Abstract
Analysis on the three dimensional structures of (α/β)8 barrel proteins provides ample light to understand the factors that are responsible for directing and maintaining their common fold. In this work, the hydrophobically enriched clusters are identified in 92% of the considered (α/β)8 barrel proteins. The residue segments with hydrophobic clusters have high thermal stability. Further, these clusters are formed and stabilized through long-range interactions. Specifically, a network of long-range contacts connects adjacent β-strands of the (α/β)8 barrel domain and the hydrophobic clusters. The implications of hydrophobic clusters and long-range networks in providing a feasible common mechanism for the folding of (α/β)8 barrel proteins are proposed.
INTRODUCTION
One of the most frequent and regular domain structures of globular proteins is the (α/β)8 barrel fold constituted by eight parallel β-strands surrounded by eight α-helices (Chothia, 1988; Farber and Petsko, 1990; Farber, 1993). It is also one of the most abundant folds in the three super kingdoms of eukaryotes, bacteria, and archaea (Wolf et al., 1999). A distinctive character of these proteins is that despite possessing a common fold, they do not show similarity at the sequence level. Recently, the structural, functional, and evolutionary characteristics of (α/β)8 barrel proteins have been reviewed in detail (Pujadas and Palau, 1999; Pujadas, 2002).
The unique three-dimensional structure and stable secondary structures of globular proteins are determined by the interactions of amino acid residues among themselves along the polypeptide chain as well as with the surrounding medium. The interactions between amino acid residues have been classified into short-, medium-, and long-range, based on their distance of separation (Gromiha and Selvaraj, 1997a). This classification has been used successfully to address the problem of protein folding and sequence recognition (Gromiha and Selvaraj, 1999; 2001; Miyazawa and Jernigan, 1999). During the process of protein folding, the cooperative, noncovalent, and long-range interactions among residues provide stability to resist the local tendency for unfolding. Dosztanyi et al. (1997) have identified clusters of such residues as stabilization centers. Further, most of these centers are found at buried positions, and have hydrophobic and aromatic side chains. Poupon and Mornon (1999) showed the correspondence between hydrophobic positions of a given fold that constitute a folding nucleus, which is considered to play a key role in protein folding (Dobson and Karplus, 1999), and amino acid residues responsible for specific interactions. The role of each amino acid residue toward different noncovalent interactions has been delineated in our earlier works (Ponnuswamy and Gromiha, 1994; Gromiha and Selvaraj, 1999).
In (α/β)8 barrel proteins, we have identified similar and identical tertiary clusters and analyzed the importance of medium- and long-range interactions for the stabilization of (α/β)8 barrel fold (Selvaraj and Gromiha, 1998a,b; Kannan et al., 2001). It is of interest to understand the influence of long-range interactions in the mechanism underlying the common tertiary fold of this class of proteins. In the present work, we identified the hydrophobic clusters in (α/β)8 barrel proteins and characterized the importance of medium- and long-range interactions in the formation of these centers. Further, we observed a network of long-range contacts in β-strands belonging to (α/β)8 barrel proteins. Based on these results, we propose a common mechanism for the folding of this class of proteins.
MATERIALS AND METHODS
Protein structural database
We set up a database of representative (α/β)8 barrel proteins from the information available in the literature (Farber and Petsko, 1990; Holm and Sander, 1993; Sergeev and Lee, 1994) for which high-resolution structures (resolution <2.5 Å) are available. The PDB codes of the 36 selected proteins are, 1ALD, 1BKS, 1BTM, 1BYB, 1CDG, 1CTN, 1DHR, 1DIK, 1FCB, 1GHR, 1GOX, 1MLI, 1MNS, 1MUC, 1NAL, 1NAR, 1NIP, 1PII, 1PKY, 1RUS, 1SCU, 1TIM, 1TSY, 1YPI, 2ACQ, 2BNH, 2CMD, 2DRI, 2TAA, 2TMD, 3CBH, 3ICD, 3MIN, 4ENL, 5XIA, and 8RUC. In this data set, the average sequence identity is less than 10% between two proteins. We have calculated the structural alignment using combinatorial extension algorithm (Shindyalov and Bourne, 1998) and the average RMSD is estimated to be 4.5 Å between two proteins. It may be noted that the seven codes 1BNH, 1DHR, 1NIP, 1SCU, 1TSY, 2CMD, and 3ICD are classified as α/β sandwich or roll by CATH database (Orengo et al. 1999). The atomic coordinates of all the proteins were obtained from the Protein Data Bank (Berman et al., 2000). We have used the DSSP algorithm (Kabsch and Sander, 1993) and the information available in the Protein Data Bank for the assignment of secondary structures.
Computational methods
Delineation of hydrophobic domains and key residue in hydrophobic clusters
The amino acid residues in a protein molecule are represented by their α-carbon atoms and each residue is assigned with the hydrophobicity index obtained from thermodynamic transfer experiments (Nozaki and Tanford, 1971; Jones, 1975). The surrounding hydrophobicity is defined as the sum of hydrophobic indices of various residues that appear within 8-Å radius limit from a given residue (Manavalan and Ponnuswamy, 1977, 1978; Ponnuswamy, 1993; Ponnuswamy and Gromiha, 1993). It has been shown that the influence of each residue over the surrounding medium extends effectively only up to 8 Å (Manavalan and Ponnuswamy, 1977) and this limit is sufficient to characterize the hydrophobic behavior of amino acid residues (Manavalan and Ponnuswamy, 1978) and to accommodate both the local and nonlocal interactions (Gromiha and Selvaraj, 2000; Jiang et al., 2002). Further, 8-Å limit has been used in several studies, such as, to understand the folding rate of two-state proteins (Debe and Goddard, 1999; Gromiha and Selvaraj, 2001), protein stability upon mutations (Gromiha et al., 1999), thermal stability of proteins (Gromiha, 2001; Gromiha and Thangakani, 2001), and to determine the transition state structures of two-state protein mutants (Gromiha and Selvaraj, 2002).
The local regions that are comprised of a cluster of residues with high surrounding hydrophobicity (equal to or greater than twice the average value for all the residues in a protein) are considered as hydrophobic domains; the residue of the highest surrounding hydrophobicity within a domain is taken as the key residue in hydrophobic cluster (Ponnuswamy and Prabhakaran, 1980).
Computation of medium- and long-range contacts in the hydrophobic clusters of (α/β)8 barrel proteins
For each residue in the (α/β)8 barrel proteins, we computed the residues coming within a sphere of 8-Å radius as described in previous section. For a given residue, the composition of surrounding residues is analyzed in terms of the location at the sequence level and the contributions from <±3 residues are treated as short-range contacts, ±3 or ±4 residues as medium-range contacts, and >±4 residues are treated as long-range contacts (Gromiha and Selvaraj, 1997a; 2000).
Computation of thermal unfolding character in hydrophobic clusters
We followed the method of Ponnuswamy's group to compute the thermal unfolding behavior of amino acid residues in globular proteins (Ponnuswamy et al., 1982; Muthusamy et al., 2000). It has been shown that the set of amino acid residues Asp, Cys, Glu, Lys, Leu, Arg, Trp, and Tyr enhance the stability and the set of residues Ala, Gly, Gln, Ser, Thr, and Val decrease the stability. Accordingly, the amount of stabilizing (X1) and destabilizing (X2) groups (in %) were obtained for each residue within the 8-Å radius volume. The thermal unfolding for a residue is calculated using the equation, Tm = 64.462 + 0.894 X1 − 0.591 X2.
RESULTS AND DISCUSSIONS
Identification of hydrophobic clusters
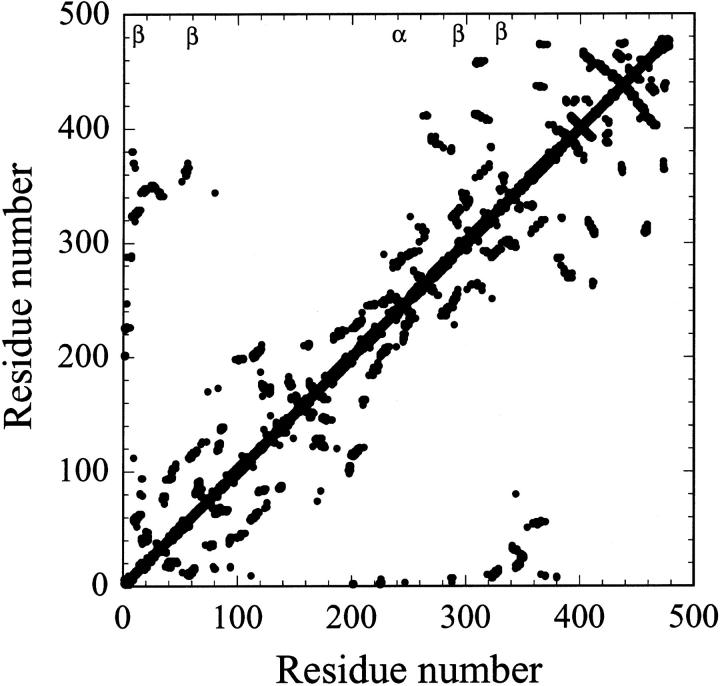
The key residues identified in the hydrophobic clusters of 36 (α/β)8 barrel proteins along with their location, surrounding hydrophobicity, and the number of medium- and long-range contacts, are presented in Table 1. We observed that, except for 1RUS, 1MUC, and 2BNH, all the (α/β)8 barrel proteins contain hydrophobic clusters, which contribute a good packing of the protein core. There is no direct relationship between number of residues and number of hydrophobic clusters. The protein 2TMD has the highest of seven hydrophobic clusters, followed by 2TAA and 1CDG, that have five hydrophobic clusters each. Among these three proteins, all the hydrophobic clusters in Taka amylase (2TAA) fall within the (α/β)8 barrel domain and the contact map (within a spatial distance of 8 Å) for 2TAA is shown in Fig. 1. This figure shows the presence of four hydrophobic clusters in β-strands and one in α-helix. Interestingly, the faraway residues contribute toward the formation of hydrophobic clusters (Y12, I60, T291, and I326) in β-strands, whereas medium-range contacts are responsible for T239 in α-helix.
TABLE 1.
Long-range interactions in the hydrophobic clusters of (α/β)8 barrel proteins
| PDB code | Residue number | Residue name | Hp | Nm | Nl | Nt | % long |
|---|---|---|---|---|---|---|---|
| 1ALD | 145 | A | 32.18 | 1 | 10 | 15 | 66.67 |
| 1BKS | 174 | T | 28.67 | 2 | 10 | 16 | 62.50 |
| 1BTM | 122 | P | 32.98 | 0 | 10 | 14 | 71.43 |
| 208 | Q | 32.00 | 0 | 12 | 16 | 75.00 | |
| 229 | G | 29.63 | 0 | 10 | 14 | 71.43 | |
| 1BYB | 53 | G | 32.26 | 2 | 8 | 14 | 57.14 |
| 103 | I | 29.00 | 1 | 10 | 15 | 66.67 | |
| 286 | V | 30.28 | 0 | 10 | 14 | 71.43 | |
| 1CDG | 19 | Q | 31.36 | 0 | 8 | 12 | 66.67 |
| 74 | I | 28.51 | 1 | 8 | 13 | 61.54 | |
| 132 | V | 29.96 | 0 | 9 | 13 | 69.23 | |
| 223 | I | 27.61 | 3 | 7 | 14 | 50.00 | |
| 607 | T | 29.78 | 0 | 13 | 17 | 76.47 | |
| 1CTN | 166 | Y | 26.70 | 1 | 8 | 13 | 61.54 |
| 250 | G | 29.59 | 2 | 9 | 15 | 60.00 | |
| 1DHR | 5 | V | 25.61 | 1 | 9 | 14 | 64.29 |
| 74 | A | 25.08 | 0 | 11 | 15 | 73.33 | |
| 221 | Q | 24.93 | 0 | 10 | 14 | 71.43 | |
| 1DIK | 661 | G | 28.13 | 2 | 9 | 15 | 60.00 |
| 737 | T | 32.37 | 0 | 12 | 16 | 75.00 | |
| 1FCB | 33 | V | 30.63 | 1 | 10 | 15 | 66.67 |
| 277 | L | 27.69 | 1 | 8 | 13 | 61.54 | |
| 1GHR | 234 | G | 26.91 | 0 | 9 | 13 | 69.23 |
| 1GOX | 249 | I | 26.91 | 1 | 11 | 16 | 68.75 |
| 1MLI | 54 | S | 24.98 | 0 | 9 | 13 | 69.23 |
| 1MNS | 33 | V | 28.48 | 0 | 11 | 15 | 73.33 |
| 1NAL | 38 | G | 26.42 | 0 | 9 | 13 | 69.23 |
| 1NAR | 221 | V | 28.34 | 2 | 8 | 14 | 57.14 |
| 1NIP | 9 | G | 26.30 | 0 | 12 | 16 | 75.00 |
| 33 | V | 27.06 | 0 | 11 | 15 | 73.33 | |
| 149 | I | 26.74 | 0 | 9 | 13 | 69.23 | |
| 1PII | 86 | S | 27.74 | 0 | 10 | 14 | 71.43 |
| 1PKY | 219 | S | 31.10 | 0 | 12 | 16 | 75.00 |
| 368 | I | 26.70 | 1 | 11 | 16 | 68.75 | |
| 1SCU | 45 | V | 29.93 | 0 | 10 | 14 | 71.43 |
| 201 | L | 28.48 | 1 | 10 | 15 | 66.67 | |
| 211 | C | 26.88 | 1 | 9 | 14 | 64.29 | |
| 260 | G | 28.64 | 0 | 12 | 16 | 75.00 | |
| 1TIM | 124 | A | 30.93 | 0 | 11 | 15 | 73.33 |
| 162 | A | 30.18 | 0 | 10 | 14 | 71.43 | |
| 1TSY | 231 | A | 28.49 | 4 | 5 | 13 | 38.46 |
| 1YPI | 61 | G | 28.74 | 0 | 11 | 15 | 73.33 |
| 162 | A | 30.20 | 0 | 10 | 14 | 71.43 | |
| 2ACQ | 156 | I | 28.67 | 1 | 11 | 16 | 68.75 |
| 259 | V | 29.35 | 0 | 10 | 14 | 71.43 | |
| 2CMD | 115 | G | 27.04 | 0 | 8 | 12 | 66.67 |
| 2DRI | 5 | A | 26.71 | 0 | 10 | 14 | 71.43 |
| 61 | L | 30.05 | 1 | 9 | 14 | 64.29 | |
| 85 | V | 27.09 | 0 | 11 | 15 | 73.33 | |
| 2TAA | 12 | Y | 31.98 | 0 | 10 | 14 | 71.43 |
| 60 | I | 26.90 | 0 | 8 | 12 | 66.67 | |
| 239 | T | 28.07 | 3 | 9 | 16 | 56.25 | |
| 291 | T | 32.91 | 0 | 12 | 16 | 75.00 | |
| 326 | I | 27.58 | 0 | 9 | 13 | 69.23 | |
| 2TMD | 23 | R | 31.26 | 1 | 10 | 15 | 66.67 |
| 62 | S | 28.25 | 1 | 10 | 15 | 66.67 | |
| 104 | L | 29.88 | 0 | 11 | 15 | 73.33 | |
| 179 | F | 28.80 | 3 | 9 | 16 | 56.25 | |
| 283 | V | 27.98 | 4 | 7 | 15 | 46.67 | |
| 354 | R | 27.73 | 4 | 7 | 15 | 46.67 | |
| 483 | V | 29.47 | 1 | 9 | 14 | 64.29 | |
| 3CBH | 85 | V | 28.28 | 0 | 10 | 14 | 71.43 |
| 3ICD | 28 | P | 35.26 | 0 | 10 | 14 | 71.43 |
| 96 | A | 30.98 | 1 | 8 | 13 | 61.54 | |
| 3MIN | 147 | A | 30.78 | 0 | 12 | 16 | 75.00 |
| 367 | A | 34.99 | 0 | 11 | 15 | 73.33 | |
| 391 | H | 27.56 | 0 | 10 | 14 | 71.43 | |
| 4ENL | 148 | V | 27.99 | 0 | 12 | 16 | 75.00 |
| 244 | G | 27.96 | 0 | 10 | 14 | 71.43 | |
| 394 | Q | 29.40 | 0 | 13 | 17 | 76.47 | |
| 5XIA | 46 | A | 26.36 | 3 | 9 | 16 | 56.25 |
| 180 | P | 26.24 | 0 | 13 | 17 | 76.47 | |
| 197 | G | 24.69 | 4 | 6 | 14 | 42.86 | |
| 212 | L | 27.40 | 0 | 13 | 17 | 76.47 | |
| 8RUC | 46 | G | 26.37 | 4 | 8 | 16 | 50.00 |
| 316 | D | 27.05 | 0 | 13 | 17 | 76.47 |
No hydrophobic clusters were identified in the proteins 1RUS, 1MUC, and 2BNH. Hp, surrounding hydrophobicity; Nm, Nl, and Nt are, respectively, number of medium-range, long-range, and total (sum of short-, which is usually 4; medium-; and long-range) contacts (%long, Nl × 100.0/Nt).
FIGURE 1.
The 8-Å contact map for Taka amylase. β shows the presence of hydrophobic clusters at Y12, I60, T291, and I326 in β-strands; and α indicates the location of T239 in α-helix.
The analysis on the relative occurrence of hydrophobic and hydrophilic residues as key residues in hydrophobic clusters showed that 74% of residues are hydrophobic. Our further analysis with the data set of Kannan et al. (2001) also showed similar results that 72% of the residues are hydrophobic. Further, the identified hydrophobic clusters are structurally conserved in different proteins. This observation indicates the crucial role played by these residues in the formation of hydrophobic clusters, which direct the cooperative folding of (α/β)8 barrel proteins.
Recently, Nagano et al. (2002) have made an extensive analysis of the sequence, structure and function of proteins that adopt the TIM barrel fold. They classified the proteins belonging to the (α/β)8 barrel fold into 18 families on the basis of homology. We have examined the conservation of hydrophobic clusters identified in the present work within these groups. Among the 36 considered proteins, 2TAA, 1BYB, 1CTN, 1NAR, and 1CDG belong to the Glycosidase family. Within this family, hydrophobic clusters have been identified in the first beta strands (β1) of 2TAA, 1BYB, and 1CDG. In both 2TAA and 1CDG another hydrophobic cluster is identified in the second beta-strand (β2). Further, both 2TAA and 1NAR have a hydrophobic cluster at the seventh beta-strand (β7). These observations suggest that the hydrophobic clusters may be conserved within homologous family members.
Role of long-range interactions in hydrophobic clusters
We have estimated the relative contribution of medium- and long-range interactions for the key residue in the hydrophobic cluster of (α/β)8 barrel proteins and the results are included in Table 1. The percentage of long-range contacts for each key residue is obtained by dividing the number of long-range contacts to the total number of contacts. Interestingly, we observed that the long-range interactions have the highest contribution, 38.5 to 76.5% for each protein, and the average contribution is 65.4%. This result indicates the vital role of long-range interactions to form the hydrophobic clusters and to stabilize the proteins. Further, the long-range interactions contribute an appreciably higher percentage in the hydrophobic clusters of β-strands or turn regions near the strands, and there is no significant contribution from medium-range interactions. However, the medium-range interactions play a dominant role in the hydrophobic clusters formed by α-helices. This observation is consistent with the previous results that α-helices are influenced by medium-range interactions and β-strands are dominated by long-range interactions (Gromiha and Selvaraj, 1998).
Occurrence of hydrophobic clusters in secondary structures
It is generally observed that in (α/β)8 barrel proteins, the α-helical segments connecting two β-strands are amphipathic and the hydrophobic face of the helix interacts with the β-strands (Pujadas and Palau, 1999). The secondary structure content of all the 36 (α/β)8 barrel proteins is presented in Table 2. We observed that 38% of the residues are in α-helix, 17% in β-strand, and 45% in coil region. Our analysis on the location of key residues in hydrophobic clusters in different secondary structures reveals that 74% of the residues fall in the β-strands of barrel fold. Similar result (71%) is observed with the data set of Kannan et al. (2001). Interestingly, 16% of hydrophobic clusters are identified in the coil region and only 10% are found in α-helical segments. We also computed the random probability for each key residue in the cluster to belong to these secondary structures. The values are 31% in helix, 23% in strand, and 46% in coil (Table 2). Further, we observed from Table 2 that the locations of more than 70% of the residues with high random probability are different from that in three-dimensional structure. This result demonstrates the importance of the location of key residues in different secondary structures for the folding and stability of (α/β)8 barrel proteins.
TABLE 2.
Secondary structure for the residues in hydrophobic clusters and percentage of helix, strand and coil in (α/β)8 barrel proteins
| PDB code | Secondary structure content (%)
|
Hydrophobic cluster | Secondary structure | Random probability
|
||||
|---|---|---|---|---|---|---|---|---|
| Helix | Strand | Coil | Helix | Strand | Coil | |||
| 1ALD | 40.8 | 13.8 | 45.5 | A145 | S | 59.5 | 9.5 | 31.0 |
| 1BKS | 52.2 | 14.1 | 33.7 | T174 | S | 33.3 | 33.3 | 33.3 |
| 1BTM | 46.6 | 14.3 | 39.0 | P122 | S | 38.5 | 7.7 | 53.8 |
| Q208 | S | 42.9 | 7.1 | 50.0 | ||||
| G229 | S | 13.6 | 9.1 | 77.3 | ||||
| 1BYB | 32.7 | 10.6 | 56.7 | G53 | H | 18.9 | 8.1 | 73.0 |
| I103 | C | 40.7 | 22.2 | 37.0 | ||||
| V286 | C | 21.9 | 28.1 | 50.0 | ||||
| 1CDG | 19.1 | 28.1 | 52.8 | Q19 | S | 30.8 | 23.1 | 46.2 |
| I74 | S | 24.3 | 51.4 | 24.3 | ||||
| V132 | S | 12.2 | 55.1 | 32.7 | ||||
| I223 | C | 24.3 | 51.4 | 24.3 | ||||
| T607 | S | 12.5 | 39.3 | 48.2 | ||||
| 1CTN | 21.9 | 25.7 | 52.4 | Y166 | S | 25.0 | 35.0 | 40.0 |
| G250 | S | 11.7 | 23.3 | 65.0 | ||||
| 1DHR | 37.3 | 23.7 | 39.0 | V5 | S | 27.8 | 44.4 | 27.8 |
| A74 | S | 25.9 | 29.6 | 44.4 | ||||
| Q221 | S | 75.0 | 12.5 | 12.5 | ||||
| 1DIK | 44.8 | 16.6 | 38.7 | G661 | C | 17.8 | 17.8 | 64.4 |
| T737 | S | 21.2 | 26.9 | 51.9 | ||||
| 1FCB | 32.6 | 11.9 | 55.5 | V33 | S | 28.6 | 31.0 | 40.5 |
| L277 | S | 25.5 | 11.8 | 62.7 | ||||
| 1GHR | 34.0 | 17.0 | 49.0 | G234 | C | 15.2 | 15.2 | 69.7 |
| 1GOX | 39.7 | 12.6 | 47.7 | I249 | S | 15.4 | 42.3 | 42.3 |
| 1MNS | 40.1 | 20.7 | 39.2 | V33 | S | 32.3 | 35.5 | 32.3 |
| 1NAL | 48.5 | 12.7 | 38.8 | G38 | C | 18.5 | 18.5 | 63.0 |
| 1NAR | 36.3 | 22.1 | 41.5 | V221 | S | 36.8 | 36.8 | 26.3 |
| 1NIP | 35.3 | 14.5 | 50.2 | G9 | C | 10.7 | 14.3 | 75.0 |
| V33 | S | 32.0 | 28.0 | 40.0 | ||||
| I149 | S | 36.4 | 22.7 | 40.9 | ||||
| 1PII | 35.4 | 19.9 | 44.7 | S86 | S | 16.0 | 24.0 | 60.0 |
| 1PKY | 35.8 | 22.0 | 42.2 | S219 | S | 20.8 | 16.7 | 62.5 |
| I368 | S | 36.4 | 36.4 | 27.3 | ||||
| 1SCU | 38.7 | 23.2 | 38.1 | V45 | S | 23.7 | 52.6 | 23.7 |
| L201 | S | 36.8 | 31.6 | 31.6 | ||||
| C211 | S | 20.0 | 80.0 | 0.0 | ||||
| G260 | S | 20.9 | 9.3 | 69.8 | ||||
| 1TIM | 42.9 | 17.0 | 40.1 | A124 | S | 42.9 | 14.3 | 42.9 |
| A162 | S | 42.9 | 14.3 | 42.9 | ||||
| 1TSY | 34.8 | 20.9 | 44.3 | A231 | H | 55.0 | 10.0 | 35.0 |
| 1YPI | 37.7 | 16.2 | 46.2 | G61 | S | 4.5 | 22.7 | 72.7 |
| A162 | S | 32.0 | 8.0 | 60.0 | ||||
| 2ACQ | 33.7 | 13.3 | 53.0 | I156 | S | 50.0 | 33.3 | 16.7 |
| V259 | S | 40.0 | 28.0 | 32.0 | ||||
| 2CMD | 44.6 | 17.0 | 38.5 | G115 | S | 25.0 | 16.7 | 58.3 |
| 2DRI | 45.0 | 22.5 | 32.5 | A5 | S | 59.5 | 18.9 | 21.6 |
| L61 | S | 48.0 | 36.0 | 16.0 | ||||
| V85 | S | 31.0 | 41.4 | 27.6 | ||||
| 2TAA | 20.7 | 14.4 | 64.9 | Y12 | S | 29.4 | 17.6 | 52.9 |
| I60 | S | 33.3 | 37.0 | 29.6 | ||||
| T239 | H | 5.0 | 15.0 | 80.0 | ||||
| T291 | S | 5.0 | 15.0 | 80.0 | ||||
| I326 | S | 33.3 | 37.0 | 29.6 | ||||
| 2TMD | 29.2 | 17.0 | 53.8 | R23 | C | 34.9 | 20.9 | 44.2 |
| S62 | S | 27.8 | 16.7 | 55.6 | ||||
| L104 | S | 42.6 | 29.8 | 27.7 | ||||
| F179 | H | 36.8 | 15.8 | 47.4 | ||||
| V283 | H | 40.9 | 34.1 | 25.0 | ||||
| R354 | H | 34.9 | 20.9 | 44.2 | ||||
| V483 | S | 40.9 | 34.1 | 25.0 | ||||
| 3ICD | 37.9 | 17.6 | 44.4 | P28 | S | 5.0 | 10.0 | 85.0 |
| A96 | S | 63.2 | 10.5 | 26.3 | ||||
| 3MIN | 47.5 | 9.6 | 42.9 | A147 | S | 51.7 | 6.9 | 41.4 |
| A367 | S | 51.7 | 6.9 | 41.4 | ||||
| H391 | S | 31.3 | 12.5 | 56.3 | ||||
| 4ENL | 39.2 | 16.3 | 44.5 | V148 | S | 29.4 | 38.2 | 32.4 |
| G244 | S | 10.8 | 8.1 | 81.1 | ||||
| Q394 | S | 55.6 | 22.2 | 22.2 | ||||
| 5XIA | 44.3 | 8.1 | 47.6 | A46 | C | 67.4 | 2.2 | 30.4 |
| P180 | C | 11.8 | 0.0 | 88.2 | ||||
| G197 | H | 19.5 | 9.8 | 70.7 | ||||
| L212 | S | 61.5 | 7.7 | 30.8 | ||||
| 8RUC | 39.7 | 13.9 | 46.4 | G46 | H | 17.8 | 4.4 | 77.8 |
| D316 | C | 30.8 | 7.7 | 61.5 | ||||
H, helix; S, strand; and C, coil. 1MLI and 3CBH contain only Cα coordinates and hence the secondary structure assignments are not available in DSSP.
Hydrophobic clusters and long-range interaction network
To understand the specific role played by long-range interactions in the formation of hydrophobic clusters, we examined the possible connectivities among all the hydrophobic clusters. For each key residue in hydrophobic clusters, we identified the farthest long-range contact in terms of distance of separation between residues on both the N- and C- terminal directions. From the observed contacts, the search was further continued until either the termini are reached or no more long-range contact is encountered to provide a long-range contact network.
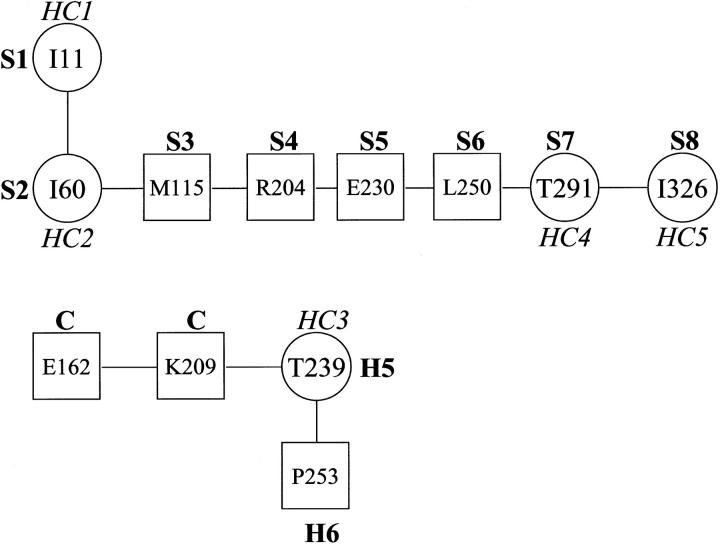
As an example, in Fig. 2 we have illustrated the long-range contact network in the hydrophobic clusters of 2TAA. This protein has five hydrophobic clusters in which four of them are in β-strands and one in α-helix of the (α/β)8 barrel domain. For the key residue at I60 in β-strand (S2), long-range contacts are observed with the hydrophobic residues, I11, Y12, F13, L14, M112, Y113, L114, and M115. At the N-terminal side, the farthermost long-range contact was observed at I11, belonging to the first strand (S1) and part of the key residue of first hydrophobic cluster (HC1). Further examination at I11 indicated no long-range contact in the same direction. In the C-terminal side of residue I60, we observed the farthest long-range contact at M115 (S3). Further search resulted in residues at R204, E230, L250, T291, and I326. Interestingly, all these residues are part of the β-strands, S4, S5, S6, S7, and S8, respectively, in which the residues T291 and I326 are themselves hydrophobic clusters of 2TAA. Thus, we observed a good pattern of long-range contact network with links to all β-strands constituting the (α/β)8 barrel domain and also among the hydrophobic clusters. A similar pattern of long-range network was also observed in other (α/β)8 barrel proteins.
FIGURE 2.
Long-range contact network in the hydrophobic clusters of Taka amylase. The circles denote hydrophobic clusters, and squares represent farthest long-range contacts. H, helix; S, strand; and C, coil.
The hydrophobic cluster (HC3) at T239 belongs to α-helical (H5) part of (α/β)8 barrel. On the N-terminal side, the network was observed at K209 and E162; no further extension was possible at E162 as it has no long-range contacts. It is noteworthy that each of these residues belong to coil regions. On the C-terminal side, the network terminates at P253 that is in α-helix (H6).
Implications for (α/β)8 barrel folding and stability
The location of hydrophobic clusters and the existence of long-range networks connecting the β-strands of the (α/β)8 barrel domain lead one to envisage the following probable scenario on (α/β)8 barrel folding and stability. In our earlier works, we have shown the dominance of short- and medium-range interactions in α-helices and long-range interactions in β-strands (Gromiha and Selvaraj, 1997a,b; 1998). In (α/β)8 barrel proteins, α-helices and β-strands alternate each other. During the process of folding, initially short- and medium-range interactions may predominate and direct the formation of α-helices. This view has been supported by several recent theoretical and experimental studies on the importance of local and nonlocal interactions in the folding of globular proteins. In molecular dynamics simulation of a small α/β protein, Sheinerman and Brooks (1998) observed that α-helix forms earlier and β-strand forms concomitantly during the process of protein folding. Further, the studies by Unger and Moult (1996) support the idea that initial formation of local substructures is important to the folding of proteins.
The formation of α-helices in (α/β)8 barrel domain will bring together adjacent β-strand forming residues in spatial proximity so that these residues can interact with each other to form β-strands, which are stabilized by both hydrogen-bonding network and hydrophobic interactions. Successive strands will also be stabilized by the enrichment of hydrophobic interactions. During the process of protein folding, Parker et al. (1996) showed through protein engineering and relaxation kinetics experiments in an α/β protein that sequence-local groups produced microdomains which is followed by the contacts between remote segments of secondary structures. Thus, the hydrophobic clusters and long-range contacts may steer the folding and stabilization of the (α/β)8 barrel fold.
Thermal unfolding character of key residues in hydrophobic clusters
We have analyzed the thermal unfolding behavior of all the residues forming hydrophobic clusters as described in the Methods section. We found that most of the residues forming hydrophobic clusters have high thermal stability and withstand higher temperature. Thus these residues help to maintain the strong interactions with other residues and provide stability to the native state. It might be due to high packing effect, as packing is one of the most important characters for the enhanced stability of thermophilic proteins (Gromiha et al., 1999b).
We have examined the thermal stability of several (α/β)8 barrel proteins using the thermodynamic database for proteins and mutants developed by us and available in web (Gromiha et al., 2000). We found that Taka amylase has the highest thermostability of 40.8 kcal/mol (Ooi and Oobatake, 1988) followed by 1TIM and 1WSY. Also, we observed a direct relationship between stability (ΔG) and number of hydrophobic clusters.
CONCLUSIONS
The analysis on the crystal structures of (α/β)8 barrel proteins revealed the presence of hydrophobic clusters in most of the studied proteins. The residues in hydrophobic clusters are mainly influenced by long-range interactions and these residues have high thermal stability, which may help to resist the local tendency for unfolding. Further, a network of long-range interactions is observed to link through β-strands of (α/β)8 barrel domain. In essence, the hydrophobic clusters and network of long-range contacts pave the way for the folding and stabilization of (α/β)8 barrel fold.
Acknowledgments
We thank the referees for constructive comments. The authors thank A. Mary Thangakani for reading of the manuscript; and M.M.G. thanks Dr. Yutaka Akiyama and Dr. Makiko Suwa for encouragement.
References
- Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia, C. 1988. Protein structure: the 14th barrel rolls out. Nature. 333:598–599. [DOI] [PubMed] [Google Scholar]
- Debe, D. A., and W. A. Goddard. 1999. First principles prediction of protein folding rates. J. Mol. Biol. 294:619–625. [DOI] [PubMed] [Google Scholar]
- Dobson, C. M., and M. Karplus. 1999. The fundamentals of protein folding: bringing together theory and experiment. Curr. Opin. Struct. Biol. 9:92–101. [DOI] [PubMed] [Google Scholar]
- Dosztanyi, Z., A. Fiser, and I. Simon. 1997. Stabilization centers in proteins: identification, characterization and predictions. J. Mol. Biol. 272:597–612. [DOI] [PubMed] [Google Scholar]
- Farber, G. K. 1993. An ( α/β) barrel full of evolutionary trouble. Curr. Opin. Str. Biol. 3:409–412. [Google Scholar]
- Farber, G. K., and G. A. Petsko. 1990. The evolution of α/β barrel enzymes. Trends Biochem. Sci. 15:228–233. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M. 2001. Important inter-residue contacts for enhancing the thermal stability of thermophilic proteins. Biophys. Chem. 91:71–77. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 2002. Important amino acid properties for determining the transition state structures of two-state protein mutants. FEBS Lett. 526:129–134. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 2001. Comparison between long-range interactions and contact order in determining the folding rates of two-state proteins: application of long-range order to folding rate prediction. J. Mol. Biol. 310:27–32. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 2000. Inter-residue interactions in the structure, folding and stability of proteins. Recent Res. Devel. Biophys. Chem. 1:1–14. [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 1999. Importance of long range interactions in protein folding. Biophys. Chem. 77:49–68. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 1998. Protein secondary structure prediction in different structural classes. Protein Eng. 11:249–251. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 1997a. Influence of medium and long range interactions in different structural classes of globular proteins. J. Biol. Phys. 23:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha, M. M., and S. Selvaraj. 1997b. Influence of medium and long range interactions in ( α/β)8 barrel proteins. J. Biol. Phys. 23:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha, M. M., and A. M. Thangakani. 2001. Role of medium- and long-range interactions to the stability of the mutants of T4 lysozyme. Prep. Biochem. Biotech. 31:217–227. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., M. Oobatake, H. .H. Kono, H. Uedaira, and A. Sarai. 1999a. Role of structural and sequence information in the prediction of protein stability changes: comparison between buried and partially buried mutations. Protein Eng. 12:549–555. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., M. Oobatake, and A. Sarai. 1999b. Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophys. Chem. 82:51–67. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., J. An, H. Kono, M. Oobatake, H. Uedaira, P. Prabakaran, and A. Sarai. 2000. ProTherm, version 2.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 28:283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123–138. [DOI] [PubMed] [Google Scholar]
- Jiang, Z., L. Zhang, J. Chen, A. Xia, and D. Zhao. 2002. Effect of amino acid on forming residue-residue contacts in proteins. Polymer. 43:6037–6047. [Google Scholar]
- Jones, D. D. 1975. Amino acid properties and side-chain orientation in proteins: a cross correlation approach. J. Theor. Biol. 50:167–183. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., and C. Sander. 1993. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- Kannan, N., S. Selvaraj, M. M. Gromiha, and S. Vishveshwara. 2001. Clusters in (α/β)8 barrel proteins: implications for protein structure, function and folding: a graph theoretical approach. Prot. Struct. Funct. Genet. 43:103–112. [DOI] [PubMed] [Google Scholar]
- Manavalan, P., and P. K. Ponnuswamy. 1978. Hydrophobic character of amino acid residues in globular proteins. Nature. 275:673–674. [DOI] [PubMed] [Google Scholar]
- Manavalan, P., and P. K. Ponnuswamy. 1977. A study of the preferred environment of amino acid residues in globular proteins. Arch. Biochem. Biophys. 184:476–487. [DOI] [PubMed] [Google Scholar]
- Miyazawa, S., and R. L. Jernigan. 1999. Evaluation of short-range interactions as secondary structure energies for protein fold and sequence recognition. Proteins. 36:347–356. [PubMed] [Google Scholar]
- Muthusamy, R., M. M. Gromiha, and P. K. Ponnuswamy. 2000. On the thermal unfolding character of globular proteins. J. Prot. Chem. 19:1–8. [DOI] [PubMed] [Google Scholar]
- Nagano, N., C. A. Orengo, and J. M. Thornton. 2002. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321:741–765. [DOI] [PubMed] [Google Scholar]
- Nozaki, Y., and C. Tanford. 1971. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J. Biol. Chem. 246:2211–2217. [PubMed] [Google Scholar]
- Ooi, T., and M. Oobatake. 1988. Intermolecular interactions between protein and other molecules including hydration effects. J. Biochem. 104:440–444. [DOI] [PubMed] [Google Scholar]
- Orengo, C. A., F. M. Pearl, J. E. Bray, A. E. Todd, A. C. Martin, L. Lo Conte, and J. M. Thornton. 1999. The CATH Database provides insights into protein structure/function relationships. Nucleic Acids Res. 27:275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, M. J., R. B. Sessions, I. G. Badcoe, and A. R. Clarke. 1996. The development of tertiary interactions during the folding of a large protein. Fold. Des. 1:145–156. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy, P. K. 1993. Hydrophobic characteristics of folded proteins. Prog. Biophys. Mol. Biol. 59:57–103. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy, P. K., and M. M. Gromiha. 1994. On the conformational stability of folded proteins. J. Theor. Biol. 166:63–74. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy, P. K., and M. M. Gromiha. 1993. Prediction of transmembrane helices from hydrophobic characteristics of proteins. Int. J. Pept. Protein Res. 42:326–341. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy, P. K., and M. Prabhakaran. 1980. Properties of nucleation sites in globular proteins. Biochem. Biophys. Res. Commun. 97:1582–1590. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy, P. K., R. Muthusamy, and P. Manavalan. 1982. Amino acid composition and thermal stability of globular proteins. Int. J. Biol. Macromol. 4:186–190. [Google Scholar]
- Poupon, A., and J. P. Mornon. 1999. Predicting the protein folding nucleus from sequences. FEBS Lett. 452:283–289. [DOI] [PubMed] [Google Scholar]
- Pujadas, G. 2002. TIM barrel research in the genomic and proteomic era. In Recent Research Developments in Protein Folding, Stability and Design. M. M. Gromiha, and S. Selvaraj, editors. Research Signpost, Trivandrum, India. pp37–55.
- Pujadas, G., and J. Palau. 1999. TIM barrel fold: structural, functional and evolutionary characteristics in natural and designed molecules. Biologia Bratislava. 54:231–254. [Google Scholar]
- Selvaraj, S., and M. M. Gromiha. 1998a. An analysis of amino acid clustering pattern in ( α/β)8 barrel proteins. J. Prot. Chem. 17:407–415. [DOI] [PubMed] [Google Scholar]
- Selvaraj, S., and M. M. Gromiha. 1998b. Importance of long-range interactions in ( α/β)8 barrel fold. J. Prot. Chem. 17:691–697. [DOI] [PubMed] [Google Scholar]
- Sergeev, Y., and B. Lee. 1994. Alignment of beta-barrels in (β/α)8 proteins using hydrogen-bonding pattern. J. Mol. Biol. 244:168–182. [DOI] [PubMed] [Google Scholar]
- Sheinerman, F. B., and C. L. Brooks 3rd. 1998. Molecular picture of folding of a small alpha/beta protein. Proc. Natl. Acad. Sci. USA. 95:1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov, I. N., and P. E. Bourne. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11:739–747. [DOI] [PubMed] [Google Scholar]
- Unger, R., and J. Moult. 1996. Local interactions dominate folding in a simple protein model. J. Mol. Biol. 259:988–994. [DOI] [PubMed] [Google Scholar]
- Wolf, Y. I., S. E. Brenner, P. A. Bash, and E. V. Koonin. 1999. Distribution of protein folds in the three superkingdoms of life. Genome Res. 9:17–26. [PubMed] [Google Scholar]