1 specific for leader sequence 1 was used. In contrast, the LLS 2 specific probe detected a 1.2 kb RNA corresponding to vlp1B. recurrentis A1 message only in RNA extracted from isolate A1. Of note, probe LLS2 detected a message of about 800 b only in isolate A17 RNA. These results demonstrate that there is a silent and expressed copy of vlp1 in B. recurrentis A1; sequence 2 in figure 2 representing the expressed copy of vlp1B. recurrentis A1.
Figure 3.
vlp1B. recurrentis A1 is organised into silent and expressed copies. Total RNA from isolates A1 and A17 were separated on an agarose gel, blotted to a membrane and hybridised with specific probes. A: 715 bp vlp1; B: LLS1-specific probe; C: LLS2-specific probe
Discussion
The dominant surface antigen of Borrelia is a variant protein encoded by a large repertoire of genes, only one of which is expressed at anyone time. In contrast to other borreliae, the study of antigenic variation in B. recurrentis has been hampered by the lack of an animal model of infection. Isolates can only be recovered from patients and further characterised in vitro. Nevertheless, as the first step to better understand this phenomenon, we report the first complete characterisation of vlp1B. recurrentis A1 gene from a clinical isolate. This gene has common features with variant protein genes from other Borrelia species but has some intriguing features.
In isolate A1, which expresses the Vlp1B. recurrentis A1 protein [13,15], there are two copies of vlp1B. recurrentis A1, one on a 54 kbp plasmid and the other on a 24 kbp plasmid. This is reinforced by the observation of hybridisation with these two plasmids and the 715 bp vlp1 probe in other A1 group members (isolates A2–A4) [15]. The third plasmid, which hybridised at lower stringency, may carry an A1-like, but different vlp gene. In isolate A17, which expresses a different form of variant protein, vlp1B. recurrentis A17 is found only on the 54 kbp plasmid. Sequence analysis shows that absolute identity between the two vlp1 copies in isolate A1 starts 5 nucleotides upstream of the unique cysteine codon. This cysteine is the N-terminal amino acid of the mature Vlp1B. recurrentis and is the attachment point for the lipid modification of the variant protein [17,20]. Upstream of this region the two copies are divergent but both sequences fit the lipoprotein leader consensus sequence. Using oligonucleotides probes specific for these two leader sequences we demonstrate by Northern analysis that only the vlp1B. recurrentis A1 copy present on the 24 kbp plasmid is expressed in isolate A1. This result suggests that silent vlp1B. recurrentis A1, carried on a 54 kbp plasmid, may have undergone a duplicative translocation to the active expression site on a 24 kbp plasmid. Of note, 12 the 18 B. recurrentis clinical isolates described to date, one of which is isolate A17, possess a plasmid migrating in the region of 24 to 31 kbp [15].
This organisation is similar to that of other relapsing fever borreliae in which expressed and archived variant protein genes are located on different plasmids [6,21,22]. In these borreliae, the use of an animal model of infection has allowed the precise delineation of events taking place during an antigenic switch. In B. hermsii, antigenic variation occurs by replacement of one variable gene with another downstream from a telomeric promoter. In B. turicatae, gene conversion is more extensive, involving 10 or more kilobases downstream of the promoter giving rise to changes in the size of the plasmid carrying the expression site [22]. However, a third of B. recurrentis clinical isolates do not possess a plasmid migrating in the 24 kbp region [15]. Hence, it is possible that this plasmid was lost during in vitro cultivation or alternatively, as these isolates express a major antigen [15], there may exist an expression site located on a different plasmid. Indeed, activation of an alternative expression site has already been described in B. hermsii[23], utilised for expression of the vector transmission associated protein Vtp33 [24].
Based on the sequence alignment between silent and expressed vlp1B. recurrentis A1, it is tempting to speculate that the recombination site having given rise to expressed vlp1 is located where the two sequences become identical; that is within the open reading frame itself. Such a model would be supported by the finding that the probe specific for the lipoprotein leader sequence of the expressed vlp1B. recurrentis recognises a message of around 800 bases in RNA purified from isolate A17. The size of this message is in agreement with the theoretical size of the major antigen, Vsp17B. recurrentis A17, expressed by isolate A17 as determined previously by protein gel electrophoresis [15]. Further analysis of sequences from other variant protein genes should answer this point.
It is not clear how a rapid and successful antigenic switch is achieved in either B. recurrentis or B. hermsii[25] since for both species there is an approximately threefold excess of the expression plasmid compared to the silent plasmid. Clearly, maintenance and copy number for the two plasmids are differentially controlled and this may provide an opportunity to specifically modulate the copy number of the silent plasmid prior to a switching event.
The upstream region of both expressed and silent vlp1B. recurrentis A1 possesses potential promoter sequence, whose way of functioning remains speculative. Firstly, the expressed vlp1 contains an unusually long polydT tract that starts 167 nucleotides from the putative initiating codon. Shorter polydT tracts closer to the initiating codon (adjacent to the -35 region of the promoter) of two lipoprotein genes of other Borrelia species have been shown to have a role in transcriptional regulation [26]. It may be that this region replaces the UP element since such a consensus sequence could not be found in these promoters [26]. Unlike the expressed vlp1B. recurrentis A1 where there is a nearly perfect UP element consensus sequence there exists a long polydT tract. Silencing of the archived variant protein gene copies in B. hermsii is due to the absence of a promoter upstream of these genes [6], whereas in B. turicatae some silent variant protein genes lack promoter features while others lack both a promoter signature and an initiation codon [22]. Unexpectedly, we find that the silent copy of vlp1B. recurrentis A1 contains all the features necessary for expression. Although the upstream sequences of silent and expressed vlp1B. recurrentis A1 are dissimilar, both copies have a consensus lipoprotein leader sequence and putative characteristic features of a constitutive promoter. These observations are based on the assumption that promoters in B. recurrentis are similar to those in Escherichia coli. However, only mRNA corresponding to the expressed copy could be detected in isolate A1. It is possible that the promoter present in the silent copy is not recognised as functional. An alternative interpretation is that in B. recurrentis transcription of the silent copy of the vlp1B. recurrentis A1 gene is actively repressed. Such a silencing mechanism to prevent inappropriate expression of a variant protein gene has not been reported previously for Borreliae [6,27,28] and may be unique to B. recurrentis. Active repression of surface antigen gene expression has been described in trypanosomes and is achieved by telomere silencing [29] and further investigation is required to determine whether a similar process operates here. In that context, it would be interesting to pinpoint the location of the silent vlp1B. recurrentis A1 gene relative to plasmid ends.
Conclusions
The clinical isolate of B. recurrentis, A1, was demonstrated to contain two copies of a vlp1 gene on plasmids of 54 and 24 kbp. In a further isolate, A17, not expressing this gene, the vlp1 gene was only present on a 54 kbp plasmid. Northern blotting confirmed the expression site was located on the 24 kbp plasmid. The cloning and sequencing of the vlp1B. recurrentis A1 loci have highlighted the broad similarities in the organisation of variant protein genes in relapsing fever Borreliae. However this work has also uncovered some putative features that may be unique to B. recurrentis and that warrant future studies. Unlike other relapsing fever Borrelia studied to date, the upstream sequences of both silent and expressed genes contain features necessary for expression, suggesting the possibility of control of transcription through a silencing mechanism.
Authors' contributions
VV carried out the majority of the experimental work reported for A1 and A17. SC obtained the isolates during field investigation and independently located bands carrying A1-like genes in all 18 clinical isolates. IS undertook some of the experimental investigations. VV did the initial drafting of the manuscript with assistance from DK, SC and DW. These authors also assisted in the conception and design of the study.
Competing interests
None declared.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was supported by the Medical Research Council (UK) and the Wellcome Trust.
Contributor Information
Vincent Vidal, Email: vfvidal@yahoo.fr.
Sally Cutler, Email: s.cutler@vla.defra.gsi.gov.uk.
Ian G Scragg, Email: i.g.scragg@dundee.ac.uk.
David JM Wright, Email: d.j.wright@ic.ac.uk.
Dominic Kwiatkowski, Email: dominic.kwiatkowski@paediatrics.ox.ac.uk.
References
- Barbour AG, Tessier SL, Stoenner HG. Variable major proteins of Borrelia hermsii. J Exp Med. 1982;156:1312–1324. doi: 10.1084/jem.156.5.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoenner HG, Dodd T, Larsen C. Antigenic variation of Borrelia hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitten T, Barbour AG. Juxtaposition of expressed variable antigen genes with a conserved telomere in the bacterium Borrelia hermsii. Proc Natl Acad Sci U S A. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH, Simon MI, Barbour AG. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- Barbour AG. Antigenic variation of a relapsing fever Borrelia species. Annu Rev Microbiol. 1990;44:155–171. doi: 10.1146/annurev.mi.44.100190.001103. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Burman N, Carter CJ, Kitten T, Bergström S. Variable antigen genes of the relapsing fever agent Borrelia hermsii are activated by promoter addition. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- Donelson JE. Mechanisms of antigenic variation in Borrelia hermsii and African trypanosomes. J Biol Chem. 1995;270:7783–7786. doi: 10.1074/jbc.270.14.7783. [DOI] [PubMed] [Google Scholar]
- Restrepo BI, Carter CJ, Barbour AG. Activation of a vmp pseudogene in Borrelia hermsii: an alternate mechanism of antigenic variation during relapsing fever. Mol Microbiol. 1994;13:287–299. doi: 10.1111/j.1365-2958.1994.tb00423.x. [DOI] [PubMed] [Google Scholar]
- Johnson WD., Jr . Borrelias species (relapsing fever). In: Mandell GL, Bennett JE, Dolin R, editor. In Principles and practice of infectious diseases. 4. New York: Churchill Livingstone; 1994. pp. 2141–2143. [Google Scholar]
- Orloski K, Tharmaphornpilas P, O'Leary D, Ryan M, Schreifer M, Shoo R, et al. Epidemic louse-borne relapsing fever, Rumbek County, Sudan 1998–9. Am J Trop Med & Hyg. 1999;61:223–4. Abstract 173. [Google Scholar]
- Fekade D, Knox K, Hussein K, Melka A, Lalloo DG, Coxon RE, Warrell DA. Prevention of Jarisch-Herxheimer reactions by treatment with antibodies against tumor necrosis factor alpha. N Engl J Med. 1996;335:311–315. doi: 10.1056/NEJM199608013350503. [DOI] [PubMed] [Google Scholar]
- Negussie Y, Remick DG, DeForge LE, Kunkel SL, Eynon A, Griffin GE. Detection of plasma tumor necrosis factor, interleukins 6, and 8 during the Jarisch-Herxheimer Reaction of relapsing fever. J Exp Med. 1992;175:1207–1212. doi: 10.1084/jem.175.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal V, Scragg IG, Cutler SJ, Rockett KA, Fekade D, Warrell DA, Wright DJ, Kwiatkowski D. Variable major lipoprotein is a principal TNF-inducing factor of louse-borne relapsing fever. Nat Med. 1998;4:1416–1420. doi: 10.1038/4007. [DOI] [PubMed] [Google Scholar]
- Cutler SJ, Fekade D, Hussein K, Knox KA, Melka A, Cann K, Emilianus AR, Warrell DA, Wright DJ. Successful in-vitro cultivation of Borrelia recurrentis. Lancet. 1994;343:242. doi: 10.1016/S0140-6736(94)91032-4. [DOI] [PubMed] [Google Scholar]
- Cutler SJ, Moss J, Fukunaga M, Wright DJ, Fekade D, Warrell D. Borrelia recurrentis characterization and comparison with relapsing-fever, Lyme-associated, and other Borrelia spp. Int J Syst Bacteriol. 1997;47:958–968. doi: 10.1099/00207713-47-4-958. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Venter JC, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- Klein P, Somorjai RL, Lau PC. Distinctive properties of signal sequences from bacterial lipoproteins. Protein Eng. 1988;2:15–20. doi: 10.1093/protein/2.1.15. [DOI] [PubMed] [Google Scholar]
- Estrem ST, Gaal T, Ross W, Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scragg IG, Kwiatkowski D, Vidal V, Reason A, Paxton T, Panico M, Dell A, Morris H. Structural Characterisation of the Inflammatory Moiety of a Variable Major Lipoprotein of Borrelia recurrentis. J Biol Chem. 2000;275:937–941. doi: 10.1074/jbc.275.2.937. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Restrepo BI. Antigenic variation in vector-borne pathogens. Emerg Infect Dis. 2000;6:449–457. doi: 10.3201/eid0605.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington PM, Cadavid D, Bunikis J, Norris SJ, Barbour AG. Extensive interplasmidic duplications change the virulence phenotype of the relapsing fever agent Borrelia turicatae. Mol Microbiol. 1999;34:1120–1132. doi: 10.1046/j.1365-2958.1999.01675.x. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Carter CJ, Sohaskey CD. Surface protein variation by expression site switching in the relapsing fever agent Borrelia hermsii. Infect Immun. 2000;68:7114–7121. doi: 10.1128/IAI.68.12.7114-7121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Hinnebusch BJ. Bloodstream-versus tick-associated variants of a relapsing fever bacterium. Science. 1998;280:1938–1940. doi: 10.1126/science.280.5371.1938. [DOI] [PubMed] [Google Scholar]
- Kitten T, Barbour AG. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics. 1992;132:311–32. doi: 10.1093/genetics/132.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Zuckert WR, Barbour AG. The extended promoters for two outer membrane lipoprotein genes of Borrelia spp. uniquely include a T-rich region. Mol Microbiol. 1999;33:41–51. doi: 10.1046/j.1365-2958.1999.01443.x. [DOI] [PubMed] [Google Scholar]
- Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1988;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/S0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Blundell PA, Dirks Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Garon CF. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–537. [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Cold Spring Harbor. 2. Cold Spring Harbor Press; 1989. Molecular Cloning: A Laboratory Manual, [Google Scholar]