Abstract
Background
Leukocyte Immunoglobulin-like Receptor-1 (LIR-1) and LIR-2 (also known as ILT2 and ILT4 respectively) are highly related cell surface receptors that bind a broad range of class I MHC molecules with low (μM) affinities. Expressed on monocytic cells and macrophages, both molecules transmit inhibitory signals after binding ligands. In addition to binding host class I MHC, the LIR-1 molecule, which is also expressed on lymphoid tissues, binds with a high (nM) affinity to UL18, a class I MHC homolog encoded by Human Cytomegalovirus (HCMV). In comparison, LIR-2 binds UL18 only weakly (μM KD). To understand how HCMV preferentially targets the more broadly expressed LIR-1 molecule, we determined the crystal structure of a ligand-binding fragment of LIR-2, and compared this to the existing high-resolution crystal structure of LIR-1.
Results
Recombinant LIR-2 (domains 1 and 2) was produced in E. coli and crystallized using streak seeding to optimize the crystal morphology. A data set complete to 1.8 Å was collected at 100 K from a single crystal in the P41212 spacegroup. The structure was solved by molecular replacement, using a search model based on the LIR-1 structure.
Conclusions
The overall structure of LIR-2 D1D2 resembles both LIR-1, and Killer Inhibitory Receptors, in that the A strand in each domain forms hydrogen bonds to both β sheets, and there is a sharp angle between the two immunoglobulin-like domains. However, differences from LIR-1 are observed in each domain, with two key changes apparent in the ligand-binding domain, D1. The region corresponding to the residue 44–57 helix of LIR-1 adopts a topology distinct from that of both LIR-1 and the KIR structures, involving a shortened 310 helix. Secondly, the predicted UL18 binding region of LIR-1 is altered substantially in LIR-2: the 76–84 loop mainchain is displaced 11 Å with respect to LIR-1, and Tyrosine 38 adopts an alternative rotamer conformation. In summary, the structure of LIR-2 has revealed significant differences to LIR-1, including ones that may help to explain the >1000-fold lower affinity of LIR-2 for UL18.
Background
The Leukocyte Immunoglobulin-like Receptor (LIR) family (also called the ILT's, MIR's and CD85) comprises a set of immunoreceptors expressed on the surface of lymphoid and myeloid cells [1,2]. Encoded from within the Leukocyte Receptor Cluster [3] (a region of human chromosome 19 which also encompasses the loci for the related Killer Inhibitory Receptor (KIR) and Leukocyte-Associated Immunoglobulin-like Receptor (LAIR) families), the LIRs are highly similar to one another (63–84% amino acid identity in the extracellular region). All except LIR-4 (which appears to encode a soluble molecule) encode type-1 transmembrane proteins, containing either two or four immunoglobulin superfamily (IgSF) domains in their extracellular regions. One subset of cell surface LIR molecules (LIR-1, -2, -3, -5, and -8) possesses Intracellular Tyrosine-based Inhibitory Motifs (ITIMs) in the cytoplasmic domain. Once phosphorylated, these sequences mediate association with intracellular SH2-domain-containing phosphatases, permitting transmission of inhibitory signals to the effector cell. Another group (LIR-6, -7, ILT7, -8, -11) includes short cytoplasmic domains containing no known signaling motifs. These molecules have a charged amino acid (arginine) within their transmembrane regions, enabling them to transmit activatory signals by associating with the adaptor molecule FcεRIγ.
The best characterized members of the LIR family are LIR-1 (ILT-2) and LIR-2 (ILT-4), both of which are inhibitory receptors with four extracellular IgSF domains, and cytoplasmic ITIM motifs mediating association with the phosphatase SHP-1 [4-7]. Both LIR-1 and LIR-2 bind to class I MHC molecules and are expressed on monocytes and dendritic cells [5]. However, unlike LIR-2, LIR-1 is also expressed on lymphoid tissues, namely all B-cells, as well as some T-cells and subsets of NK cells [5]. In vitro experiments in a range of immune effector cells, including monocytes, macrophages, dendritic cells and (for LIR-1) B-cells, have shown that upon cross-linking with antibodies, LIR-1 and LIR-2 can each abrogate activatory signals resulting in a potent inhibition of effector functions [5-7]. Similarly, engagement of class I MHC ligands by LIR-1 expressed on NK and T-cells has been shown to potently inhibit cytotoxicity [6,8].
Following the discovery that LIR-1 is the receptor for the Human Cytomegalovirus (HCMV) class I MHC homolog UL18 [4], both LIR-1 and LIR-2 were shown to bind a broad range of endogenous class I MHC molecules [5-7,9,10], with affinities in the micromolar (μM) range [11]. Domain-swapping experiments established that binding to both UL18 and class I MHC proteins is mediated by interaction between surfaces on the membrane distal (D1) domain of LIR-1 or LIR-2 and the α3 domain of either class I MHC or UL18 [11]. Despite the high sequence identity between LIR-1 and LIR-2 (82% in the extracellular region), and the similar mode and affinity of their interactions with endogenous class I MHC, the UL18 protein distinguishes effectively between the two receptors, binding LIR-1 with a nanomolar (nM) affinity, over 1000-fold more strongly than it binds to LIR-2 [11]. Recently, a crystal structure of the two membrane-distal domains of LIR-1 (D1-D2) indicated both similarities to and differences from the KIR receptor family [11]. A parallel mutagenesis study resulted in the identification of a group of residues on LIR-1 domain 1 that were proposed to form part of the binding site for UL18 [11]. In order to further investigate the structural features of the LIR family of immunoreceptors, and in particular the ability of UL18 to preferentially target the LIR-1 molecule, we determined the crystal structure of a ligand-binding fragment of LIR-2.
Results and Discussion
Production, characterization and crystallisation of LIR-2
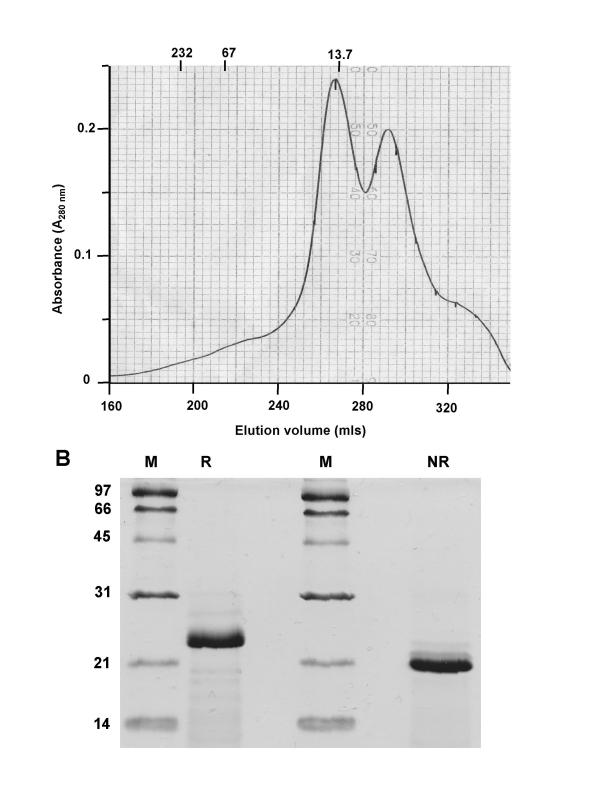
The N-terminal two domains of LIR-2 (residues 1–197) were produced in a manner similar to LIR-1, by overexpression in E. coli, followed by purification from inclusion bodies, and renaturation by dilution refolding [11]. Size-exclusion of refolded material (Figure 1A) indicated a peak of protein eluting before the 13.7 kDa marker protein, and subsequent SDS-PAGE analysis (Figure 1B) confirmed this contained predominantly one major band corresponding to the expected molecular weight of the LIR-2 D1-2 fragment (22 kDa). Under non-reducing conditions, this band migrated as one species of a higher mobility (Figure 1B), suggesting a single combination of disulphide bonds was formed during refolding. Peak fractions from the size-exclusion purification were collected, concentrated, and used directly for crystallization experiments. Initial trials yielded microcrystals, and following optimization of the conditions, streak seeding was used to grow single crystals of a size (approximately 0.16 mm × 0.16 mm × 0.05 mm) sufficient for data collection.
Figure 1.
Purification and characterization of soluble LIR-2 D1D2. A: Size exclusion chromatography of refolded LIR-2 D1D2. Refolded LIR-2 D1D2 elutes from a Superdex 200 column at just before a 13.7 kDa marker protein, consistent with the protein behaving as a compact monomer in solution. The elution volumes of 232, 67 and 13.7 kDa standard proteins are indicated. B: SDS-PAGE analysis of refolded LIR-2. Identical samples of size-exclusion purified LIR-2 D1D2 (~5 μg) were loaded onto a 15% acrylamide SDS gel, under either reducing (R) or non-reducing (NR) conditions. The molecular weights of marker proteins (M) are shown in kilodaltons. Under reducing conditions, LIR-2 migrates in a position consistent with its predicted molecular weight of 22 kDa. Under non-reducing conditions, LIR-2 D1D2 migrates as a single band of higher mobility, suggesting formation of the appropriate disulfide bonds. No higher molecular weight cross-linked species are observed.
Crystal structure of LIR-2
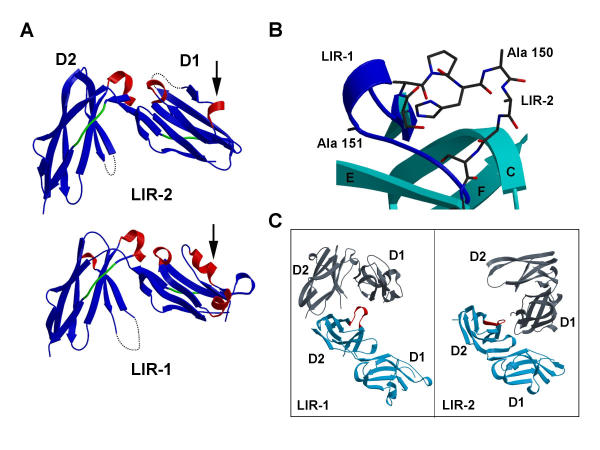
The crystal structure of LIR-2 D1D2 was determined in the space group P41212 to 1.8 Å by molecular replacement using the LIR-1 D1D2 structure [12]. Overall, the structure closely resembles that of LIR-1 in that it consists of two Ig-like domains related by an approximately orthogonal angle, forming a highly bent structure (Figure 2A). As with LIR-1, each domain is composed primarily of β strands arranged into two antiparallel sheets, with a KIR-like folding topology, whereby the first strand of each domain forms hydrogen bonds to both the B and G strands, thereby bridging the two β sheets [12].
Figure 2.
LIR-2 D1D2 Crystal structure. A: Ribbon diagram of the structure LIR-2 D1D2 and comparison with the LIR-1 D1D2 structure. Dashed lines indicate disordered loops. Arrows indicate the single turn of 310 helix observed in LIR-2 D1 (residues 52–55), in comparison to the longer 310 helical segment (residues 44–57) in the same region of LIR-1 D1. 410 and 310 helices are shown in red, Polyproline II helices in green. The root mean square deviation between the two structures is 3.05 Å for domain 1 and 2.31 Å for D2 for all carbon alpha atoms. B: Conformational change in D2 146–154 loop relative to LIR-1. The LIR-2 loop is shown in ball-and-stick representation. The LIR-1 loop is shown in blue as a ribbon diagram. The conformational difference is maximal at residue Ala 150 (equivalent to Ala 151 in LIR-1), where the Cα positions differ by 14 Å. C: Crystal contacts involving the D2 146–154 loop for LIR-2 and LIR-1 (147–155). Both LIR-1 (left panel) and LIR-2 (right panel) are shown in cyan with the loop region highlighted in red, and the equivalent neighboring molecule shown in grey.
The structure of LIR-1 D1D2 revealed helical regions interspersed with β structure in each of the two domains [12]. In particular, the region of each domain corresponding to the C' strand of KIRs contained some helical structure, a feature that was not observed in the related KIR and Fc-receptor families or other IgSF domains. In each domain, LIR-2 contains significantly less helical structure than LIR-1 in this C' strand region. In D1, a single helical turn displaying a 310 hydrogen bonding pattern is observed over residues 52–55, whereas the preceding region (residues 48–50) adopts a β-strand conformation as observed in KIR receptors [13-15]. In contrast, this entire region of LIR-1 D1 (residues 44–57) adopts a helical conformation, interrupted only by a proline at position 51 [12]. This change in topology is accompanied by a significant shift in the LIR-2 main chain relative to LIR-1 D1 over residues 48–50, a conformation stabilized by main chain hydrogen bonding with the C strand. Such an arrangement, with retention of the C' strand followed by a single turn of 310 helix, is similar to that observed in LIR-1 D2 [12].
In the region corresponding to the C' strand of D2, topological differences from LIR-1 are also observed. The single turn of 310 helix observed in LIR-1 (residues 148–150) is absent in LIR-2. Instead, this and the adjacent D strand (151–153) are replaced by a region incorporating a hydrogen bonded turn. As a result of these rearrangements, the two structures differ substantially over residues 146–154 (equivalent to 147–155 in LIR-1), with a maximum divergence around Ala 150 (equivalent to Ala 151 in LIR-1), where the Cα positions are shifted by ~14 Å (Figure 2B). The turn region is stabilized by two internal hydrogen bonds (between the carbonyl oxygen of His 149 and the main chain nitrogen of Gly 152; also between the side chain hydroxyl of Ser 153 and the carbonyl oxygen of Arg 155) and by four main chain hydrogen bonds to residues connecting the D2 B and C strands (Ser 123, Phe 127, Gly 128, Phe 130). This conformational change between the structures is unexpected as the two proteins are identical in sequence over this region, and most probably reflects the substantially different packing in the LIR-1 and LIR-2 crystals (Figure 2C). In the LIR-2 crystals, the 146–154 turn appears relatively unconstrained, packing loosely against the D1 domain of a neighboring molecule, and forming a single ≤ 3.6 Å contact from Pro 148 to Gln 27. In contrast, the analogous neighboring molecule is oriented differently in the LIR-1 structure, packing more closely, and forming direct contacts to the 147–155 region from both the D1 and D2 domains (Figure 2C). Furthermore, an additional D2-D2 crystal contact specific to the LIR-2 crystal appears to be incompatible with the LIR-1 conformation.
In contrast with these changes, regions of polyproline II helix observed in the F-G loop regions of LIR-1 are preserved in both LIR-2 domains. These left-handed helices are characterized by a three residue repeat and phi, psi angles near -75°, 145° [16], and include the sequences WSELS (D1) and WSSPS (D2), both variations of the WSXWS sequence motif of haematopoietic receptors [17]. In each case, main chain nitrogen and side chain serine hydroxyls in positions n and n + 3 form hydrogen bonds to the main chain of the F strand, a feature also observed for cytokine receptors [18-21], KIRs [13-15] and fibronectin III repeats [22].
LIR-2 interdomain interface
LIR-2 maintains a similar interdomain angle to LIR-1 (88% versus 90% for LIR-1 in the same space group), consistent with conservation of the majority (8/13) of the interdomain contact residues. However, superimposition of the D1 domains of LIR-1 and LIR-2 reveals a slight lateral shift in the orientation of D2, such that residues at the tip of D2 distal to the interdomain interface are shifted by approximately 5 Å. The most significant amino acid change at the LIR-2 interface involves the introduction of a larger Met residue for Val at residue 94 (equivalent to residue 95 in LIR-1). This causes a reorientation of Trp 67 and an accompanying shift in the main chain of 2 Å at this position. The Ala to Thr alteration at residue 70 appears to stabilize this shift, by introducing a hydrogen bond between the Thr hydroxyl group and the carbonyl oxygen of Trp 67.
Conformational differences from LIR-1 in the UL18-binding region of domain 1
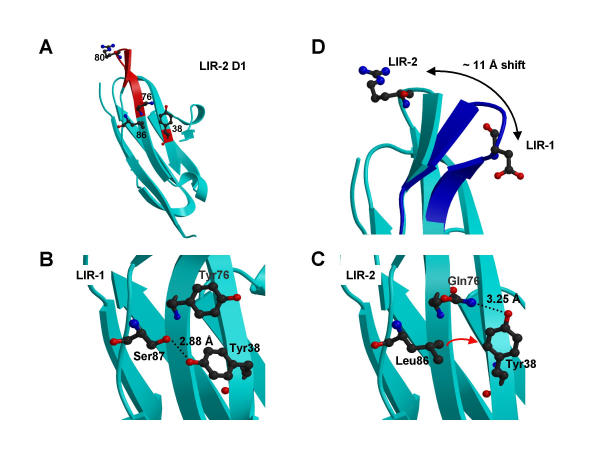
A mutagenesis study of LIR-1 identified residues in the membrane-distal tip of the A'CC'FG face of D1 as important for UL18-binding [12]. This region (Figure 3A) is distant from the D1D2 elbow region that KIRs use to contact class I MHC molecules [13,23]. Mutation of LIR-1 Tyr 38 to Ala reduced UL18 binding affinity by ~18-fold, and a triple mutant (Y76A/D80A/R84A) showed a similar (~20-fold) reduction in affinity [12]. The analogous residues in LIR-2 (Tyr 38, Gln 76, Arg 80 and Trp 83) either exhibit an altered conformation, a different side chain, or both. Although Tyr 38 is conserved in both structures, the sidechain positions differ: in LIR-1, Tyr 38 is a solvent-exposed residue on the C strand, which points towards the D1 RSESS motif connecting the F and G strands and is within hydrogen-bonding distance of the hydroxyl oxygen of Ser 87 (Figure 3B). In LIR-2 the residue equivalent to Ser 87 is a leucine, and as a result Tyr 38 reorients such that it points towards the N-terminus of the C strand with its hydroxyl group within hydrogen-bonding distance of the amino group of Gln 76 (Figure 3C). The other residues implicated in LIR-1 binding to UL18 (equivalent to LIR-2 residues 76, 80, and 83) are located in the F to G turn (residues 76–84). In LIR-2, this region contains a one residue deletion relative to LIR-1, and two glycine to non-glycine changes (G78 and G83, corresponding to Y78 and R82 in LIR-2), and undergoes a significant conformational shift relative to LIR-1. Whereas this region protrudes from the A'CC'FG face of D1 in LIR-1, in LIR-2 it is oriented closer to residues 28–32 connecting the B and C strands (Figure 3D). In each case the conformation is stabilized by three main chain hydrogen bonds within the turn region, and for LIR-2 the shift is accompanied by an additional hydrogen bond between the carbonyl oxygen of Tyr 78 and the main chain NH group of Gln 33. The conformational shift in this region culminates at residue 80, where the Cα positions are greater than 11 Å apart (Figure 3D).
Figure 3.
Alteration of the UL18-binding site in LIR-2 D1. A. Structure of LIR-2 D1. The region of LIR-2 analogous to the proposed UL18-binding site in LIR-1. Main chain regions of the 76–84 F-G loop and Tyr 38, both of which implicated in LIR-1/UL18 interaction, are indicated in red. Side chains for LIR-2 residues 38 (Tyr), 76 (Gln), 80 (Arg) and 86 (Leu) are also shown. B. Conformation of Tyr 38 in LIR-1. Likely hydrogen bonds in (B) and (C) are shown as dotted lines. C. Conformation of Tyr 38 in LIR-2. The Ser (87) to Leu (86) change would cause a steric clash with the LIR-1 conformation of Tyr 38, and forces a side chain reorientation towards Gln 76. In LIR-1, substitution of Gln 76 for Tyr would similarly prevent adoption of the LIR-2 Tyr 38 orientation due to steric hindrance. D. Conformational shift of the 76–84 loop region. The LIR-1 loop (dark blue) protrudes from the side of the domain
In LIR-1, the region identified as the UL18 binding site is involved in crystal contacts to a symmetry-related molecule [12]. Similarly, the 76–84 loop of LIR-2 is involved in crystal contacts to another D1D2 protein, although in LIR-2 contacts are made to residues on the F and G strands of the D2 domain of a neighbouring molecule, whereas in LIR-1 they are to the same UL18-binding surface on the D1 domain of the neighbouring molecule. Nevertheless, the fact that in each structure this region of the molecule is involved in crystal contacts, and the relatively large total surface area involved (810–870 Å2 for LIR-1 and 877 Å2 for LIR-2 compared to an average of 570 Å2 for non-specific crystal contacts [24]) may reflect the fact that this surface is well-suited to protein-protein interactions and is therefore used for both ligand binding and crystal packing.
Conclusions
The structure of LIR-2 D1D2 reported here closely resembles that of the highly related LIR family member, LIR-1. Both proteins are composed of two IgSF domains related by a nearly orthogonal angle to create a highly bent structure. The folding topologies of the two LIR-2 domains closely resemble their LIR-1 counterparts, although a reduction in helical content is observed in the LIR-2 domains. The D1-D2 interdomain angle is comparable to that of LIR-1 D1D2, in both cases being maintained by a set of conserved hydrophobic interactions at the interdomain interface.
Despite these similarities, the HCMV class I MHC homolog UL18 binds LIR-1 with an ~2 nM affinity, whereas UL18 binds LIR-2 with an ~14 μM affinity [12]. Furthermore, whereas a LIR-1 Fc fusion protein bound to cell surface UL18, no binding was observed between cell surface UL18 and a LIR-2 Fc fusion protein [25]. The structure of LIR-2 D1D2 reveals changes from LIR-1 in both domains, some of which could potentially provide a molecular basis for this discrimination. One of the most obvious changes involves a conformational difference in the region of D2 residues 146–154, but the sequence of this region is conserved between LIR-1 and LIR-2 [25], and the conformational change appears to be mediated by different intermolecular contacts in the LIR-1 and LIR-2 crystals. Consequently, while the two conformations observed may well indicate this region of the receptors displays a degree of flexibility in solution, it is unlikely to explain discrimination between LIR-1 and LIR-2 by UL18. In contrast, changes in D1 are more likely to significantly affect binding to UL18, since previous experiments suggest that D1 is the main site of interaction with UL18 (and class I MHC) [11]. Likely to be especially relevant are those changes observed in the predicted UL18-binding site, namely the reorientation of Tyr 38 and the 11 Å conformational change in the 76–84 region of the D1 F to G loop. Mutation of Tyr 38 to Ala reduced LIR-1/UL18 binding by ~18 fold [12]. In LIR-1, the hydroxyl oxygen of Tyr 38 is oriented towards the RSESS motif connecting the F and G strands, and hydrogen bonds to Ser 87. However, in LIR-2 a Ser to Leu change at this position forces a reorientation of the Tyr 38 side chain (Figure 3B), towards the N-terminus of the C strand, a conformation stabilized by hydrogen bonding to the amino group of Gln 76.
Another potentially important alteration in the UL18-binding site is a conformational change in the 76–84 F-G loop, mutation of which reduced UL18 binding by LIR-1 by ~20 fold [12]. One notable feature of the LIR-1 and LIR-2 structures is that in each case the 76–84 loop is involved in crystal contacts, and thus the structure in the crystal could represent only one of many conformations occupied in solution. Nevertheless, the changes observed in this region evident in the LIR-1 and LIR-2 crystal structures may well reflect energetic preferences for distinct loop conformations. It is notable that the LIR-1 loop is one residue longer and contains two glycine residues absent in the LIR-2 sequence, and therefore the LIR-2 loop may be considerably restricted in conformation relative to LIR-1. While amino acid differences in this loop could in themselves be sufficient to reduce UL18 binding, the main chain conformation adopted by LIR-2 might preclude formation of favorable contacts between this region and UL18, thereby contributing to a lower affinity for the viral ligand. In contrast, both receptors bind class I MHC proteins with similar (μM) affinities, and it may be that energetically significant contacts with this region are enhanced in the UL18-LIR-1 interaction relative to class I MHC interactions with either LIR-1 or LIR-2, where they may be either minimal or absent. Crystal structures of either receptor in complex with UL18 or class I MHC should help resolve some of these issues.
Materials and Methods
Protein expression
PCR was used to amplify the sequence encoding domains 1 and 2 of LIR-2 (residues 1–198 of the mature protein) from cDNA kindly provided by David Cosman. LIR-2 D1D2 was expressed in E. coli from the pET23a vector, purified from inclusion bodies essentially as described [26], and solubilized in 8 M Guanidine Hydrochloride, 50 mM Tris pH 8.0. Solubilized protein was renatured by dilution into a standard refolding buffer (400 mM L-Arginine, 100 mM Tris, 10 mM EDTA, 0.5 mM reduced Gluathione, 5 mM oxidized Glutathione, pH 8.3). The final concentration of protein was approximately 50 μg/ml. Renatured protein was concentrated, then purified by size exclusion chromatography using a Pharmacia Superdex 200 column, into 150 mM NaCl, 20 mM Tris pH 8.0). Purifed protein was concentrated and quantified by absorption measurements at 280 nm. An extinction coefficient (280 nm) for LIR-2 of 46785 M-1cm-1 was calculated using the method of Pace et al[27].
Crystallization, data collection and processing
All crystals were grown at 23°C. Initial microcrystals of LIR-2 were obtained by vapor diffusion in 1:1 μl hanging drops of 6.1 mg/ml protein equilibrated with a reservoir solution of 0.1 M Hepes pH 7.5, 10% Isopropanol, 20% PEG 4 K. Crystals of a quality sufficient for data collection were obtained by streak seeding from existing crystals into 5:5 μl sitting drops of 1.9 mg/ml protein equilibrated overnight with 50 mM MES pH 6.5, 5% Isopropanol, 10% PEG 4000, using a chinchilla whisker. A 1.8 Å native data set was collected at 100 K from a single crystal using a Quantum CCD Reseach Detector at the Stanford Synchrotron Radiation Laboratory beamline 9–2 (λ = 0.999 Å). The crystal was soaked in cryoprotectant solutions consisting of mother liquor incorporating 8.5%, 17% (10 minutes each), and then 25% Ethylene Glycol (30 minutes), prior to immersion in liquid nitrogen. Diffraction due to ice formation was reduced significantly by the use of flash annealing [28]. Data were processed and scaled using DENZO and SCALEPACK [29].
Structure solution, refinement and analysis
The structure of LIR-2 D1D2 was solved by molecular replacement using AmoRe [30]. A rotation and translation function was found using the 1.8 Å LIR-2 data set and the 2.1 Å structure of LIR-1 D1D2 as the search model (correlation coefficient 35%, R factor 49%). Rigid body refinement (20–4 Å) resulted in an Rcryst of 47% and an Rfree of 49%. Solvent flattened maps calculated to 2.5 Å were used for initial rebuilding. Anisotropy and bulk solvent corrections were applied and the model partially refined using individual temperature (B) factors with CNS [31] (Rcryst = 31%, Rfree = 32 %), after which cycles of real space and reciprocal space refinement were carried out, followed by addition of water molecules in the final stages of refinement. Residues at the N terminus of the fragment (1–2) and within two loops (42–46 and 136–141) are not seen in the electron density and were omitted from the model, and side chains for residues 3, 11, 47, 51, 56, 57 and 166 and 167 are disordered and were truncated to alanines. Disulphide bonds are observed between residues 26 and 75, 121 and 173, and 133 and 143. For analysis of interdomain angles, contacts and buried surface areas, D1 was defined as residues 1–97 and D2 was defined as residues 98–197, as for LIR-1 [12]. Interdomain contact residues were identified using the program CONTACT [32], and defined as residues containing an atom within 3.6 Å of the partner domain. Buried surface areas were calculated using SURFACE [32] with a 1.4 Å probe radius. Interdomain angles were calculated using the program Dom_angle [33], which determines the angle between the long axes of adjacent domains that are approximated by ellipsoids calculated from the coordinates. Figures 2 and 3 were prepared with Molscript [34] and Raster3D [35].
Authors' contributions
BEW expressed, purified and produced high quality crystals of LIR-2, carried out data collection and processing, structure solution, refinement and analysis and also wrote the manuscript. LMT carried out the latter stages of refinement and aided structural analyses. TLC produced initial microcrystals of LIR-2. APH carried out molecular cloning of the LIR-2 expression construct. APW provided help during data collection and processing, and critical scientific discussions. PJB conceived of the study, participated in its design and coordination, and provided financial support.
Table 1.
Data Collection and Refinement Statistics for LIR-2 D1D2
| Space Group | P41212 |
| Unit Cell Dimensions (Å) | 62.505, 62.505, 106.845 |
| Data Collection | |
| Resolution (Å) | 34.1–1.8 (2.0–1.8) |
| Number of Observed Reflections | 155116 |
| Number of Independent Reflections | 20290 |
| aCompleteness (%) | 99.35 (100) |
| bRmerge (%) | 4.0 (33.0) |
| I/σ (I) | 43.4 (5.9) |
| Refinement | |
| Resolution (Å) | 54.2–1.8 |
| Reflections in working set | 19241 (1405) |
| Reflections in test set | 991, equivalent to 4.9% |
| cRcryst (%) 23. | 23.4 |
| dRfree> (%) | 27.3 |
| Number of atoms | |
| Protein | 1559 |
| 184 of 198 residues | |
| Water | 161 |
| Average B factor | 33.87 Å2 |
| Anisotropic B correction | |
| B11 = B22 = 0.37 Å2, B33 = -0.74 Å2 | |
| Model Geometry | |
| Rms deviation from ideality Bond lengths | |
| Bond lengths (Å) | 0.021 |
| Bond angles (deg) | 1.858 |
| Ramachandran plot quality | |
| Most favoured Region of Ramachandran plot (%) | 91.90% |
| Additionally allowed Protein | 8.10% |
| Generously allowed | 0.0% |
| Disallowed | 0.0% |
aCompleteness = (number of independent reflections/total theoretical number) bRmerge (I) = (Σ|I(i) - <I(h)>|/ΣI(i)), where I(i) is the ith observation of the intensity of the hkl reflection and <I> is the mean intensity from multiple measurements of the h,k,l reflection. cRcryst (F) = Σh||Fobs(h)| - |Fcalc(h)||/Σh|Fobs(h)| and |Fcalc(h)| are the observed and calculated structure factor amplitudes for the h, k, l reflection. dRfree is calculated over reflections in a test set not included in atomic refinement. Figures in parentheses apply to data in the highest resolution shell.
Acknowledgments
Acknowledgements
We thank Drs Z.A. Hamburger, C. A. O'Callaghan and W.L. Martin for assistance with crystallographic software and valuable scientific discussions; D. Cosman for LIR-2 cDNA; S. Rowland-Jones for the kind gift of chinchilla whiskers; and members of the Bjorkman laboratory for critical reading of the manuscript. BEW is supported by a Wellcome Trust Traveling Fellowship (Grant code GR059939MF). Requests for coordinates should be addressed to BEW.
Contributor Information
Benjamin E Willcox, Email: b.willcox@bham.ac.uk.
Leonard M Thomas, Email: thomasle@its.caltech.edu.
Tara L Chapman, Email: taralynn@chem.ucsd.edu.
Astrid P Heikema, Email: Jeco.Astrid@tiscali.nl.
Anthony P West, Jr, Email: apwest@its.caltech.edu.
Pamela J Bjorkman, Email: bjorkman@its.caltech.edu.
References
- Cella M, Nakajima H, Facchetti F, Hoffmann T, Colonna M. ILT receptors at the interface between lymphoid and myeloid cells. Curr Top Microbiol Immunol. 2000;251:161–6. doi: 10.1007/978-3-642-57276-0_20. [DOI] [PubMed] [Google Scholar]
- Borges L, Cosman D. LIRs/ILTs/MIRs, inhibitory and stimulatory Ig-superfamily receptors expressed in myeloid and lymphoid cells. Cytokine Growth Factor Rev. 2000;11:209–17. doi: 10.1016/S1359-6101(00)00007-1. [DOI] [PubMed] [Google Scholar]
- Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–60. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–82. doi: 10.1016/S1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- Fanger NA, Cosman D, Peterson L, Braddy SC, Maliszewski CR, Borges L. The MHC class I binding proteins LIR-1 and LIR-2 inhibit Fc receptor-mediated signaling in monocytes. Eur J Immunol. 1998;28:3423–34. doi: 10.1002/(SICI)1521-4141(199811)28:11<3423::AID-IMMU3423>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells [see comments]. J Exp Med. 1997;186:1809–18. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O'Callaghan CA, Dunbar R, Ogg GS, Cerundolo V, Rolink A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096–100. [PubMed] [Google Scholar]
- Vitale M, Castriconi R, Parolini S, Pende D, Hsu ML, Moretta L, Cosman D, Moretta A. The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: analysis of LIR-1 + NK cell clones. Int Immunol. 1999;11:29–35. doi: 10.1093/intimm/11.1.29. [DOI] [PubMed] [Google Scholar]
- Navarro F, Llano M, Bellon T, Colonna M, Geraghty DE, Lopez-Botet M. The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur J Immunol. 1999;29:277–83. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Allan DS, Colonna M, Lanier LL, Churakova TD, Abrams JS, Ellis SA, McMichael AJ, Braud VM. Tetrameric complexes of human histocompatibility leukocyte antigen (HLA)-G bind to peripheral blood myelomonocytic cells. J Exp Med. 1999;189:1149–56. doi: 10.1084/jem.189.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman TL, Heikema AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral MHC homolog UL18. Immunity. 1999;11:603–11. doi: 10.1016/S1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2). Immunity. 2000;13:727–36. doi: 10.1016/S1074-7613(00)00071-6. [DOI] [PubMed] [Google Scholar]
- Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–60. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- Maenaka K, Juji T, Stuart DI, Jones EY. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure Fold Des. 1999;7:391–8. doi: 10.1016/S0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci U S A. 1999;96:3864–9. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei AA, Sternberg MJ. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993;229:472–93. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci U S A. 1990;87:6934–8. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–12. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- Somers W, Ultsch M, De Vos AM, Kossiakoff AA. The X-ray structure of a growth hormone-prolactin receptor complex. Nature. 1994;372:478–81. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Johnson DL, Middleton SA, Mulcahy LS, Wrighton NC, Dower WJ, Jolliffe LK, Wilson IA. Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 A. Science. 1996;273:464–71. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- Bravo J, Staunton D, Heath JK, Jones EY. Crystal structure of a cytokine-binding region of gp130. Embo J. 1998;17:1665–74. doi: 10.1093/emboj/17.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Wang YM, Bieber AJ, Bjorkman PJ. Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 A. Neuron. 1994;12:717–31. doi: 10.1016/0896-6273(94)90326-3. [DOI] [PubMed] [Google Scholar]
- Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–43. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- Janin J. Specific versus non-specific contacts in protein crystals. Nat Struct Biol. 1997;4:973–4. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- Borges L, Hsu ML, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–6. [PubMed] [Google Scholar]
- Willcox BE, Gao GF, Wyer JR, O'Callaghan CA, Boulter JM, Jones EY, van der Merwe PA, Bell JI, Jakobsen BK. Production of soluble alphabeta T-cell receptor heterodimers suitable for biophysical analysis of ligand binding. Protein Sci. 1999;8:2418–23. doi: 10.1110/ps.8.11.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–23. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JI, Hol WG. A flash-annealing technique to improve diffraction limits and lower mosaicity in crystals of glycerol kinase. Acta Crystallogr D Biol Crystallogr. 1998;54:479–80. doi: 10.1107/S0907444998004697. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMORE – an automated package for molecular replacement. Acta Crystallogr. 1994;A 50:157–63. [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D 50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Su XD, Gastinel LN, Vaughn DE, Faye I, Poon P, Bjorkman PJ. Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion. Science. 1998;281:991–5. doi: 10.1126/science.281.5379.991. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–50. doi: 10.1107/S0021889891004399. [DOI] [Google Scholar]
- Merritt EA, Murphy MEP. Raster3D Version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. 1994;D 50:869–73. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]